Abstract
The decay-accelerating factor (DAF) contains four complement control protein repeats (CCPs) with a single N-linked glycan positioned between CCPs 1 and 2. In previous studies we found that the classical pathway regulatory activity of DAF resides in CCPs 2 and 3 while its alternative pathway regulatory activity resides in CCPs 2, 3 and 4. Molecular modelling of the protein predicted that a positively charged surface area on CCPs 2 and 3 (including KKK125–127) and nearby exposed hydrophobic residues (L147F148) on CCP3 may function as ligand-binding sites. To assess the roles of the N-linked glycan and the above two sets of amino acids in the function of DAF, we mutated N61 to Q, KKK125–127 to TTT and L147F148 to SS. Following expression of the mutated cDNAs in Chinese hamster ovary cells, the glycosylphosphatidylinositol (GPI)-anchored mutant proteins were affinity purified and their functions were assessed. In initial assays, the proteins were incorporated into sheep and rabbit erythrocytes and the effects of the mutations on regulation of classical and alternative C3 convertase activity were quantified by measuring C3b deposition. Since DAF also functions on C5 convertases, comparative haemolytic assays of cells bearing each mutant protein were performed. Finally, to establish if spatial orientation between DAF and the convertases on the cell surface played any role in the observed effects, fluid-phase C3a generation assays were performed. All three assays gave equivalent results and showed that the N-linked glycan of DAF is not involved in its regulatory function; that L147F148 in a hydrophobic area of CCP3 is essential in both classical and alternative pathway C3 convertase regulation; and that KKK125–127 in the positively charged pocket between CCPs 2 and 3 is necessary for the regulatory activity of DAF on the alternative pathway C3 convertase but plays a lesser role in its activity on the classical pathway enzyme.
Introduction
The decay-accelerating factor (DAF or CD55) is a 70 000 MW membrane protein that protects cells from activation of autologous complement on their surfaces. It acts to accelerate the decay of the classical and alternative C3 and C5 convertases, the central amplification enzymes of the cascade.1 The functional sites of DAF are contained within four ∼61 amino acid long repeating units initially termed short consensus repeats (SCRs) and more recently designated complement control protein repeats (CCPs).2,3 The four repeats are suspended above the surface membrane on an S/T-rich cushion that is in turn linked to the plasma membrane by a glycosylphosphatidylinositol (GPI) anchor.4 In previous studies,5 we mapped classical pathway C3 convertase regulatory activity of DAF to CCPs 2 and 3. This contrasted with its alternative pathway C3 convertase regulatory activity, which we mapped to CCPs 2, 3 and 4. Subsequently,6 we built a molecular model of the protein which predicted possible contact sites through which DAF interacts with components of the C3 convertases.
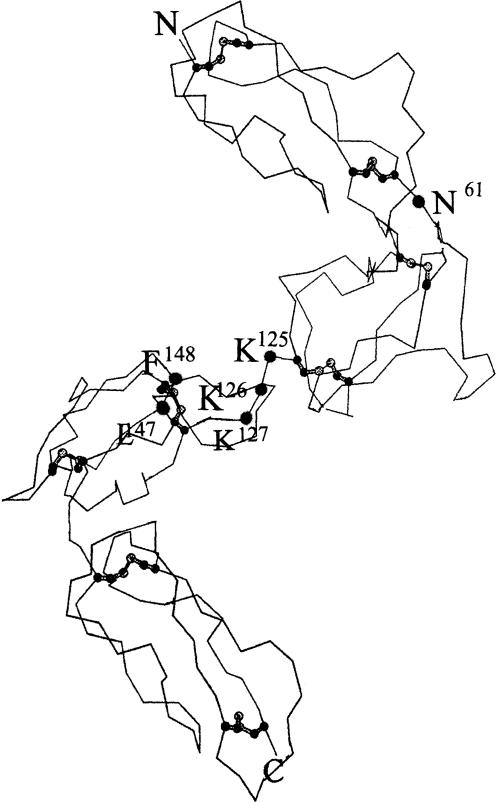
According to the model, the four CCPs of DAF are arranged in a helical fashion.6 Among the regions predicted to participate in contact with the convertases is a positively charged surface area extending from CCP2 into a groove between CCPs 2 and 3 that contains three sequential K residues at positions 125–127 (see Fig. 1). A second region predicted to be involved is a nearby hydrophobic pocket on CCP3 containing two exposed hydrophobic residues (L147F148). A further site that potentially could be relevant to contact between DAF and the C3 convertases based on conformation and/or rotational freedom of the molecule is the N-linked glycan of DAF, located at residue N61.
Figure 1.
Predicted sites of the mutations made in DAF protein. Molecular model of the DAF CCP units as described.6The putative positions of the mutated amino acids are indicated.
In the present study we assayed the effects of changing amino acids at the above positions within the DAF molecule. We mutated amino acids KKK125–127 to TTT to eliminate the positive charge, L147F148 to SS to eliminate the hydrophobicity, and N61 to Q to abolish the glycosylation signal. We performed three types of analyses, two to assess the effects of the changes on the regulation by DAF of the convertases on the same cell membrane, conditions under which DAF functions physiologically, and a third to ascertain if spatial orientation on the cell surface was involved in the measured effects.
Materials and methods
Preparation of cDNAs encoding DAF mutant proteins
Human DAF cDNA mutants were made using the Transformer Site-Directed Mutagenesis Kit (Clontech, Palo Alto, CA). Briefly, mutagenic and selection primers were annealed to pBTKSII containing DAF 13 : 2 cDNA,7 and second-strand DNA was synthesized. DAF 13 : 2 in pBTKSII was placed in reverse orientation relative to the positive strand. The mutagenic primers used were:
N61 → Q: 5′ CCTCGCAGCTACGCTGGCAGAACTC-TTC 3′
KKK125−127 → TTT: 5′ GGGCATGATGTCGTTGTAC-AAAATTCGACTGC 3′
L147F148 → SS: 5′ GGTTGCACCAGATGATATGCCACCTGG-3′.
Site-directed mutagenesis reaction 1 employed the phosphorylated selection primer ScaI/StuI, 5′ P-GTGACTGGTGAGGCCTCAACCAAGTC 3′ (Clontech) while reactions 2 and 3 employed the phosphorylated selection primer AlwNI/SpeI, 5′ P-GCAGCCACTAGTAACAGGATT 3′ (Clontech).
The plasmid was transformed into mismatch repair-deficient Escherichia coli strain BMH 71–18 mut S and amplification of the mutated plasmid was achieved by digestion with the selection enzyme followed by a second transformation into non-mutant E. coli strain DH5a.
All substitutions were confirmed by sequencing. For stable transfection into Chinese hamster ovary (CHO)-K1 cells, each DAF mutant cDNA was excised with EcoRI and cartridged into the EcoRI site of expression plasmid pEE14.5
Transfections and purification of recombinant DAF (rDAF) proteins
For the preparation of transfectants, 10 μg of DNA, preincubated with 30 μl of lipofectin (Life Technologies, Gaithersberg, MD) in 1 ml of serum-free Dulbecco's minimal essential medium (DMEM) (Life Technologies) for 1 hr at 25°, was added to CHO-K1 cells grown to 60% confluency in DMEM/10% fetal bovine serum (FBS) in 100-mm2 plates.5 Following incubation at 37° for 4 hr, serum-free DMEM was replaced with complete medium. After 24 hr, glutamine-free DMEM/10% FBS containing 25 μm methionine sulphoximine (MSX) was added, and resistant colonies were selected over the next 4 weeks.
Following isolation with cloning cylinders, individual colonies were expanded in DMEM/10% FBS containing 25 μm MSX, the cells were stained with anti-DAF monoclonal antibodies (mAbs) IA10 or IIH6 (5 μg/ml) followed by fluorescein isothiocyanate (FITC)-labelled goat anti-murine F(ab′)2, and the fluorescence was quantified by flow cytometry. Cells exhibiting the highest surface levels of each recombinant DAF (rDAF) protein were further expanded in DMEM/10% FBS containing 25–100 μm MSX.
For isolation of each rDAF protein, transfectants were grown to confluency in 100-mm plates. In a typical isolation, 10–15 plates were prepared. Growth medium was removed from each plate, 1 ml phosphate-buffered saline (PBS) containing 30 μl of protease inhibitor mix8 and 60 mmN-octyl-B-d-glucopyranoside (Sigma, St Louis, MO) were added, and the plates were rocked for 15 min at 4°. Lysates were then removed from the plates and centrifuged for 5 min at 1000 g, the initial supernatant was collected and centrifuged several times, and the final supernatant was aliquoted and stored at −70°.
Cell extracts containing rDAF proteins were added to cyanogen bromide–Sepharose beads coupled to anti-DAF mAb IA10 (50 μl beads per 1 μg protein4). The mixtures were rotated overnight at 4°, and the beads were washed twice with 1 ml of 0·1% 3-[(3-cholamidopropyl)dimethyl-ammonio]-1-propane sulfonate (CHAPS), 0·5 m NaCl, 1 m Tris, pH 7·4. The beads were then eluted five times (for 3 min each) with an equal volume of 0·1% CHAPS, 0·05 m triethylamine, 50 mm Tris, pH 11·2 and the eluates were immediately neutralized with a 1 : 20 volume of glycine-saturated 0·5 m Tris, pH 6·0 containing 0·1% CHAPS. Affinity-purified proteins were dialysed against PBS with six buffer changes, aliquoted and stored at −70°.
Quantification of rDAF concentrations
DAF protein concentrations were quantified by two-site immunoradiometric assay (IRMA).9 For this assay, the wells of 96-well plates were coated at 4° for 18 hr with anti-DAF mAB IA10 (which binds to CCP1) in PBS containing 0·02% azide. Following blocking with PBS containing 1% bovine serum albumin (BSA) and 0·02% azide for 1 hr at 25° and washing three times with the same buffer, 25-μl aliquots of samples or serial dilutions of the purified human erythrocyte (Ehu) DAF standard were added in triplicate wells. Following incubation for 18 hr at 4° and a further three washes with the same buffer containing 0·05% Tween-20, 25 μl [1 × 105−2 × 105 counts per minute (c.p.m.)] of 125I-labelled anti-DAF mAb IIH6 (which binds to CCP4) was added to each well.10 After 2 hr incubation at 25°, the wells were washed four times with the same buffer, cut out, and counted. The concentrations of DAF in the samples were calculated by comparison of counts in the sample wells to counts in the wells that received DAF standards.
Western blot analyses
Proteins were electrophoresed on 7·5% sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS–PAGE) gels under non-reducing conditions. Separated proteins were transferred to Immobilon polyvinylidene difluoride 0·45-μm membranes (Millipore, Inc., Bedford, MA) for 90 min at 150 V using a Transblot apparatus (Bio-Rad, Inc., Hercules, CA) and blocked for 18 hr at 4° with 5% BSA. Blocked filters were incubated with 5 μg/ml IA10 mAb. After washing three times with 0·1 m Tris, pH 7·6 buffered saline containing 0·1% Tween-20, filters were further incubated with horseradish peroxidase-conjugated sheep anti-mouse immunoglobulin G (IgG), and the protein bands were developed using an enhanced chemiluminescence kit (Amersham, Inc., Chicago, IL).
Complement buffers, complement components and labelling
Isotonic veronal-buffered saline (DGVB2+), used in most studies, consisted of 2·5 mm veronal, 72·7 mm NaCl pH 7·5, 0·1% gelatin, 2·5% dextrose and 1 mm MgCl2/0·15 mm CaCl2. Isoionic veronal buffer (GVB2+) contained 145 mm NaCl and lacked dextrose. Metal-chelating veronal buffer (GVB-E) lacked dextrose and divalent cations but contained 10 mm ethylenediaminetetraacetic acid (EDTA). GVB/Mg2+/ethylenebis(oxyethyelenitrilo)tetra-acetic acid (EGTA) buffer contained 1 mm MgCl2 and 1 mm EGTA in place of EDTA.
Rabbit anti-sheep haemolysin was purchased from BioWhittaker (Walkersville, MD). Guinea-pig C1, human C4, and C5-depleted human serum (C5D) were purchased from Quidel (San Diego, CA). Human C2 and C1s1,5 and C3, factor B (B), and factor D (D) 11 were prepared as described. C3(H2O), i.e. C3 hydrolysed at the thioester, was separated from native C3 by fast protein liquid chromatography (FPLC) on mono S.12
C3 was labelled with Iodogen (Pierce Inc., Rockford, IL) as described by the manufacturer.
DAF incorporation studies
For incorporation, varying concentrations of DAF proteins were incubated with 107 cells in 200 μl DGVB2+ for 45 min at 37°. Cells then were washed twice and resuspended in the appropriate assay buffer. To measure the amount of incorporated DAF, following incubation of cells with rDAF protein and washing, replicate samples were resuspended in 1 mlGVB-E, washed twice and resuspended in 100 μl GVB-E. A 100-μl volume of GVB-E containing a saturating amount of 125I-labelled IA10 (1 × 105−2 × 105 c.p.m.) was added and the cell suspension was incubated for 15 min on ice. Cells were then washed twice, transferred to new tubes, washed again and the pellet was counted. The number of molecules incorporated per cell was calculated by assuming a 1 : 1 ratio of binding to cell-associated DAF of 125I-labelled IA10 mAb (which has high affinity for a single epitope on CCP1). Using the specific activity (c.p.m./molecule) of the labelled mAb, the number of molecules of cell-associated DAF was calculated and divided by the cell number (107) to yield the number of DAF molecules per cell.
C3b deposition assays
For studies of classical pathway activation, 107 sheep erythrocytes (Esh) sensitized with rabbit haemolysin (EshA) were preincubated with each rDAF protein or with buffer containing the same detergent concentration. Following washing, the alternatively treated EshA were resuspended in 100 μl of 1 : 200 C5D in GVB2+ containing 125I-labelled C3 (50 ng/ml). After 10 min incubation at 37°, the reaction was stopped by the addition of 1 ml of ice-cold GVB-E and the cells were washed. The number of C3b molecules deposited was calculated by measuring the radioactivity associated with the cells and dividing by the specific activity.
Alternative pathway-mediated C3b deposition was measured in a similar fashion using rabbit erythrocytes (Erab). Following incorporation of rDAF proteins as above, washed cells were incubated for 10 min at 30° in GVB/Mg2+-EGTA containing 1 : 16 C5D and 50 ng/ml 125I-labelled C3. Cells were then washed with GVB-E, transferred to new tubes and, after counting, C3b deposition was calculated as above.
Haemolytic assays
For studies of classical pathway activation, rDAF proteins were incorporated into 107 EshA as described above. Following washes and centrifugation, cells were resuspended in 200 μl GVB2+ containing 1 : 100 diluted normal human serum and the mixtures were incubated for 30 min at 37°. Ice-cold GVB-E (1·3 ml) was added and the remaining cells were pelleted. Lysis was determined by measuring the optical density at 412 nm (OD412) of the supernatant. Inhibition of haemolysis was calculated by comparing the lysis of cells containing incorporated rDAF to that of cells incubated with detergent-containing buffer alone.
For studies of alternative pathway activation, DAF proteins were incorporated into Erab. Following washing, cells were resuspended in 200 μl of a 1 : 16 dilution of normal human serum in GVB/Mg2+-EGTA and incubated for 30 min at 37°. Reactions were stopped by the addition of 1·3 ml of ice-cold GVB-E, the OD412 was measured, and inhibition of haemolysis was calculated as described for the classical pathway.
Fluid-phase C3a assays
C3a generation was quantified using a C3a immunoassay kit (Quidel, San Diego, CA). For studies of classical pathway-mediated C3a generation, 20-μl volumes of C4 (0·16 ng/ml), C2 (1·4 ng/ml) and C3 (1·4 ng/ml) were mixed.13 C3 activation was initiated by adding 100 μl of C1s (30 ng/ml) and placing the mixture at 37°. For studies of alternative pathway-mediated C3a generation, 100 μl volumes of C3 (H2O) (0·9 ng/ml), B (1 ng/ml) and C3 (1·4 ng/ml) were mixed.11 C3 generation was initiated by adding 100 μl of D (20 ng/ml) and incubating as above. The rDAF proteins were added to the classical and alternative pathway mixtures prior to the addition of C1s and D, respectively. Inhibition of C3a generation was calculated by comparing the concentration of C3a generated in samples in which DAF proteins were added to that of the sample containing detergent buffer alone.
For all functional studies, fold differences were determined by utilizing 50% of the maximum inhibition observed in each assay and comparing the concentration of the mutant protein to the concentration of native protein that gave the same level of inhibition. For proteins that did not reach 50% inhibition, the effective concentration was extrapolated. The differences are expressed as x-fold decreases of inhibition and represent the average of three experiments with that mutant for the given assay.
Results
Properties of the recombinant DAF proteins
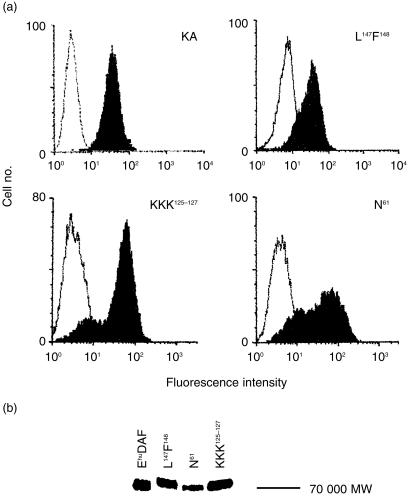
CHO cell transfectants expressing each mutant DAF protein were selected by flow cytometry following staining with anti-DAF mAb IA10, a mAb specific for CCP1 that was not mutagenized. As shown in Fig. 2a, transfectants expressing the three mutated DAF proteins exhibited DAF expression levels approximating that of endogenous DAF on K562 cells. Staining of each transfectant with mAb IIH6 directed against CCP4 gave equivalent results and the ratio of fluorescence intensity between that observed with IIH6 and IA10 was identical to that measured with K562 cells (data not shown).
Figure 2.
Properties of the DAF mutant proteins. (a) Transfected CHO-K1 cells were stained with IA10 mAb (5 μg/ml) followed by goat–anti-mouse IgG FITC conjugate (5 μg/ml) and analysed in a FACscan flow cytometer (Becton Dickinson). Positive transfectants are shown for each of the transfectants along with K562 (KA) cells used as a positive stain control. The open histograms are isotype controls. (b) Detergent extracts of each transfectant and purified Ehu DAF were loaded onto SDS–PAGE gels and transferred to Immobilon. Proteins were detected with IA10 mAb. All proteins migrated at an apparent MW of ∼70 000.
Figure 2(b) shows Western blot analyses of the DAF proteins isolated from the transfectants using mAb IA10 as a probe. Similar to endogenous Ehu DAF, each of the rDAF proteins migrated with the predicted apparent MW of ∼70 000 with the N61 mutant slightly smaller in size.
Effects of the mutations on cell-associated regulatory function of DAF
For testing how each mutation affected the function of DAF on the cell surface, two types of assays were performed: the C3b deposition assays in C5D, and the lytic assays in whole serum.
In the first set of studies, native rDAF and each mutant rDAF protein were incorporated into EshA or Erab and the ability of each to inhibit C3b deposition was compared. In several of the initial experiments, following incorporation and washing of cells incubated with each rDAF protein, duplicate sets of tubes were incubated with 125I-labelled mAb IA10 to determine the number of DAF molecules incorporated. These measurements revealed that at 100 ng/ml of DAF added, 240 molecules were incorporated per cell and that each rDAF protein incorporated into EshA and Erab with equivalent efficiency (Table 1).
Table 1.
Number of DAF molecules incorporated per cell
| No. of molecules incorporated | ||||
|---|---|---|---|---|
| Protein concentration added (ng/ml) | DAF 1–4 | N | LF | KKK |
| 11 | 82 | 66 | 60 | 53 |
| 33 | 139 | 139 | 147 | 94 |
| 100 | 270 | 236 | 237 | 218 |
| 300 | 375 | 432 | 480 | 556 |
The number of DAF molecules incorporated per cell was calculated as described in the Materials and Methods. The mean values taken from three separate experiments in which each DAF uptake was measured in triplicate are shown. The numbers given are for protein incorporated into the membranes of EshA. For each concentration of protein added, no significant differences among the proteins were seen (P > 0·05). Similar numbers were obtained for incorporation into Erab (data not shown).
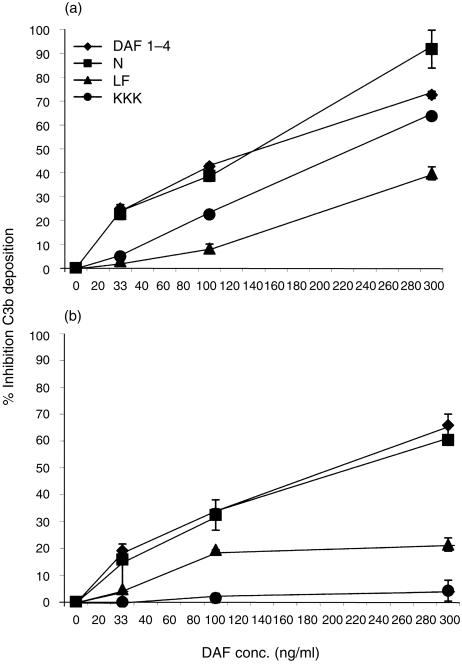
The results of the C3b deposition studies are shown in Fig. 3. In the case of the classical pathway (Fig. 3a), removal of the N-linked glycan of DAF (located between CCPs 1 and 2) had no significant effect on the protein's ability to inhibit C3b deposition. In contrast, altering KKK125–127 in the charged region between CCPs 2 and 3 reduced the protein's inhibitory efficiency approximately three-fold. More markedly, mutation of the hydrophobic L147F148 residues, located on CCP3, reduced the protein's activity approximately five-fold.
Figure 3.
Effects of mutations on the ability to inhibit C3b deposition. (a) Classical pathway: rDAF proteins were incorporated into EshA as described in the Materials and Methods. The per cent inhibition was calculated by comparing the number of C3b molecules deposited on cells treated with detergent-containing buffer alone to cells containing the incorporated DAF proteins. (b) Alternative pathway: DAF proteins were incorporated into Erab as above. Percentage inhibition of C3b deposition was calculated as described for (a). Results in both panels are expressed as the means of three replicate experiments ± the standard deviations.
The four rDAF proteins next were incorporated into Erab and their inhibitory activities on alternative pathway-mediated C3b deposition were compared (Fig. 3b). Again, mutant N61 functioned with an efficiency equivalent to that of native DAF. As also seen with the classical pathway, the L147F148 mutant displayed a substantial decrease of inhibitory activity. In contrast to findings with the classical pathway, however, mutation of the KKK125–127 sequence making up part of the CCP2–3 charged region completely abolished the activity of DAF.
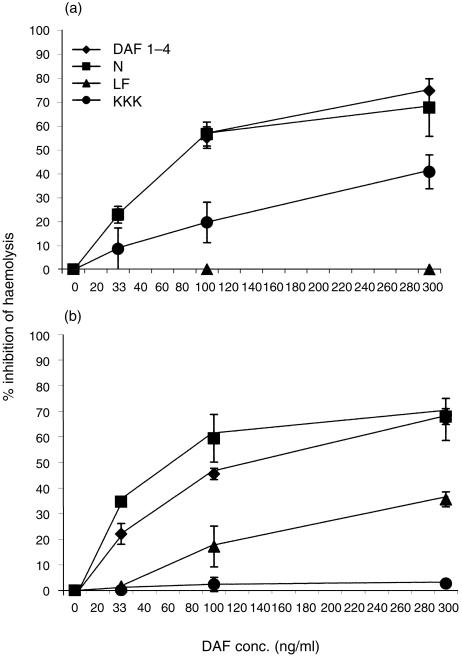
To ascertain if the combined effects on C5 and C3 convertases paralleled those on the C3 convertases alone, the rDAF proteins were incorporated into EshA or Erab and their abilities to inhibit whole serum-mediated haemolysis were quantified. The results of these assays are shown in Fig. 4.
Figure 4.
Effects of the mutations on the ability of DAF to inhibit haemolysis. (a) Classical pathway: increasing concentrations of native or mutant rDAF proteins were incorporated into sensitized Esh. Percentage inhibition was calculated by comparison to cells incubated with detergent-containing buffer. (b) Alternative pathway: increasing concentrations of native or mutant rDAF proteins were incorporated into Erab. Lysis was developed and percentage inhibition calculated by comparison to cells incubated with the same buffer alone. In both panels the data are the means of three replicate experiments ± the standard deviations.
As seen in Fig. 4(a), in accordance with the C3b deposition assays, removal of the N-linked glycan of DAF had no negative effect on the protein's ability to inhibit classical pathway-mediated haemolysis. Also as observed in the C3b deposition assays, altering KKK125–127 in the CCP2–3 charged region had a partial (four-fold) effect in the protein's regulatory efficiency. Likewise, as in the previous assay, mutation of the hydrophobic L147F148 residues reduced the protein's inhibitory effect on haemolysis more profoundly (> nine-fold).
When the proteins were incorporated into Erab and their effects on alternative pathway-mediated haemolysis were studied (Fig. 4b), mutant N61 once again functioned with an efficiency equivalent to that of native DAF. Similarly, the L147F148 mutant displayed a significant decrease (approximately four-fold) in activity. Likewise, as previously seen, the KKK125–127 mutant exhibited a complete loss of function.
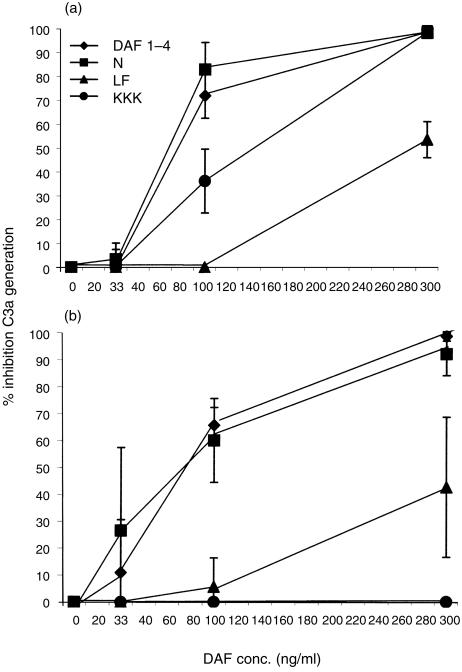
Ability of the mutant DAF proteins to regulate fluid-phase C3 convertase complexes
To determine whether the effects of the mutations on the function of DAF depended on its spatial relationship to the convertases on the membrane, the isolated DAF proteins were assayed for their abilities to inhibit classical and alternative pathway-mediated cleavage of C3 (C3a generation) in the fluid phase.
As shown in Fig. 5(a), C3a generated by the classical pathway C3 convertase was inhibited by native DAF protein and by the N61 mutant with equivalent efficiency. Similarly, as observed for classical pathway-mediated C3b deposition and haemolysis of EshA, mutation of KKK125–127 reduced the function of DAF partially (∼2·5-fold) while mutation of L147F148 reduced the activity of DAF more profoundly (∼4·5-fold).
Figure 5.
Effects of the mutations on the ability of DAF to regulate fluid-phase C3 convertases. (a) Classical pathway: increasing concentrations of rDAF proteins were added to mixtures of purified C4, C2 and C3. Following activation by addition of C1s, reactions were stopped by the addition of EDTA and the amount of C3a generated was measured by ELISA. (b) Alternative pathway: increasing concentrations of rDAF proteins were added to mixtures of C3(H2O), factor B and C3. The mixtures then were activated with D, after which the reaction was stopped by the addition of EDTA. As in Figs 3 and 4, the results in both panels are the means of three replicate experiments ± the standard deviations.
In assays measuring C3a generation by the fluid-phase alternative pathway C3 convertase (Fig. 5b), again native DAF and mutant N61 exhibited equivalent inhibitory activities. Likewise, as observed for C3b deposition onto and haemolysis of Erab, the L147F148 mutant exhibited a four-fold reduction of inhibitory function. Again, in marked contrast to classical pathway C3a generation, the KKK125–127 mutant completely abrogated the inhibitory activity of DAF.
Discussion
These studies constitute a first attempt to identify motifs that are functionally important within the DAF molecule. We examined three sites based on our molecular model of the protein.6 Another criterion was that these three sites are conserved or are conservative replacements in guinea-pig14 and rat15 DAF. We found that the absence of the N-linked glycan of DAF at position N61 does not affect its regulatory activity in either the classical or alternative pathways. In contrast, disruption of KKK125–127 in the positively charged region between CCPs 2 and 3 has a moderate effect on the regulation by DAF of the classical pathway convertase while it completely abolishes its alternative pathway regulatory activity. Strikingly, disruption of the hydrophobic L147F148 residues on CCP3 markedly decreases DAF regulatory activity on both the classical and alternative pathway C3 convertases, more so on the former than the latter.
We utilized three different assay systems to directly measure the effects of the mutations on the C3 convertases on the cell surface, to compare the effects on the C3 convertases to those on lysis in whole serum and to determine if orientation between DAF and the convertases imposed on the cell surface plays any role in the observed effects since the interaction of DAF with the convertases is intrinsic in the same membrane. The same patterns of inhibition were seen in the C3b deposition and C3a generation assays. Based on this result, we can conclude that the mutations directly affect the ability of the protein to interact with the classical and alternative C3 convertases independently of membrane constraints. In general, the pattern for the haemolytic assays paralleled those of the other two assays, although the L147F148 mutation had a greater effect on the inhibitory activity of DAF in the classical pathway. Since DAF regulates both C3 and C5 convertases, one possibility is that the additive effects of this mutation, which disrupts the hydrophobic pocket on CCP3, affects the combined regulation by DAF of the classical pathway C3 and C5 convertases to a greater extent than the KKK125–127 mutation.16
In principle, the results of the cell incorporation studies could be influenced by different incorporation efficiencies of the proteins. However, all proteins displayed the same incorporation efficiencies when tested for cell-associated DAF by using radiolabelled mAb IA10 (Table 1). Moreover, when tested in the cell-free fluid-phase assay system, the proteins behaved in a comparable manner to that seen for the incorporated proteins.
We were uncertain as to what effect the removal of the N-linked glycan of DAF would have. On one hand, previous studies,5 using CCP deletion mutants showed that a Δ1 construct composed of CCPs 2, 3 and 4 without the N-linked glycan had similar decay-accelerating function to wild-type DAF. On the other hand, removal of this glycan but not CCP1 might cause CCP1 to move spatially closer to CCP2, impeding CCP2 in its function on the convertases. Our studies indicate that the N-linked glycan-free DAF is fully functional. In other work17 it has been shown that deletion of the N-linked glycan(s) in mouse DAF similarly does not alter its function. This contrasts with findings for other intrinsic cell surface regulators. Studies of human membrane cofactor protein (MCP), which contains three N-linked glycans, have shown that removal of either of the N-linked glycans located on CCPs 2 and 4 reduces the co-factor activity of the protein.18
We show here that disruption of amino acids L147 and F148 in a potential hydrophobic pocket on DAF CCP3 significantly reduces the ability of DAF to accelerate the decay of both the classical and alternative C3 convertases, more so the former than the latter. It is noteworthy that recent studies of CR1 have shown that its decay-accelerating activity is located on CCPs 1–3 of long homologous repeat (LHR) 1 and that mutation of F82 to V in CCP2, a position comparable to F148 in DAF, similarly markedly reduces its activity in both pathways.16
An unexpected finding was that disruption of KKK125–127 comprising part of a positively charged groove located between CCPs 2 and 3 affected the inhibitory function of DAF much more in the alternative pathway than the classical pathway. As mentioned in the Introduction, previous studies have shown that while DAF CCP4 is essential for regulation of the alternative pathway, CCPs 2 and 3 are essential to inhibit both pathways. The finding in the present study that the charged area between CCPs 2 and 3 is more important for inhibiting the alternative pathway C3 convertase than the classical pathway enzyme suggests that different sites on DAF are involved in regulating the two enzymes.
As with most mutational analyses, we cannot exclude the possibility that conformational changes in the rDAF proteins could play a role in some of the observed effects. However, in flow cytometric assays, reactivities equivalent to those with native DAF were observed with IA10 and IIH6 as well as six other mAbs (2D2-1, 2D2-2, 2D2-3, 2D2-5, 2D2-7 and 2D2-8,19 data not shown). IA10 and IIH6 do not react with reduced DAF protein and thus are dependent upon secondary structure arising from intra-CCP folding. Moreover, mutant KKK125–127 retained classical pathway activity while mutant L147F148 retained some alternative pathway activity. In other studies of point mutants as well as three to five amino acid long substitutions in other CCP-containing C3 regulatory proteins it similarly has not been possible to rigorously address the issue of subtle structural changes.16,20,21 Of relevance in our study, the KKK125–127 mutant involving intra-CCP residues could affect the CCP2–3 spatial relationship as well as charge. With respect to this mutant, however, more recent studies (L. Kuttner-Kondo, L. Mitchell, D. Hourcade and M.E. Medof, unpublished results) have shown that point substitutions of K126 and K127 to A similarly inhibit alternative pathway function.
The findings that both the charged region and hydrophobic region are important for the interaction of DAF with the C3 convertases support the hypothesis that they are externally exposed and involved in function. Further studies are in progress towards mapping other regions of DAF of functional significance in C3 convertase interactions. When a crystal or nuclear magnetic resonance (NMR)-based structure of the molecule is available, these data should contribute to an understanding of the precise manner in which DAF interacts with these central amplification enzymes of the classical and alternative pathways.
Acknowledgments
This investigation was supported by National Institutes of Health grant R01 AI23598 and core grant EY11373. The authors thank Sara Cechner for manuscript preparation, Dr M. Shoham for help in preparing Fig. 1, and Dr Y. Fukuoka for critical review.
Glossary
Abbreviations
- C5D
C5-depleted human serum
- CCPs
complement control protein repeats
- DAF
decay-accelerating factor
- DMEM
Dulbecco's minimal essential medium
- Ehu
human erythrocyte
- Erab
rabbit erythrocyte
- Esh
sheep erythrocyte
- FBS
fetal bovine serum
- GPI
glycosylphosphatidylinositol
- IRMA
immunoradiometric assay
- MSX
methionine sulphoximine
- S/T
serine/threonine
REFERENCES
- 1.Nussenzweig V. Inhibition of complement activation on the surface of cells after incorporation of decay-accelerating factor (DAF) into their membranes. J Exp Med. 1984;160:1558–78. doi: 10.1084/jem.160.5.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Medof ME, Lublin DM, Holers VM, Ayers DJ, Getty RR, Leykam JF, Atkinson JP, Tykocinski ML. Cloning and characterization of cDNAs encoding the complete sequence of decay-accelerating factor of human complement. Proc Natl Acad Sci USA. 1987;84:2007–11. doi: 10.1073/pnas.84.7.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caras IW, Davitz MA, Rhee L, Weddell G, Martin Dw., Jr Cloning of decay accelerating factor suggests novel use of splicing to generate two proteins. Nature. 1987;325:545–9. doi: 10.1038/325545a0. [DOI] [PubMed] [Google Scholar]
- 4.Medof ME, Walter EI, Roberts WL, Haas R, Rosenberry TL. Decay-accelerating factor of complement is anchored to cells by a C-terminal glycolipid. Biochemistry. 1986;25:6740–7. doi: 10.1021/bi00370a003. [DOI] [PubMed] [Google Scholar]
- 5.Brodbeck WG, Liu D, Sperry J, Mold C, Medof ME. Localization of classical and alternative pathway regulatory activity within the decay-accelerating factor. J Immunol. 1996;156:2528–33. [PubMed] [Google Scholar]
- 6.Kuttner-Kondo L, Medof ME, Brodbeck W, Shoham M. Molecular modeling and mechanism of action of human decay-accelerating factor. Prot Eng. 1996;9:1143–9. doi: 10.1093/protein/9.12.1143. [DOI] [PubMed] [Google Scholar]
- 7.Stafford HA, Tykocinski ML, Lublin DM, Holers VM, Rosse WF, Atkinson JP, Medof ME. Normal polymorphic variations and transcription of the decay-accelerating factor (DAF) gene in normal and paroxysmal nocturnal hemoglobinuria (PNH) cells. Proc Natl Acad Sci USA. 1988;85:880–4. doi: 10.1073/pnas.85.3.880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ratnoff WD, Knez JJ, Prince GM, Okada H, Lachmann PJ, Medof ME. Structural properties of the glycoplasmanylinositol anchor phospholipid of the complement membrane attack complex inhibitor CD59. Clin Exp Immunol. 1992;87:415–21. doi: 10.1111/j.1365-2249.1992.tb03012.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kinoshita T, Medof ME, Silber R, Nussenszweig V. Distribution of decay-accelerating factor in the peripheral blood of normal individuals and patients with paroxysmal nocturnal hemoglobinuria. J Exp Med. 1985;162:75–92. doi: 10.1084/jem.162.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coyne KE, Hall SE, Thompson ES, et al. Mapping of epitopes, glycosylation sites, and complement regulatory domains in human decay accelerating factor. J Immunol. 1992;149:2906–13. [PubMed] [Google Scholar]
- 11.Wang RH, Phillips G, Jr, Medof ME, Mold C. Activation of the alternative complement pathway by exposure of phosphatidylethanolamine and phosphatidylserine on erythrocytes from sickle cell disease patients. J Clin Invest. 1993;92:1326–35. doi: 10.1172/JCI116706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pangburn MK. A fluorimetric assay for native C3. The hemolytically active form of the third component of human complement. J Immunol Methods. 1987;102:7–14. doi: 10.1016/s0022-1759(87)80003-0. [DOI] [PubMed] [Google Scholar]
- 13.Seya T, Holers VM, Atkinson JP. Purification and functional analysis of the polymorphic variants of the C3b/C4b receptor (CR1) and comparison with C4b-binding protein (C4bp), and decay accelerating factor (DAF) J Immunol. 1985;135:2661–7. [PubMed] [Google Scholar]
- 14.Nonaka M, Miwa T, Okada N, Nonaka M, Okada H. Multiple isoforms of guinea pig decay-accelerating factor (DAF) generated by alternative splicing. J Immunol. 1995;155:3037–48. [PubMed] [Google Scholar]
- 15.Hinchliffe SJ, Spiller OB, Rushmere NK, Morgan BP. Molecular cloning and functional characterization of the rat analogue of human decay-accelerating factor (CD55) J Immunol. 1998;161:5695–703. [PubMed] [Google Scholar]
- 16.Krych-Goldberg M, Hauhart RE, Subramanian VB, Yurcisin BMII, Crimmins DL, Hourcade DE, Atkinson JP. Decay accelerating activity of complement receptor type 1 (CD35) J Biol Chem. 1999;274:31160–8. doi: 10.1074/jbc.274.44.31160. 10.1074/jbc.274.44.31160. [DOI] [PubMed] [Google Scholar]
- 17.Lin F, Fukuoka Y, Medof ME. N-linked glycans in mouse decay accelerating factor are not involved in its regulatory activity in the classical or alternative pathway. Molec Immunol. 1999;36:284. (Abstract) [Google Scholar]
- 18.Liszewski MK, Leung MK, Atkinson JP. Membrane cofactor protein: importance of N- and O-glycosylation for complement regulatory function. J Immunol. 1998;161:3711–8. [PubMed] [Google Scholar]
- 19.Brodbeck WG, Medof ME. Use of recombinant DAF proteins to localize the epitopes recognized by monoclonal anti-CD55. Trans Clin Biol. 1997;4:125–6. doi: 10.1016/s1246-7820(97)80026-2. [DOI] [PubMed] [Google Scholar]
- 20.Krych M, Hourcade D, Atkinson JP. Sites within the complement C3b/C4b receptor important for the specificity of ligand binding. Proc Natl Acad Sci USA. 1991;88:4353–7. doi: 10.1073/pnas.88.10.4353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hourcade DE, Wagner LM, Oglesby TJ. Analysis of the short consensus repeats of human complement factor B by site-directed mutagenesis. J Biol Chem. 1995;270:19716–22. doi: 10.1074/jbc.270.34.19716. 10.1074/jbc.270.34.19716. [DOI] [PubMed] [Google Scholar]