Abstract
Immunoglobulin E (IgE)‐dependent mechanisms play a pivotal role in mediating allergic disease. Previously, VH‐Cε transcripts from blood or spleen of atopic asthmatics have been analysed for VH gene usage and patterns of somatic mutation. An over‐representation of the minor VH5 family has been observed, consistent with a superantigen drive. As local mucosal events in IgE production may be more significant in the disease process, we have analysed VH‐Cε transcripts from a bronchial biopsy of a patient with severe asthma. VH5 predominance was confirmed with 10 of 30 unique clones derived from this family. Repeated sequences, some with intraclonal variation, revealed clonal expansion and continuing mutational activity at the site. Unexpectedly, three unmutated VH‐Cε sequences were found, indicating that isotype switching to IgE can occur without mutation. Detection of a sister clone with extensive mutations was again consistent with local mutational activity. Evidence for local isotype switching was obtained by identification of clonally related immunoglobulin M (IgM), immunoglobulin G (IgG) and immunoglobulin E (IgE) sequences. However, in contrast to findings in blood, no IgG4 transcripts clonally related to IgE were detected, suggesting that the balance between synthesis of IgG4 and IgE may differ between systemic and local sites. These data confirm a VH5 bias in IgE, and support the concept that IgE‐synthesizing B cells arise via local differentiation.
Introduction
Immunoglobulin E (IgE) antibodies are known mediators of allergic disease, including allergic asthma.1,2 Allergen can cross‐link IgE that is bound to its high‐affinity receptor (FcεRI) on the surface of mast cells or basophils, resulting in the release of mediators that lead to the symptoms of Type I hypersensitivity.3 The presence of high‐ and low‐affinity receptors has been reported on many cell types in the bronchial mucosa of asthmatics, with an increased number of FcεRI‐expressing cells being found in asthmatics.4 IgE has the potential to mediate inflammation in the airways by enhancing the release of proinflammatory mediators from activated cells.5–7 IgE‐mediated antigen presentation is another potential way by which IgE is involved in the inflammatory processes of asthma and atopy.8,9 The central role of IgE in both the early and late responses has been confirmed by studies with non‐anaphylactogenic anti‐IgE monoclonal antibody (mAb) that binds to free IgE and to IgE on B cells. Treatment of mild asthmatics with this mAb inhibited the late reaction by 60% and also suppressed the early response.10
Allergen‐specific IgE has been detected in nasal and respiratory secretions,11,12 with a recent study finding IgE specific for house dust mite (HDM) in the sputum of HDM‐sensitive asthmatics, but not in healthy control subjects.13 However, the origin of IgE‐secreting cells is unknown, although IgE‐positive B cells have been identified in local tissue.14,15 It is unclear whether such cells have been recruited from lymphoid tissue or are induced to undergo isotype switching within the mucosal site: recent data supports the latter possibility.14–16 As locally synthesized IgE may be important in responses to exogenous antigen, the origin and nature of IgE‐expressing B cells at local sites of disease is of interest.
Immunogenetic analysis allows us to identify B‐cell clones that have undergone isotype switching to IgE. It is then possible to analyse the nature and mutational patterns of VH genes used. During genetic recombination, one VH gene from a germline repertoire of 51, in combination with D and JH genes, is joined to a C‐region gene (initially immunoglobulin M [IgM]) to give rise to functional genes that can encode the H chain of antibody. A preferential usage of the minor VH5 family by IgE was previously observed in the peripheral blood and spleen of atopic asthmatics17,18 and also in peripheral blood from patients with atopic dermatitis.19 Bias in VH gene usage can indicate an influence of superantigen (SAg), which binds VH via the conserved framework region (FWR) outside the conventional binding sites in the complementarity‐determining region (CDR).20 One suggestion is that allergens, and perhaps parasitic antigens, are acting in this manner.17 In order to focus on events at the site of disease, we studied a bronchial biopsy from a severe asthmatic. We report clear predominance of VH5 usage. Analysis of B‐cell clones also indicated that somatic mutation and isotype switching are occurring in the local environment.
Materials and methods
Background of the patient
The patient was a 32‐year‐old male who had suffered with asthma from birth. He has a severe form of perennial asthma and is highly allergic to HDM, grass pollen, cat, dog and feather, with a very high serum IgE level of 5000 IU/ml. At the time of the study his forced expiratory volume in 1 second (FEV1) was 1·72, which is 49% of predicted. He has daily symptoms with morning chest tightness and significant effect on his physical activity. In addition, at the time of biopsy he had nocturnal symptoms every night, an indicator of poorly controlled asthma. His treatment consisted of budesonide aerosol (2 mg twice daily) delivered by a nebuliser, 10 mg daily of oral prednisolone and regular twice‐daily nebulised salbutamol (2·5 mg).
Bronchoscopy procedure
The patient agreed to participate in the study, which was approved by the Joint Southampton University and Hospital Ethics Committee. Following premedication with 2·5 mg of salbutamol and 0·5 mg of ipratropium bromide, delivered by nebuliser, and intravenous atropine (0·6 mg) the patient underwent bronchoscopy according to the guidelines issued by the National Institutes of Health,21 as previously reported,22 using lignocaine for topical anaesthesia. A fibreoptic bronchoscope was introduced and two biopsies of subcarinae were taken using alligator forceps. The biopsies were snap‐frozen and stored in liquid nitrogen until analysis.
Preparation of cDNA and amplification of VH genes
The biopsies were homogenized with 200 μl of RNAzol and the RNA was then extracted as previously described.23 cDNA was then prepared with oligo dT primer and a first‐strand cDNA synthesis kit (Pharmacia, Uppsala, Sweden). Superscript II was used as the reverse transcriptase enzyme (Gibco BRL, Life Technologies Inc., Paisley, Strathclyde, UK). Four separate nested polymerase chain reactions (PCRs) were carried out for amplification of VH 1–7 families in combination with Cε. The products were cloned as previously described17 and sequenced using an ABI 377 automated sequencer (Applied Biosystems, Foster City, CA), with M13 forward and reverse primers. The VH primers had previously been checked for any inherent bias for the VH5 family by a nested PCR using IgM primers in place of IgE primers. No asymmetry in VH family usage was obtained (Fig. 1; reference 18).
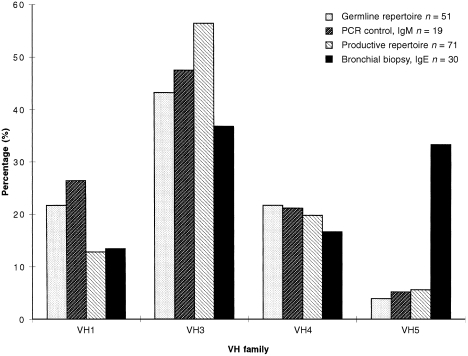
Figure 1.
Graph showing a significant over‐representation of the VH5 family by immunoglobulin E (IgE) in the bronchial biopsy when compared with the germline repertoire,38 a polymerasec chain reaction (PCR) control for immunoglobulin M (IgM)18 and the productive repertoire from normal B cells.26
Investigation of VH‐Cε related clones
Identification of CDR3–FWR4–CH sequences
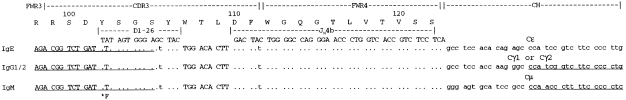
For two VH5‐Cε sequences (clones 1 and 2), clonally related transcripts were sought by the use of CDR3‐specific oligonucleotides. A nested PCR, specific for VH5 in combination with IgM, immunoglobulin G (IgG)23 and IgG4,24 was carried out as previously described.25 followed by amplification with 5′ CDR3‐specific primers for IgE clones 1 (5′‐AGACGGGCTGACTATAGGGGGA‐3′) or 2 (5′‐AGACGGTCTGATTTTAG‐TGGGA‐3′), with the inner primers for IgM, IgG or IgG4. A control PCR was also carried out, of the CDR3‐specific primer with the inner IgE primer. Products obtained were then purified, cloned and sequenced. The nucleotide sequences of the CDR3‐CH transcripts obtained were compared with that of the IgE clones to check for clonal relatedness (Fig. 2).
Figure 2.
Immunoglobulin M (IgM)‐, immunoglobulin G (IgG)‐ and immunoglobulin E (IgE)‐related clones with a common CDR3/FWR4 sequence. The nucleotide sequences of CDR3/FWR4/CH transcripts are aligned to D‐segment and JH germline genes. Primer sequences are underlined, the Cε primer is 3′ to the sequence shown.
Results
VH family utilization in IgE
In the bronchial biopsy, we analysed VH gene usage in IgE by using a nested PCR. Sequences from 30 distinct B cells were obtained, indicated by the unique CDR3 regions of the VH‐Cε transcripts. To determine VH family usage, sequences were aligned to the closest germline V gene (Fig. 1). The VH5 family was significantly over‐represented (P < 0·001 by χ2 analysis) with 10 of 30 (33%) clones derived from the two members V5–51 and VHVMW (a polymorphic variant of VH32). To assess for reproducibility, the PCRs were carried out as four separate reactions and the VH5 family was evident in all PCRs. To control for primer bias, we compared this pattern with that obtained for IgM using the same VH primers (Fig. 1), where only one of 19 VH5‐encoded sequences were obtained.18Figure 1 also compares these results with the VH repertoire of productive rearrangements from individual normal B cells, where VH5 was used in four of 71 B cells (5·6%; reference 26).
Analysis of VH5‐Cε gene sequences
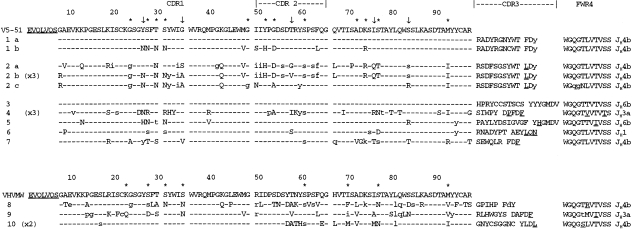
Sequences obtained are shown in Fig. 3, with unique CDR3s indicating clonal relationships (GenBank accession nos: AF110479–AF110491). Groups 1 and 2 appear to be each derived from a single B cell that has undergone further diversification. The similarity between the CDR3s of the two groups is intriguing, and it raises the possibility that they are all derived from the same original B cell. This is underlined by the sharing of several replacement mutations between 1b and Group 2 clones. However, there are too many differences between the CDR3 sequence to confirm a common clonal origin, and the possibility remains that they represent two parallel, independent clones, possibly converging on one epitope.
Figure 3.
VH5‐immunoglobulin E (IgE) amino acid sequences from bronchial biopsy aligned to closest germline genes. Clones 1a, b, and 2a, b, c are sets of related clones. Identical repeated sequences were found for clones 2b, 4 and 10, with number of repeats shown in brackets. Upper case: replacement mutation; lower case: silent substitution. *Indicates sites that have previously been reported as hot‐spots of mutation.23 ↓Indicates frequently mutated sites not previously reported.
As mRNA from the low numbers of IgE+ B cells was limited, we considered the possibility of PCR artefacts arising from cross‐over events. In view of the fact that separate PCR reactions were carried out, and that the sequences obtained did not show patterns consistent with cross‐over, we thus eliminated this possibility.
The finding of repeated identical sequences both within Group 2 and in other single sequences (4 and 10) is consistent with clonal expansion. However, an alternative explanation may be that plasma cells with increased levels of mRNA were present. Identical repeats occurred not only in VH5‐derived sequences, but were seen in IgE encoded by VH1 (one instance), VH3 (two instances) and VH4 (one instance) (data not shown). It is unlikely, because of the high serum level of IgE in this patient and the large number of different clones obtained, that the repeated sequences indicate the presence of a restricted set of B cells in the mucosa.
There was a generally high level of somatic mutation, both in the VH5‐Cε sequences (mean = 8·2%) and in the other VH families (6·5%, data not shown). Analysis of the VH5‐derived sequences showed that mutations were dispersed throughout the sequences (Fig. 3), with no significant clustering of replacement mutations in the CDR, characteristic of antigen selection.27 However, hot‐spots of mutational activity were evident, as described previously for VH5,23 and are indicated by an asterisk in Fig. 3. The incidence was similar to that previously seen in IgE, with particularly prominent sites at Ser31 and Ser77, known to be susceptible codons. Other sites were also frequently mutated, which had not been observed previously. These are shown by the arrows in Fig. 3, for example Tyr27. However, none of these sites have identical repeated substitutions, which would be indicative of antigen selection,28 and the sample size is fairly small, preventing any significant conclusions to be made about additional hot‐spot incidence.
Clones 1a and 3 had no apparent somatic mutations, a feature that appears to be unique to local tissue, and has not been found in 124 VH‐Cε sequences from blood or spleen.17,18,23–25 Interestingly, 1b shares the CDR3 of 1a, indicative of the same B cell of origin, but has accumulated 11 replacement mutations (Fig. 3), probably as a result of local somatic mutation events. Clones in Group 2 share the CDR3 sequence, characteristic of a common B‐cell origin, but there is a considerable level of intraclonal variation, with both shared and distinct mutations evident (Fig. 3). This is again consistent with local somatic mutation.
Presence of clonally related Cμ and Cγ transcripts
We focused on two VH5‐Cε clones, 1 and 2, to look for transcripts of alternative isotypes related to the IgE clones already obtained. By using a CDR3‐specific 5′ primer, together with 3′ primers based in Cμ or Cγ, we obtained IgM‐ and IgG‐related transcripts with a common CDR3 for clone 2 (Fig. 2). In the case of clone 1, the PCRs were negative and no alternative transcripts were detected. Control PCRs for CDR3‐Cε transcripts on material from the nested VH5‐CH PCR products were negative, eliminating the possibility that the IgM and IgG transcripts were actually PCR contaminants derived from the IgE clone by a PCR cross‐over event.
The clonal relationship of the alternative isotypes obtained from clone 2 is evident from the identical CDR3‐FWR4 sequences (see Fig. 2), when compared with the CDR3/FWR4 of the IgE clone from the end of the 5′‐CDR3 primer onwards. Four features underline their shared origin: the common N‐additions at positions 107, 108 and 109; common D‐segment alignments of the same length and reading frame; a common JH gene usage; and the common silent nucleotide substitutions in amino acids 105 (Ser, AGC→AGT) and 111 (Phe, TAC→TAT), Fig. 2. The related clones were derived from either clone 2a or 2b, not 2c, as seen by comparison of the FWR4 sequence of the clones (Fig. 3).
The IgG subclass of the transcripts obtained was either IgG1 or IgG2, the consensus IgG primer used being too far upstream to distinguish between these two. No evidence for IgG3 or IgG4 sequences was obtained. In addition, an IgG4‐specific PCR was carried out, but CDR3‐Cγ4 transcripts were not obtained. The upstream sequences were sought (VH5‐CDR3), as previously described,25 but we were unable to obtain these. The primers amplified the appropriate VH5‐Cε sequence when tested. However, with the IgM and IgG PCRs there was cross‐priming of the CDR3‐specific primers with VH‐CH transcripts that had similar, but not identical, CDR3s. These may have been present in greater frequency than the clones we were seeking.
Discussion
The first striking result from this analysis of VH‐Cε transcripts from a bronchial mucosal biopsy of an atopic asthmatic is the predominance of the small VH5 family. This is confirmatory of our previous observations of a bias towards VH5 usage in IgE from the peripheral blood and spleen of HDM‐allergic asthmatics, although the bias found here is more evident than that observed in the peripheral blood.18 This finding supports the concept that there may be a common allergen acting as a B‐cell SAg. As SAgs are usually derived from pathogens, it raises the possibility that a pathogen‐derived SAg may preferentially recruit VH5‐expressing B cells that are initially expressing IgM. These are subsequently induced to switch to IgE expression in the appropriate cytokine environment.
The pattern of somatic mutation observed in these VH5‐Cε transcripts is similar to that observed previously in clones from the blood and spleen.17,18 The same hot‐spots are observed in these sequences as in VH5 clones reported previously,23 with a few additional sites that are commonly mutated. Two of these new sites are in the CDR, and the others are adjacent to, or within, the susceptible serine motif AGC/T, so their presence is not surprising.28
An unexpected finding is the absence of somatic mutation in three of the IgE clones, two of which are from the VH5 family. It appears that isotype switching to IgE in B cells can be reached locally with either a complete absence of somatic mutation or with a high level. Unmutated IgE sequences presumably arise from the isotype switching of germline‐encoded IgM antibody to IgE before somatic mutation and affinity maturation. In one of the sets of expanded clones, the unmutated clone has a mutated sister clone. Therefore, it would appear that somatic mutation is occurring locally, as it is unlikely that the unmutated clone has travelled to a lymph node to undergo somatic mutation and then migrated back to the exact same local sites. Further support for local somatic mutation is seen in the second set of related clones that have distinct, as well as shared, mutations. This is indicative of clonal expansion at a local site with ongoing somatic mutation. This pattern has also been detected in B cells from the peripheral blood, where there are generally far fewer IgE‐expressing B cells than obtained in this biopsy.18,29
Local isotype switching also appears to be occurring, with IgM‐, IgG‐ and IgE‐related clones being found together at this local site. This is demonstrated by the presence of identical nucleotide sequences of CDR3/FWR4 transcripts with IgM, IgG1/2 or IgE C regions. This contrasts with our previous results from the blood where IgG4‐ and IgE‐related clones were detected together, with no evidence of other IgG subclasses.25 Co‐expression of IgG4 and IgE may be expected, as interleukin (IL)‐4 and IL‐13 induce switching to both isotypes.30,31 The different pattern at the local site can be explained by the influence of other cytokines on isotype transcript levels. It is known that higher IL‐4 : interferon‐γ (IFN‐γ) levels enhance IgE production, while increased IFN‐γ expression favours IgG4 production,32,33 and this may be reflected at the mRNA level. Therefore, the absence of IgG4 at a disease site may be a result of local cytokine expression, with high IL‐4 : IFN‐γ ratios inducing greater IgE production. Consistent with this possibility, a T helper 2 (Th2)‐dominant cytokine pattern has been reported in the bronchial mucosa of asthmatics with elevated IL‐4 and IL‐13 levels.34,35 In contrast, the IgE‐expressing B cells in the systemic circulation may have arisen in sites, possibly the spleen, where the IL‐4 : IFN‐γ ratio is lower, favouring production of IgE and IgG4.
Recent studies provide evidence to suggest that class switching to IgE occurs at local sites of disease. Increases in Iε germline transcripts, in addition to Cε transcripts and IL‐4, have been detected in nasal mucosal biopsies of hayfever patients following allergen challenge, while the number of B cells remained constant.14 Similar results have been obtained in sinus mucosal biopsies from atopic patients with chronic sinusitis.15 A different group has used PCR detection of Sμ/Sε switch circle DNA as evidence for in vivo IgE isotype switching in ragweed‐sensitive individuals following nasal challenge with ragweed and diesel exhaust particles.16 The accumulated evidence for local somatic mutation and isotype switching events is surprising, given that there is little evidence for germinal centre (GC) structures at the mucosal site.14 Although dependence on the GC is not absolute,36 the high degree of mutational activity will probably require an environment where CD40 ligand (CD40L), cytokines and antigen are present.37 At least in patients with asthma, the cellular milieu at the mucosal surface appears to be capable of providing this.
In summary, it appears that at least some IgE‐expressing B cells arise within a local site of disease. Our hypothesis from the immunogenetic analysis is that IgM‐expressing B cells, particularly those expressing the VH5 family, are stimulated by allergen and undergo local differentiation, accumulation of mutations and isotype switching to IgE. The relationship with IgE‐expressing B cells in blood and spleen is not yet apparent, but local cytokine influences clearly direct B cells in this site where disease is manifest.
Acknowledgments
This work was funded by the National Asthma Campaign. We would like to thank Zadie Davies for her expert technical assistance.
Glossary
Abbreviations
- CDR
complementary‐determining region
- FWR
framework region
- SAg
superantigen
References
- 1.Burrows B, Martinez FD, Halonen M, Barbee RA, Cline MG. Association of asthma with serum IgE levels and skin‐test reactivity to allergens. N Engl J Med. 1989;320:271. doi: 10.1056/NEJM198902023200502. [DOI] [PubMed] [Google Scholar]
- 2.Sunyer J, Anto JM, Sabria J, et al. Relationship between serum IgE and airway responsiveness in adults with asthma. J Allergy Clin Immunol. 1995;95:699. doi: 10.1016/s0091-6749(95)70175-3. [DOI] [PubMed] [Google Scholar]
- 3.Holgate ST, Burns GB, Robinson C, Church MK. Anaphylactic‐ and calcium‐dependent generation of prostaglandin D2 (PGD2), thromboxane B2, and other cyclooxygenase products of arachidonic acid by dispersed human lung cells and relationship to histamine release. J Immunol. 1984;133:2138. [PubMed] [Google Scholar]
- 4.Humbert M, Grant JA, Taborda‐barata L, et al. High‐affinity IgE receptor (FcepsilonRI) ‐bearing cells in bronchial biopsies from atopic and nonatopic asthma. Am J Respir Crit Care Med. 1996;153:1931. doi: 10.1164/ajrccm.153.6.8665058. [DOI] [PubMed] [Google Scholar]
- 5.Fuller RW, Morris PK, Richmond R, et al. Immunoglobulin E‐dependent stimulation of human alveolar macrophages: significance in type 1 hypersensitivity. Clin Exp Immunol. 1986;65:416. [PMC free article] [PubMed] [Google Scholar]
- 6.Broide DH, Gleich GJ, Cuomo AJ, et al. Evidence of ongoing mast cell and eosinophil degranulation in symptomatic asthma airway. J Allergy Clin Immunol. 1991;88:637. doi: 10.1016/0091-6749(91)90158-k. [DOI] [PubMed] [Google Scholar]
- 7.Campbell AM, Vachier I, Chanez P, et al. Expression of the high‐affinity receptor for IgE on bronchial epithelial cells of asthmatics. Am J Respir Cell Mol Biol. 1998;19:92. doi: 10.1165/ajrcmb.19.1.2648. [DOI] [PubMed] [Google Scholar]
- 8.Pirron U, Schlunck T, Prinz JC, Rieber EP. IgE‐dependent antigen focusing by human B lymphocytes is mediated by the low‐affinity receptor for IgE. Eur J Immunol. 1990;20:1547. doi: 10.1002/eji.1830200721. [DOI] [PubMed] [Google Scholar]
- 9.Stingl G, Maurer D. IgE‐mediated allergen presentation via Fc epsilon RI on antigen‐ presenting cells. Int Arch Allergy Immunol. 1997;113:24. doi: 10.1159/000237499. [DOI] [PubMed] [Google Scholar]
- 10.Fahy JV, Fleming HE, Wong HH, et al. The effect of an anti‐IgE monoclonal antibody on the early‐ and late‐phase responses to allergen inhalation in asthmatic subjects [see comments] Am J Respir Crit Care Med. 1997;155:1828. doi: 10.1164/ajrccm.155.6.9196082. [DOI] [PubMed] [Google Scholar]
- 11.Crimi E, Scordamaglia A, Crimi P, Zupo S, Barocci S. Total and specific IgE in serum, bronchial lavage and bronchoalveolar lavage of asthmatic patients. Allergy. 1983;38:553. doi: 10.1111/j.1398-9995.1983.tb04139.x. [DOI] [PubMed] [Google Scholar]
- 12.Diaz‐sanchez D, Tsien A, Fleming J, Saxon A. Combined diesel exhaust particulate and ragweed allergen challenge markedly enhances human in vivo nasal ragweed‐specific IgE and skews cytokine production to a T helper cell 2‐type pattern. J Immunol. 1997;158:2406. [PubMed] [Google Scholar]
- 13.Nahm DH, Park HS. Analysis of induced sputum for studying allergen‐specific IgE antibodies in airway secretion from asthmatic patients [see comments] Clin Exp Allergy. 1998;28:686. doi: 10.1046/j.1365-2222.1998.00291.x. [DOI] [PubMed] [Google Scholar]
- 14.Durham SR, Gould HJ, Thienes CP, et al. Expression of epsilon germ‐line gene transcripts and mRNA for the epsilon heavy chain of IgE in nasal B cells and the effects of topical corticosteroid. Eur J Immunol. 1997;27:2899. doi: 10.1002/eji.1830271123. [DOI] [PubMed] [Google Scholar]
- 15.Ghaffar O, Durham SR, Al‐ghamdi K, et al. Expression of IgE heavy chain transcripts in the sinus mucosa of atopic and nonatopic patients with chronic sinusitis. Am J Respir Cell Mol Biol. 1998;18:706. doi: 10.1165/ajrcmb.18.5.3030. [DOI] [PubMed] [Google Scholar]
- 16.Fujieda S, Diaz‐sanchez D, Saxon A. Combined nasal challenge with diesel exhaust particles and allergen induces in vivo IgE isotype switching. Am J Respir Cell Mol Biol. 1998;19:507. doi: 10.1165/ajrcmb.19.3.3143. [DOI] [PubMed] [Google Scholar]
- 17.Snow RE, Chapman CJ, Frew AJ, Holgate ST, Stevenson FK. Analysis of Ig VH region genes encoding IgE antibodies in splenic B lymphocytes of a patient with asthma. J Immunol. 1995;154:5576. [PubMed] [Google Scholar]
- 18.Snow RE, Chapman CJ, Holgate ST, Stevenson FK. Immunogenetics of human IgE. Hum Antibodies Hybridomas. 1996;7:157. [PubMed] [Google Scholar]
- 19.van der Stoep N, van der Linden J, Logtenberg T. Molecular evolution of the human immunoglobulin E response: high incidence of shared mutations and clonal relatedness among epsilon VH5 transcripts from three unrelated patients with atopic dermatitis. J Exp Med. 1993;177:99. doi: 10.1084/jem.177.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Silverman GJ. Human antibody responses to bacterial antigens: studies of a model conventional antigen and a proposed model B cell superantigen. Int Rev Immunol. 1992;9:57. doi: 10.3109/08830189209061783. [DOI] [PubMed] [Google Scholar]
- 21.Workshop summary and guidelines: investigative use of bronchoscopy, lavage, and bronchial biopsies in asthma and other airway diseases. J Allergy Clin Immunol. 1991. p. 808. [DOI] [PubMed]
- 22.Djukanovic R, Wilson JW, Lai CKW, Holgate ST, Howarth PH. The safety aspects of fiberoptic bronchoscopy, bronchoalveolar lavage, and endobronchial biopsy in asthma. Am Rev Respir Dis. 1991;143:772. doi: 10.1164/ajrccm/143.4_Pt_1.772. [DOI] [PubMed] [Google Scholar]
- 23.Snow RE, Chapman CJ, Frew AJ, Holgate ST, Stevenson FK. Pattern of usage and somatic hypermutation in the V (H) 5 gene segments of a patient with asthma: implications for IgE. Eur J Immunol. 1997;27:162. doi: 10.1002/eji.1830270124. [DOI] [PubMed] [Google Scholar]
- 24.Efremov DG, Batista FD, Burrone OR. Molecular analysis of IgE H‐chain transcripts expressed in vivo by peripheral blood lymphocytes from normal and atopic individuals. J Immunol. 1993;151:2195. [PubMed] [Google Scholar]
- 25.Snow RE, Chapman CJ, Holgate ST, Stevenson FK. Clonally related IgE and IgG4 transcripts in blood lymphocytes of patients with asthma reveal differing patterns of somatic mutation [In Process Citation] Eur J Immunol. 1998;28:3354. doi: 10.1002/(SICI)1521-4141(199810)28:10<3354::AID-IMMU3354>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 26.Brezinschek HP, Brezinschek RI, Lipsky PE. Analysis of the heavy chain repertoire of human peripheral B cells using single‐cell polymerase chain reaction. J Immunol. 1995;155:190. [PubMed] [Google Scholar]
- 27.Chang B, Casali P. The CDR1 sequences of a major proportion of human germline Ig VH genes are inherently susceptible to amino acid replacement. Immunol Today. 1994;15:367. doi: 10.1016/0167-5699(94)90175-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Betz AG, Neuberger MS, Milstein C. Discriminating intrinsic and antigen‐selected mutational hotspots in immunoglobulin V genes. Immunol Today. 1993;14:405. doi: 10.1016/0167-5699(93)90144-a. [DOI] [PubMed] [Google Scholar]
- 29.Janezic A, Chapman CJ, Snow RE, Hourihane JO, Warner JO, Stevenson FK. Immunogenetic analysis of the heavy chain variable regions of IgE from patients allergic to peanuts. J Allergy Clin Immunol. 1998;101:391. doi: 10.1016/S0091-6749(98)70253-2. [DOI] [PubMed] [Google Scholar]
- 30.Lundgren M, Persson U, Larsson P, et al. Interleukin 4 induces synthesis of IgE and IgG4 in human B cells. Eur J Immunol. 1989;19:1311. doi: 10.1002/eji.1830190724. [DOI] [PubMed] [Google Scholar]
- 31.Punnonen J, Aversa G, Cocks BG, et al. Interleukin 13 induces interleukin 4‐independent IgG4 and IgE synthesis and CD23 expression by human B cells. Proc Natl Acad Sci USA. 1993;90:3730. doi: 10.1073/pnas.90.8.3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carballido JM, Carballido‐perrig N, Oberli‐schrammli A, Heusser CH, Blaser K. Regulation of IgE and IgG4 responses by allergen specific T‐cell clones to bee venom phospholipase A2 in vitro. J Allergy Clin Immunol. 1994;93:758. doi: 10.1016/0091-6749(94)90256-9. [DOI] [PubMed] [Google Scholar]
- 33.Akdis M, Akdis CA, Weigl L, Disch R, Blaser K. Skin‐homing, CLA+ memory T cells are activated in atopic dermatitis and regulate IgE by an IL‐13‐dominated cytokine pattern: IgG4 counter‐ regulation by CLA‐memory T cells. J Immunol. 1997;159:4611. [PubMed] [Google Scholar]
- 34.Humbert M, Durham SR, Ying S, et al. IL‐4 and IL‐5 mRNA and protein in bronchial biopsies from patients with atopic and nonatopic asthma: evidence against ‘intrinsic’ asthma being a distinct immunopathologic entity. Relationship between IL‐4 and IL‐5 mRNA expression and disease severity in atopic asthma. Am J Respir Crit Care Med. 1996;154:1497. doi: 10.1164/ajrccm.154.5.8912771. [DOI] [PubMed] [Google Scholar]
- 35.Humbert M, Durham SR, Kimmitt P, et al. Elevated expression of messenger ribonucleic acid encoding IL‐13 in the bronchial mucosa of atopic and nonatopic subjects with asthma. J Allergy Clin Immunol. 1997;99:657. doi: 10.1016/s0091-6749(97)70028-9. [DOI] [PubMed] [Google Scholar]
- 36.Matsumoto M, Lo SF, Carruthers CJ, et al. Affinity maturation without germinal centres in lymphotoxin‐alpha‐deficient mice. Nature. 1996;382:462. doi: 10.1038/382462a0. [DOI] [PubMed] [Google Scholar]
- 37.Choe J, Kim HS, Zhang X, Armitage RJ, Choi YS. Cellular and molecular factors that regulate the differentiation and apoptosis of germinal center B cells. Anti‐Ig down‐regulates Fas expression of CD40 ligand‐stimulated germinal center B cells and inhibits Fas‐mediated apoptosis. J Immunol. 1996;157:1006. [PubMed] [Google Scholar]
- 38.Cook GP, Tomlinson IM. The human immunoglobulin VH repertoire. Immunol Today. 1995;16:237. doi: 10.1016/0167-5699(95)80166-9. [DOI] [PubMed] [Google Scholar]