Abstract
TNF-induced activation of the transcription factor NF-κB and the c-jun N-terminal kinase (JNK/SAPK) requires TNF receptor-associated factor 2 (TRAF2). The NF-κB-inducing kinase (NIK) associates with TRAF2 and mediates TNF activation of NF-κB. Herein we show that NIK interacts with additional members of the TRAF family and that this interaction requires the conserved “WKI” motif within the TRAF domain. We also investigated the role of NIK in JNK activation by TNF. Whereas overexpression of NIK potently induced NF-κB activation, it failed to stimulate JNK activation. A kinase-inactive mutant of NIK was a dominant negative inhibitor of NF-κB activation but did not suppress TNF- or TRAF2-induced JNK activation. Thus, TRAF2 is the bifurcation point of two kinase cascades leading to activation of NF-κB and JNK, respectively.
Keywords: interleukin 1, nuclear factor-κB-inducing kinase, mitogen-activated protein kinase/extracellular signal-regulated kinase kinase 1, TNF receptor-associated factor 6
Cytokine-induced activation of otherwise latent transcription factors is one of the mechanisms by which extracellular signals are transduced from the cell surface to the nucleus (1). TNF is a potent cytokine produced mainly by activated macrophages and monocytes that elicits a broad range of biological effects (for reviews, see refs. 2–4). TNF triggers the activation of two transcription factors, nuclear factor-κB (NF-κB) (5) and activator protein 1 (AP-1) (6), which regulate the expression of numerous immune and inflammatory response genes (for reviews see, refs. 7–9). Both transcription factors are activated through protein kinase cascades culminating in the phosphorylation of yet-to-be-identified IκB kinases (10) and the molecularly characterized c-jun N-terminal kinases (JNK) (11), respectively.
TNF-induced activation of both NF-κB and JNK requires TNF receptor-associated factor 2 (TRAF2) (12–16), a member of the TRAF family of proteins that associate with and transduce signals from TNF receptor family members (12, 17–28). TRAF proteins consist of a conserved C-terminal TRAF domain and an N-terminal region containing a RING-finger motif and an additional array of zinc-finger-like structures (17–19). The TRAF domain is involved in receptor association and homo- and hetero-oligomerization of TRAFs and serves as a docking site for a number of other signaling proteins (13, 17–19, 29–34). The N-terminal zinc binding domains are involved in mediating downstream signaling events (12, 19).
Of the six described TRAF proteins, TRAF5 and TRAF6 have also been implicated in NF-κB activation. TRAF5 is involved in NF-κB activation by members of the TNF receptor family similar to TRAF2 (25–28). In contrast, TRAF6 participates in NF-κB activation by interleukin 1 (IL-1) (35). TRAF6 associates with the serine-threonine kinase IRAK after the IL-1-induced activation of IRAK in the IL-1 receptor complex (35, 36). Recent studies (14–16) have shown that TRAF2-mediated activation of JNK, but not NF-κB, can be inhibited by a catalytically inactive mutant of mitogen-activated protein kinase/extracellular signal-regulated kinase kinase 1 (MEKK1) (MEKK1Δ(K432M); ref. 11), a member of the mitogen-activated protein (MAP) kinase kinase kinase (MAP3K) family. This indicates that the JNK and NF-κB signaling pathways diverge at some point downstream of TRAF2.
The NF-κB-inducing kinase (NIK) is a MAP3K-related kinase that binds TRAF2 and activates NF-κB when overexpressed (37). Kinase-inactive mutants of NIK containing its TRAF2-interacting C-terminal domain (NIK(624–947)) or lacking the two lysine residues in its catalytic domain (NIK(KK429–430AA)) behave as dominant negative inhibitors that suppress TNF- and IL-1-induced NF-κB activation (37).
Herein we show that NIK associates with other members of the TRAF family in addition to TRAF2, including TRAF1, TRAF3, TRAF5, and TRAF6. These interactions require the conserved “WKI” motif within the TRAF domain. Catalytically inactive NIK mutants inhibit NF-κB activation by TRAF5 and TRAF6 similar to TRAF2. Like TRAF2, TRAF5, and TRAF6 activate JNK when overexpressed. In contrast, whereas overexpression of NIK potently induces NF-κB activation, it does not stimulate JNK activation. In addition, catalytically inactive NIK(KK429–430AA) is a dominant negative inhibitor of NF-κB activation but does not suppress TNF- or TRAF2-induced JNK activation. Thus, TRAF2 is the bifurcation point of two kinase cascades leading to activation of NF-κB and JNK, respectively.
MATERIALS AND METHODS
Cell Culture and Biological Reagents.
Human embryonic kidney 293 cells were maintained as described (38). Recombinant human TNF was provided by Genentech. The anti-Flag epitope mAb M2 and anti-hemagglutinin (HA) epitope monoclonal antibody HA.11 were purchased from Eastman Kodak and Babco (Richmond, CA), respectively. Rabbit anti-Myc, anti-Flag, and anti-HA epitope polyclonal antibodies were from Santa Cruz Biotechnology.
cDNA Cloning and Expression Vectors.
A full-length NIK cDNA was obtained (and subsequently sequenced) by screening a human embryonic kidney 293 cell cDNA library by standard methods (39). We found one difference in the amino acid sequence of the deduced human NIK protein compared with the previously described amino acid sequence of NIK (37). This difference is at residue 25 (Ala vs. Pro). An expression vector for Myc-epitope-tagged NIK was constructed in-frame with DNA encoding an N-terminal Myc epitope in pRK (40). Expression plasmids encoding NIK(624–947) and NIK(KK429–430AA) were generated by PCR. Expression vectors for TRAFs have been described (12, 18, 25, 33, 35). The NF-κB-dependent E-selectin–luciferase reporter gene plasmid and pRSV-β-gal have also been described (12, 41). Expression vectors for MEKK1, MEKK1Δ(K432M), HA-epitope-tagged JNK1, and glutathione S-transferase (GST)–cJun(1–79) were provided by Zheng-gang Liu and Michael Karin (University of San Diego, San Diego).
Transfections and Reporter Assays.
293 cells (4 × 105 cells per well) were seeded into six-well (35 mm) plates. Cells were transfected the following day by the calcium phosphate precipitation method (42) with 0.5 μg of E-selectin–luciferase reporter gene plasmid and various amounts of each expression construct. The total DNA concentration (5 μg) was kept constant by supplementation with pRK. Cells were lysed 24 h after transfection and reporter gene activity determined with the luciferase assay system (Promega). A pRSV-βgal vector (1 μg) was used for normalizing transfection efficiencies.
Immunoprecipitations and Immunoblotting.
293 cells (2 × 106 cells) were plated on 10-cm dishes and transfected with a total of 10 μg of DNA containing various expression plasmids. After 24 h, cells were collected, and lysates were prepared and immunoprecipitated with anti-Flag epitope or anti-HA epitope monoclonal antibody or control mouse IgG1 monoclonal antibody (Sigma) as described (29). Bound proteins were fractionated by SDS/PAGE and transferred to nitrocellulose membrane. Coprecipitating proteins were detected by immunoblot analysis with anti-Myc epitope polyclonal antibodies as described (29). In all cases, expression of transfected plasmids was verified by immunoblotting of aliquots of cell lysates with anti-Flag, anti-HA, or anti-Myc epitope polyclonal antibodies.
Protein Kinase Assays.
To obtain comparable results for JNK and NF-κB activation, two wells with 293 cells received calcium phosphate/DNA precipitate from the same mixture (see Transfections and Reporter Assays, above) which also contained 0.5 μg of a HA epitope-tagged JNK1 expression vector. After 24 h, cells from one well were stimulated with TNF (100 ng/ml) for 6 h or left untreated and then collected for luciferase assays. Cells from the other well were either left untreated or stimulated with TNF for 30 min and harvested. Whole-cell lysates were prepared (43), and epitope-tagged JNK1 was immunoprecipitated with anti-HA monoclonal antibody. The activity of HA epitope-tagged JNK1 was determined by immune complex kinase assay with 1 μg of GST–cJun(1–79) as substrate (14). The amount of epitope-tagged JNK1 in each transfection was analyzed by immunoblotting of aliquots of cell lysates with anti-HA polyclonal antibodies. Only experiments with comparable amounts of HA epitope-tagged JNK1 in each sample were taken into consideration. For solid-phase JNK assays, whole-cell lysates from transiently transfected 293 cells were prepared and incubated with bacterially expressed Gst–cJun(1–79) fusion protein beads. The beads were washed and incubated in kinase buffer with [γ-32P]-ATP as described (43). Reactions were analyzed by SDS/PAGE and autoradiography.
RESULTS
Effect of NIK Mutants on TRAF-Mediated NF-κB Activation.
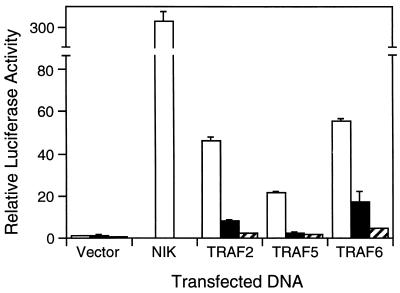
Catalytically inactive NIK mutants have previously been shown to inhibit NF-κB activation induced by TNF and IL-1 stimulation as well as TRAF2 overexpression (37). To further investigate the role of NIK in NF-κB signaling, we examined its effect on NF-κB activation by other members of the TRAF family, TRAF5 and TRAF6. NIK(624–947) and NIK(KK429–430AA) were transiently coexpressed in 293 cells with TRAF2, TRAF5, or TRAF6 in the presence of an NF-κB-dependent E-selectin reporter gene plasmid. Luciferase assays were performed 24 h later and demonstrated that both kinase-inactive NIK mutants inhibit activation of the reporter gene by TRAF2 as well as TRAF5 and TRAF6 (Fig. 1).
Figure 1.
Dominant negative effect of NIK mutants on TRAF-mediated NF-κB activation. 293 cells were transiently cotransfected with an E-selectin–luciferase reporter gene plasmid and vector control or expression vectors for wild-type NIK (1 μg) or TRAFs (1 μg) as indicated, either alone (open bars) or in combination with expression plasmids for NIK(624–947) (2 μg; solid bars) or NIK(KK429–430AA) (2 μg; hatched bars). After 24 h, cells were harvested and luciferase activities were determined and normalized based on β-galactosidase expression. Values shown are averages (mean ± SD) of one representative experiment in which each transfection was performed in duplicate.
Association of NIK with TRAF Family Members.
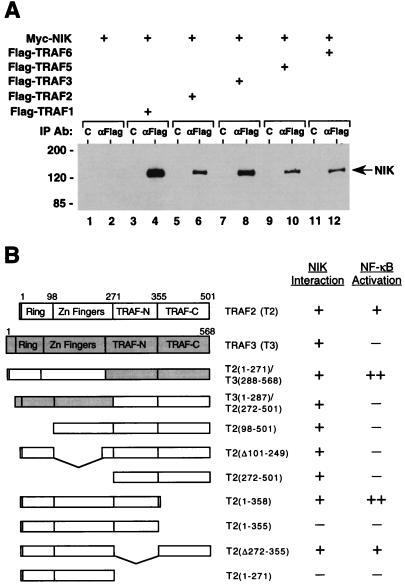
We next analyzed the interaction of NIK and TRAF proteins in overexpression-based coimmunoprecipitation assays. Flag-epitope-tagged TRAF proteins were coexpressed with Myc-epitope-tagged NIK in 293 cells. After 24 h, cell lysates were immunoprecipitated with anti-Flag monoclonal antibody and coprecipitating epitope-tagged NIK was detected by immunoblot analysis with anti-Myc polyclonal antibodies. In these experiments NIK was found to associate with all three NF-κB-inducing TRAF proteins (Fig. 2A). Thus, with the aforementioned results, this indicates that NIK acts as a common mediator of NF-κB activation by TRAF2, TRAF5, and TRAF6, thereby explaining the inhibitory effect of kinase-inactive NIK mutants on NF-κB signaling by both TNF and IL-1.
Figure 2.
Interaction of NIK with TRAF proteins. (A) Association of NIK with members of the TRAF family. 293 cells were transiently cotransfected with epitope-tagged NIK (lanes 1–12) and TRAF expression vectors (3 μg; lanes 3–12) as indicated. After 24 h, cell lysates were prepared and Flag epitope-tagged TRAF proteins were immunoprecipitated with anti-Flag epitope mAb (lanes 2, 4, 6, 8, 10, and 12) or control mAb (lanes 1, 3, 5, 7, 9, and 11). Coprecipitating Myc epitope-tagged NIK was detected by immunoblot analysis using anti-Myc epitope polyclonal antibodies. The position of molecular mass markers (in kilodaltons) is shown on the left. (B) Interaction of NIK with TRAF mutants; 293 cells were transiently cotransfected with expression vectors for Myc epitope-tagged NIK and the indicated HA epitope-tagged wild-type and mutant TRAF proteins. After 24 h, aliquots of cell lysates were immunoprecipitated with anti-HA epitope mAb or control mAb. Coprecipitating NIK was detected by immunoblot analysis with anti-Myc epitope polyclonal antibodies. +, binding of NIK to the respective TRAF proteins; −, lack of such an interaction in this assay. Also listed is the ability of the indicated TRAF proteins to activate an NF-κB-dependent reporter gene in transient transfection assays (33).
NIK was also able to associate with TRAF proteins that do not induce NF-κB activation upon overexpression including TRAF1, TRAF3 (Fig. 2A), and a chimeric TRAF protein comprising the zinc-finger domains of TRAF3 and the TRAF domain of TRAF2 (33) (Fig. 2B). The interaction of NIK with TRAF3 was verified in yeast two-hybrid assays (H.Y.S., C.H.R., and M.R., unpublished results). These findings suggest that NIK binds to the conserved TRAF domain of TRAF proteins and that the N-terminal zinc-finger domains do not contribute to this interaction.
Interaction of NIK with Mutant TRAF2 Proteins.
We also examined the interaction of NIK with various truncation mutants of TRAF2 (33). Expression vectors encoding HA-epitope-tagged TRAF2 deletion mutants were cotransfected with an expression plasmid for Myc-epitope-tagged NIK into 293 cells. Cell lysates were immunoprecipitated with anti-HA monoclonal antibody, and coprecipitating epitope-tagged NIK was again detected by immunoblot analysis with anti-Myc epitope polyclonal antibodies. In this analysis partial as well a complete deletions of the N-terminal zinc-finger regions of TRAF2 had no effect on the interaction with NIK (Fig. 2B). This confirms that the zinc-finger domains of TRAF2 are dispensible for its association with NIK.
To map the interaction of NIK within the TRAF domain, C-terminal truncation mutants of TRAF2 (33) were examined for their association with NIK (Fig. 2B). Consistent with aforementioned results, a TRAF2 mutant lacking the entire TRAF domain (TRAF2(1–271)) did not interact with NIK. However, an internal deletion of amino acids 272–355 of TRAF2 (TRAF2(Δ272–355)) and a C-terminal deletion of amino acids 359–501 (TRAF2(1–358)) were still able to associate with NIK. In contrast, removal of three additional amino acids from TRAF2(1–358) (TRAF2(1–355)) caused a loss of NIK binding. The deleted sequence WKI is highly conserved among the known members of the TRAF family (33), thereby identifying a conserved motif within the TRAF domain that is required for NIK binding. Previous analysis had shown that TRAF2(Δ272–355) and TRAF2(1–358) were still capable of inducing NF-κB activation whereas TRAF2(1–355) had lost this ability (33). Thus, the loss of NIK association with TRAF2 correlates with the loss of NF-κB inducing activity. This further supports a crucial role for NIK in NF-κB signaling by TRAF2.
Activation of JNK by TRAF Family Members.
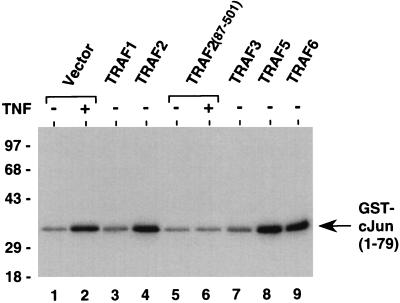
In addition to NF-κB activation, overexpression of TRAF2 induces activation of JNK, and a truncated derivative of TRAF2 without the N-terminal RING-finger domain acts as a dominant negative inhibitor of both TNF-induced NF-κB and JNK activation (refs. 14–16 and Fig. 3). To test whether other members of the TRAF family can also signal JNK activation, JNK assays were performed in 293 cells overexpressing TRAF proteins by using bacterially expressed GST–cJun(1–79) protein as substrate. TRAF5 and TRAF6, but not TRAF1 or TRAF3, induced JNK activation similar to TRAF2 (Fig. 3). Thus, all TRAF family members capable of NF-κB induction are also able to signal JNK activation.
Figure 3.
Activation of JNK by TRAF family members. 293 cells were transiently transfected with vector control (lanes 1 and 2) or TRAF expression vectors (3 μg; lanes 3–9) as indicated. After 24 h, cells were stimulated with TNF (100 ng/ml) for 30 min (lanes 2 and 6) or left untreated (lanes 1, 3, 4, and 7–9) prior to harvest. Aliquots of cell lysates were incubated with GST–cJun(1–79) fusion protein beads (1 μg). The activity of coprecipitating JNK was determined by solid-phase kinase assay. The location of the GST–cJun(1–79) fusion protein is indicated. The position of molecular mass markers (in kilodaltons) is shown on the left.
NIK Does Not Signal JNK Activation.
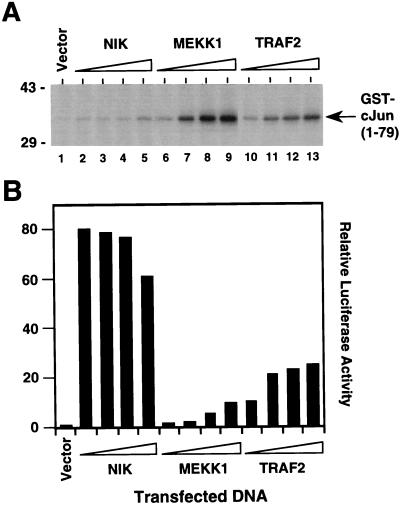
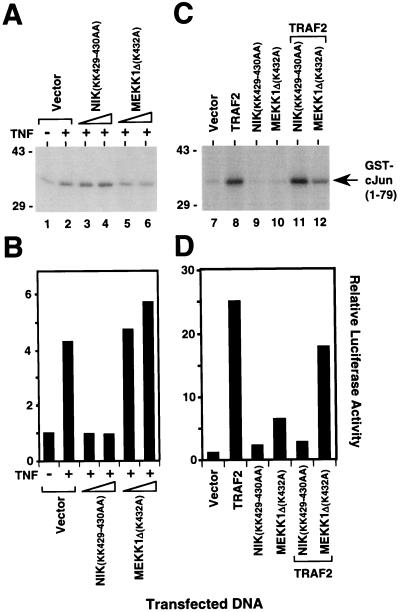
We next determined whether NIK can activate JNK. Overexpression of wild-type NIK in 293 cells did not induce activation of JNK at concentrations of transfected DNA that simultaneously gave rise to strong NF-κB activation (Fig. 4). In contrast, overexpression of MEKK1 induced strong activation of JNK at low concentrations of transfected DNA, which caused only modest activation of NF-κB (Fig. 4). TRAF2 overexpression on the other hand activated NF-κB and JNK equally well (Fig. 4). To examine whether NIK is involved in TNF-induced JNK activation, we again used kinase-inactive NIK(KK429–430AA). Whereas overexpression of this mutant NIK protein in 293 cells inhibited NF-κB activation by TNF (Fig. 5B), it did not suppress TNF-mediated JNK activation (Fig. 5B). In contrast, overexpression of catalytically inactive MEKK1Δ(K432M) inhibited JNK but not NF-κB activation induced by TNF (Fig. 5 A and B and ref. 14). The same results were obtained for TRAF2-induced JNK and NF-κB activation (Fig. 5 C and D). These findings demonstrate that NIK overexpression signals NF-κB but not JNK activation and that NIK does not mediate JNK activation by TNF. Thus, two distinct signaling cascades emanate from TRAF2 that lead to NF-κB and JNK activation, respectively.
Figure 4.
Activation of NF-κB but not JNK by NIK. 293 cells were transiently cotransfected with an E-selectin–luciferase reporter gene plasmid (lanes 1–13), an expression plasmid for HA epitope-tagged JNK1 (lanes 1–13) and vector control (lane 1) or 0.1 μg (lanes 2, 6, and 10), 0.3 μg (lanes 3, 7, and 11), 0.6 μg (lanes 4, 8, and 12), or 1 μg (lanes 5, 9, and 13) of NIK (lanes 2–5), MEKK1 (lanes 6–9), or TRAF2 expression plasmid (lanes 10–13). After 24 h, cells were collected and lysates were prepared. (A) Activity of HA epitope-tagged JNK1 was determined by immune complex kinase assay with GST–cJun(1–79) as a substrate. The location of the GST–cJun(1–79) fusion protein is indicated. The position of molecular mass markers (in kilodaltons) is shown on the left. (B) Luciferase activities corresponding to the transfections in A were determined and normalized based on β-galactosidase expression. Similar results were obtained in at least two independent experiments.
Figure 5.
Role of NIK in TNF- and TRAF2-induced NF-κB and JNK activation. Inhibition of TNF- (A and B) and TRAF2-mediated (C and D) activation of NF-κB but not JNK by NIK(KK429–430AA). 293 cells were transiently cotransfected with an E-selectin–luciferase reporter gene plasmid (lanes 1–12), an expression plasmid for HA epitope-tagged JNK1 (lanes 1–12) and vector control (lanes 1, 2, and 7), TRAF2 expression plasmid (1 μg; lanes 8, 11, and 12), and 1 μg (lanes 3, 5, 9, and 11) or 3 μg (lanes 4, 6, 10, and 12) of expression vectors for NIK(KK429–430AA) (lanes 3, 4, 9, and 11) or MEKK1Δ(K432M) (lanes 5, 6, 10, and 12). After 24 h, cells were stimulated for 30 min (A) or 6 h (B) with TNF (100 ng/ml) (lanes 2–6) or left untreated (lanes 1 and 7–12) before harvest. (A and C) Activity of HA epitope-tagged JNK1 was determined by immune complex kinase assay with GST–cJun(1–79) as a substrate. The location of the GST–cJun(1–79) fusion protein is indicated. The position of molecular mass markers (in kilodaltons) is shown on the left. (B and D) Luciferase activities corresponding to the transfections in A and C were determined and normalized based on β-galactosidase expression. Similar results were obtained in at least two independent experiments.
DISCUSSION
The recent identification of NIK provided an important link in the TNF-induced NF-κB activation pathway downstream of TRAF2. However, NIK was also found to be involved in NF-κB activation by IL-1 (37), which is mediated by TRAF6 (35). Our identification of NIK as a TRAF6- and TRAF2-interacting protein provides a unifying concept for NIK as a common mediator of TNF and IL-1 signaling.
Earlier studies have shown that the zinc-finger domains of TRAF proteins are required for signal transduction including NF-κB activation (12, 19). In particular, it has been speculated in the case of TRAF2 that its N-terminal RING-finger domain might represent a potential protein–protein interaction domain that might connect TRAF2 with subsequent steps in the signaling cascade (12). However, the present analysis demonstrates that the zinc-finger regions of TRAF2 are dispensible for its interaction with the NF-κB signal transducer NIK. Conceptually, rather than being involved in NIK binding, the RING- and zinc-finger domain of TRAF2 (and by analogy those of TRAF5 and TRAF6) may exert a regulatory function on NIK’s kinase activity or alter its substrate specificity.
Consistent with these observations, we found that the TRAF domain of TRAF2 is sufficient for association with NIK. In an attempt to map the interaction of NIK within the TRAF domain of TRAF2 more precisely, we identified a WKI motif that is required for NIK binding. This sequence motif is also essential for NF-κB induction by TRAF2 (33), further supporting an obligatory role for NIK in TRAF2-mediated NF-κB activation.
The WKI motif is conserved among TRAF family members (33). As implied by this finding, NIK associates with other members of the TRAF family in addition to TRAF2. However, although NIK appears to be dedicated to NF-κB signaling, it interacts not only with those TRAF proteins that are involved in NF-κB activation, i.e., TRAF2, TRAF5, and TRAF6, but also with TRAF1 and TRAF3 that have not been implicated in this pathway. This suggests that TRAF1 and TRAF3, although binding to NIK, are unable to activate its catalytic activity, presumably due to differences between their N-terminal regions and those of TRAF2, TRAF5, and TRAF6. This is obvious in the case of TRAF1, which does not contain a RING-finger domain (17). A similar situation has been described for the interaction of the serine-threonine kinase c-Raf-1 and members of the Ras family of GTP-binding proteins (44). Both Ras and Rap-1, a Ras-related protein that acts as an inhibitor of Ras-mediated transformation, bind with high affinity to c-Raf-1. However, the interaction with Ras stimulates c-Raf-1 activity, whereas the interaction with Rap-1 does not lead to c-Raf-1 activation. In this manner Rap-1 competes with Ras for c-Raf-1, thus inhibiting Ras signaling through c-Raf-1 (44). By analogy, it is possible that TRAF1 and TRAF3 may sequester NIK and thereby prevent its activation by other TRAFs. This may contribute to a negative regulation of TRAF-induced NF-κB activation. An implication of such a mechanism may be the existence of other (NIK-related?) kinases that specifically mediate downstream responses of other TRAF proteins (see also below). The analogy between the NIK–TRAF and Raf–Ras regulatory systems appears particularly attractive, because Raf family members function as MAP3Ks in the MAP kinase cascade (45), and therefore TRAFs act at the same level as Ras family members. However, the precise mechanisms of NIK activation after TNF and IL-1 stimulation remain to be elucidated.
The findings outlined in the present study demonstrate that the previously described bifurcation of the TNF-induced NF-κB and JNK activation pathways (14–16) occurs at the level of TRAF2. This observation supports a functional model for TRAF2 and other members of the TRAF family as adaptor proteins with docking sites for additional signaling proteins that initiate parallel downstream responses. In this respect, the MAP3K-related kinase NIK represents the initial link in the NF-κB signaling cascade. By analogy, we propose the existence of a distinct TRAF2-interacting kinase that triggers the signaling cascade culminating in JNK activation. Although the NIK-related MEKK1 may be involved in JNK activation by TNF (14), it does not bind TRAF2 directly (H.Y.S. and M.R., unpublished results). Another member of the MAP3K family, ASK1, was recently found to induce JNK activation and apoptosis in mammalian cells, and its activity is stimulated by TNF treatment (46). Indeed, whereas overexpression of Ask1 strongly activates JNK, it does not induce NF-κB activation (H.Y.S. and M.R., unpublished results). However, an overexpressed catalytically inactive mutant of Ask1 (Ask1(K709R)) does not block TNF- or TRAF2-mediated JNK activation (H.Y.S. and M.R., unpublished results), although it has been shown to suppress TNF-induced apoptosis (46). The identification of TRAF2-interacting protein kinases involved in TNF activation of JNK may provide further insights into the signaling mechanisms used by members of the TRAF family.
Acknowledgments
We thank Z.-G. Liu and M. Karin for providing various expression plasmids. G. Natoli is gratefully acknowledged for sharing results prior to publication and for helpful suggestions on the manuscript. We also thank Z. Cao and P. Baeuerle for critically reading the manuscript.
ABBREVIATIONS
- IL-1
interleukin 1
- JNK (SAPK)
c-jun N-terminal kinase
- MAP kinase
mitogen-activated protein kinase
- MAP3K
mitogen-activated protein kinase kinase kinase
- NIK
NF-κB-inducing kinase
- TNF
tumor necrosis factor
- TRAF
TNF receptor-associated factor
- MEKK1
mitogen-activated protein kinase/extracellular signal-regulated kinase kinase 1
- HA
hemagglutinin
- GST
glutathione S-transferase
References
- 1.Hill C S, Treisman R. Cell. 1995;80:199–211. doi: 10.1016/0092-8674(95)90403-4. [DOI] [PubMed] [Google Scholar]
- 2.Goeddel D V, Aggarwal B B, Bray P W, Leung D W, Nedwin G E, Palladino M A, Patton J S, Pennica D, Shepard H M, Sugarman B J, Wong G H W. Cold Spring Harbor Symp Quant Biol. 1986;51:597–609. doi: 10.1101/sqb.1986.051.01.072. [DOI] [PubMed] [Google Scholar]
- 3.Beutler B, Cerami A. Annu Rev Biochem. 1988;57:505–518. doi: 10.1146/annurev.bi.57.070188.002445. [DOI] [PubMed] [Google Scholar]
- 4.Fiers W. FEBS Lett. 1991;285:199–212. doi: 10.1016/0014-5793(91)80803-b. [DOI] [PubMed] [Google Scholar]
- 5.Osborn L, Kunkel S, Nabel G J. Proc Natl Acad Sci USA. 1989;86:2336–2340. doi: 10.1073/pnas.86.7.2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brenner D A, O’Hara M, Angel P, Chojkier M, Karin M. Nature (London) 1989;337:661–663. doi: 10.1038/337661a0. [DOI] [PubMed] [Google Scholar]
- 7.Baeuerle P A, Henkel T. Annu Rev Immunol. 1994;12:141–179. doi: 10.1146/annurev.iy.12.040194.001041. [DOI] [PubMed] [Google Scholar]
- 8.Baeuerle P A, Baltimore D. Cell. 1996;87:13–20. doi: 10.1016/s0092-8674(00)81318-5. [DOI] [PubMed] [Google Scholar]
- 9.Karin M, Liu S-g, Zandi E. Curr Opin Cell Biol. 1997;9:240–246. doi: 10.1016/s0955-0674(97)80068-3. [DOI] [PubMed] [Google Scholar]
- 10.DiDonato J A, Mercurio F, Rosette C, Wu-Li J, Suyang H, Gosh S, Karin M. Mol Cell Biol. 1996;16:1295–1304. doi: 10.1128/mcb.16.4.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Minden A, Lin A, Claret F-X, Lange-Carter C, Dérijard B, Davis R J, Johnson G L, Karin M. Science. 1994;266:1719–1723. doi: 10.1126/science.7992057. [DOI] [PubMed] [Google Scholar]
- 12.Rothe M, Sarma V, Dixit V M, Goeddel D V. Science. 1995;269:1424–1427. doi: 10.1126/science.7544915. [DOI] [PubMed] [Google Scholar]
- 13.Hsu H, Shu H-B, Pan M-P, Goeddel D V. Cell. 1996;84:299–308. doi: 10.1016/s0092-8674(00)80984-8. [DOI] [PubMed] [Google Scholar]
- 14.Liu Z-g, Hsu H, Goeddel D V, Karin M. Cell. 1996;87:565–576. doi: 10.1016/s0092-8674(00)81375-6. [DOI] [PubMed] [Google Scholar]
- 15.Natoli G, Costanzo A, Ianni A, Templeton D J, Woodgett J R, Balsano C, Levrero M. Science. 1997;275:200–203. doi: 10.1126/science.275.5297.200. [DOI] [PubMed] [Google Scholar]
- 16.Reinhard C, Shamoon B, Shyamala V, Williams L T. EMBO J. 1997;16:1080–1092. doi: 10.1093/emboj/16.5.1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rothe M, Wong S C, Henzel W J, Goeddel D V. Cell. 1994;78:681–692. doi: 10.1016/0092-8674(94)90532-0. [DOI] [PubMed] [Google Scholar]
- 18.Hu H M, O’Rourke K, Boguski M S, Dixit V M. J Biol Chem. 1994;269:30069–30072. [PubMed] [Google Scholar]
- 19.Cheng G, Cleary A M, Ye Z-s, Hong D, Lederman S, Baltimore D. Science. 1995;267:1494–1498. doi: 10.1126/science.7533327. [DOI] [PubMed] [Google Scholar]
- 20.Mosialos G, Birkenbach M, Yalamanchill R, VanArsdale T, Ware C, Kieff E. Cell. 1995;80:389–399. doi: 10.1016/0092-8674(95)90489-1. [DOI] [PubMed] [Google Scholar]
- 21.Song H Y, Donner D B. Biochem J. 1995;809:825–829. doi: 10.1042/bj3090825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sato T, Irie S, Reed J C. FEBS Lett. 1995;358:113–118. doi: 10.1016/0014-5793(94)01406-q. [DOI] [PubMed] [Google Scholar]
- 23.Gedrich R W, Gilfillan M C, Duckett C S, Van Dongen J L, Thompson C B. J Biol Chem. 1996;271:12852–12858. doi: 10.1074/jbc.271.22.12852. [DOI] [PubMed] [Google Scholar]
- 24.Lee S Y, Park C G, Choi Y. J Exp Med. 1996;183:669–674. doi: 10.1084/jem.183.2.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakano H, Oshima H, Chung W, Williams-Abbott L, Ware C F, Yagita H, Okumura K. J Biol Chem. 1996;271:14661–14664. doi: 10.1074/jbc.271.25.14661. [DOI] [PubMed] [Google Scholar]
- 26.Ishida T, Tojo T, Aoki T, Kobayashi N, Ohishi T, Watanabe T, Yamamoto T, Inoue J-i. Proc Natl Acad Sci USA. 1996;93:9437–9442. doi: 10.1073/pnas.93.18.9437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hsu H, Solovyev I, Colombero A, Elliott R, Kelley M, Boyle W J. J Biol Chem. 1997;272:13471–13474. doi: 10.1074/jbc.272.21.13471. [DOI] [PubMed] [Google Scholar]
- 28.Marsters S A, Ayres T M, Skubatch M, Gray C L, Rothe M, Ashkenazi A. J Biol Chem. 1997;272:14029–14032. doi: 10.1074/jbc.272.22.14029. [DOI] [PubMed] [Google Scholar]
- 29.Rothe M, Pan M-G, Henzel W J, Ayres T M, Goeddel D V. Cell. 1995;83:1243–1252. doi: 10.1016/0092-8674(95)90149-3. [DOI] [PubMed] [Google Scholar]
- 30.Song H Y, Rothe M, Goeddel D V. Proc Natl Acad Sci USA. 1996;93:6721–6725. doi: 10.1073/pnas.93.13.6721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheng G, Baltimore D. Genes Dev. 1996;10:963–973. doi: 10.1101/gad.10.8.963. [DOI] [PubMed] [Google Scholar]
- 32.Rothe M, Xiong J, Shu H-B, Williamson K, Goddard A, Goeddel D V. Proc Natl Acad Sci USA. 1996;93:8241–8246. doi: 10.1073/pnas.93.16.8241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takeuchi M, Rothe M, Goeddel D V. J Biol Chem. 1996;271:19935–19942. doi: 10.1074/jbc.271.33.19935. [DOI] [PubMed] [Google Scholar]
- 34.Lee S Y, Lee S Y, Choi Y. J Exp Med. 1997;185:1275–1285. doi: 10.1084/jem.185.7.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cao Z, Xiong J, Takeuchi M, Kurama T, Goeddel D V. Nature (London) 1996;383:443–446. doi: 10.1038/383443a0. [DOI] [PubMed] [Google Scholar]
- 36.Cao Z, Henzel W J, Gao X. Science. 1996;271:1128–1131. doi: 10.1126/science.271.5252.1128. [DOI] [PubMed] [Google Scholar]
- 37.Malinin N L, Boldin M P, Kovalenko A V, Wallach D. Nature (London) 1997;385:540–544. doi: 10.1038/385540a0. [DOI] [PubMed] [Google Scholar]
- 38.Hsu H, Xiong J, Goeddel D V. Cell. 1995;81:495–504. doi: 10.1016/0092-8674(95)90070-5. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 40.Schall T J, Lewis M, Koller K J, Lee A, Rice G C, Wong G H W, Gatanaga T, Granger G A, Lentz R, Raab H, Kohr W J, Goeddel D V. Cell. 1990;61:361–370. doi: 10.1016/0092-8674(90)90816-w. [DOI] [PubMed] [Google Scholar]
- 41.Schindler U, Baichwal V R. Mol Cell Biol. 1994;14:5820–5831. doi: 10.1128/mcb.14.9.5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. New York: Wiley; 1987. [Google Scholar]
- 43.Hibi A, Lin A, Smeal T, Minden A, Karin M. Genes Dev. 1993;7:2135–2148. doi: 10.1101/gad.7.11.2135. [DOI] [PubMed] [Google Scholar]
- 44.Zhang X-f, Settleman J, Kyriakis J M, Takeuchi-Suzuki E, Elledge S J, Marshall M S, Bruder J T, Rapp U R, Avruch J. Nature (London) 1993;364:308–313. doi: 10.1038/364308a0. [DOI] [PubMed] [Google Scholar]
- 45.Robinson M J, Cobb M H. Curr Opin Cell Biol. 1997;9:180–186. doi: 10.1016/s0955-0674(97)80061-0. [DOI] [PubMed] [Google Scholar]
- 46.Ichijo H, Nishida E, Irie K, ten Dijke P, Saitoh M, Moriguchi T, Takagi M, Matsumoto K, Miyazono K, Gotoh Y. Science. 1997;275:90–94. doi: 10.1126/science.275.5296.90. [DOI] [PubMed] [Google Scholar]