Abstract
The term ‘atopy’ describes the genetically determined tendency to mount immunoglobulin E (IgE) antibody responses against per se harmless antigens (allergens). In this study we investigated the usage of VH families in the formation of IgE antibodies in 10 patients suffering from mucosal and/or skin manifestations of atopy. IgE antibody reactivities to exogenous allergen sources as well as to autoallergens were determined and, by immunoabsorption, it was demonstrated that allergen-specific IgE accounted for most of the total serum IgE levels in these patients. Using primers with specificity for the VH1–6 gene families and a primer specific for the first constant region of human IgE, cDNAs coding for IgE heavy chain fragments were amplified using the reverse transcription–polymerase chain reaction (RT–PCR) from peripheral blood lymphocytes of the 10 atopic individuals. Hybridization of the heavy chain-encoding cDNAs with an IgE-specific internal oligonucleotide probe revealed a broad usage of all VH-gene families in the atopic individuals. The spectrum of VH families used in a given atopic individual was neither associated with the type or severity of clinical symptoms nor with the number of allergens recognized. The fact that allergen-specific IgE antibodies in atopic individuals originate from a broad variety of B cells thus reflects the activation of multiple B-cell clones during allergen sensitization. This finding should be borne in mind if therapeutic strategies for Type I allergy are considered that aim at a clonal elimination of allergen-specific B cells.
Introduction
Almost 20% of the population worldwide suffers from various manifestions of atopy.1 Atopy describes the tendency of a given individual to mount immunoglobulin E (IgE) antibody responses against otherwise harmless antigens (i.e. allergens).2 The central role of allergen-specific IgE antibodies for the pathogenesis of atopy is evident: allergen-induced cross-linking of effector cell (e.g. mast cell, basophil)-bound IgE antibodies leads to the rapid release of preformed mediators (e.g. histamine, leukotrienes) and thus to the immediate symptoms of atopy (e.g. allergic rhinoconjunctivitis, asthma, urticaria, anaphylactic shock).3 IgE-mediated allergen presentation causes enhanced proliferation of allergen-specific T cells as well as release of proinflammatory cytokines, leading to the chronic manifestions of atopy (e.g. atopic dermatitis, late phase of asthma).4 For many forms of atopy (allergic rhinoconjunctivitis, asthma, urticaria, IgE-mediated food allergy) the disease-eliciting allergens have been characterized at the molecular level.5–6 Sequence and structural similarities of many of the environmental allergens analysed so far explain allergen cross-reactivity at the T-cell level as well as at the B-cell level and suggest that certain atopic individuals are sensitized against a limited number of epitopes.7 Recent studies demonstrated that patients suffering from severe and chronic forms of atopy display IgE reactivity also to endogenous proteins (autoallergens).8–10 The strongly elevated levels of total serum IgE in certain patients with severe atopy11 were also attributed to a general dysregulation of IgE synthesis owing to increased interleukin-4 (IL-4) or decreased interferon-γ (IFN-γ) production, perhaps leading to the production of high levels of IgE antibodies without allergen specificity.12
While for certain infectious,13–14 as well as immunological, diseases a bias towards the usage of certain VH-gene families has been reported,15–18 rather little information is available regarding the IgE VH-gene usage in atopy. Analysis of the IgE VH-gene usage in peripheral blood lymphocytes (PBL) from three atopic dermatitis patients19 and from spleen-derived lymphocytes of an asthmatic individual20 indicated a preferential usage of the VH5-gene family. However, when the IgE VH-gene usage of PBL from two atopic dermatitis patients21 and two peanut allergic individuals22 was analysed, it became evident that VH families other than VH5 may significantly contribute to generation of the IgE antibody repertoire in atopic individuals. Neither of the previous studies provided definitive information as to whether the cDNA sequences investigated coded for allergen-specific IgE antibodies or resulted from polyclonal IgE production and whether a certain manifestation of atopy may be associated with a particular VH-gene usage. Here we investigated 10 patients suffering from mucosal and/or skin manifestations of atopy. The presence of IgE antibodies with specificity to environmental allergens and autoallergens was measured in their sera. In order to estimate the contribution of allergen-specific IgE or polyclonal IgE without antigen specificity to total serum IgE levels in these patients, immunoabsorption experiments were performed. The IgE VH-gene repertoire in all 10 atopic individuals was investigated by reverse transcription–polymerase chain reaction (RT–PCR) amplification of the IgE heavy chain-encoding cDNAs from peripheral blood mononuclear cells (PBMC), using oligonucleotide primers with specificity for the VH1–6 family and the first constant domain of IgE followed by hybridization with an internal IgE-specific oligonucleotide probe.
Materials and methods
Characterization of atopic individuals
In this study we investigated 10 unrelated atopic individuals suffering from various clinically well-defined forms of atopy, such as atopic dermatitis (AD) 23 and mucosal forms of atopy (rhinitis, conjunctivitis, asthma) (A–J,Tables 1 and 2). Atopic individuals were characterized by case history, skin-prick testing and determination of total and specific IgE antibodies, as described previously.24 A non-atopic individual was included for control purposes (K,Tables 1 and 2). Demographic, clinical and serological data of all individuals are reported in Tables 1 and 2. The presence of IgE autoantibodies in the sera was determined by IgE immunoblotting.8
Table 1.
Clinical and demographic characterization of atopic patients (A–J) and a non-atopic individuals (K)
| Gender | Age (yr) | IgE to: | Symptoms | Therapy | ||
|---|---|---|---|---|---|---|
| Patient | ||||||
| A | F | 45 | t, g, w, a, m | ad, r, c, a, fa | Corticosteroids | |
| B | F | 60 | t, g, w, a, m, f | ad, r, c, a, fa | Corticosteroids | |
| C | M | 28 | t, g, a, m, f | ad, r, c | Antihistamines | |
| D | M | 44 | t, g, w, a, m, f | ad, r, c, fa | Corticosteroids | |
| E | M | 53 | t, g, w, a, m, f | ad, r, c, fa | Corticosteroids | |
| F | F | 29 | t, g, w, a, m | r, c, a, fa | Antihistamines | |
| G | F | 27 | a, m | r, c, a | Antihistamines | |
| H | M | 36 | t, g, w, m | ad, r, c, a, fa | Antihistamines, immunotherapy | |
| I | M | 28 | t, g, w, m | r, c, fa | – | |
| J | F | 34 | t, g, a | r, c | Antihistamines | |
| Control | ||||||
| K | F | 34 | – | – | – | |
a, animal proteins; f, fungi; fa, food allergy; g, grass pollen; m, mites; t, tree pollen; w, weed pollen.
a, asthma bronchiale; ad, atopic dermatitis; c, conjunctivitis; r, rhinitis.
F, female; M, male.
Table 2.
Serological characterization of atopic persons (A–J) and of a non-atopic individual (K)
| Patients | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| IgE to: | A | B | C | D | E | F | G | H | I | J | K |
| Timothy grass | 2 | 2 | 3 | 5 | 2 | 3 | – | 4 | 5 | 4 | – |
| Birch | 2 | 5 | 3 | 4 | 5 | 4 | – | 4 | 4 | 4 | – |
| Mugwort | 2 | 2 | – | 2 | 2 | 2 | – | 2 | 3 | – | – |
| Cat | 5 | 4 | 3 | 3 | 4 | 2 | 1 | – | – | 2 | – |
| Dog | 6 | 4 | – | 1 | 5 | – | – | – | – | – | – |
| Horse | 3 | 3 | 3 | 3 | 3 | – | – | – | – | – | – |
| Guinea-pig | 2 | 3 | – | – | 3 | – | – | – | – | – | – |
| House dust mite | 3 | 3 | 2 | 5 | 3 | 2 | 4 | 3 | 2 | – | – |
| Cladosporium herbarum | – | 2 | – | 2 | 2 | – | – | – | – | – | – |
| Aspergillus fumigatus | – | 3 | 1 | 2 | 3 | – | – | – | – | – | – |
| Cod fish | 3 | – | – | 2 | – | – | – | – | – | – | – |
| Pork | 3 | 3 | – | 1 | 3 | – | – | – | – | – | – |
| Peanut | 2 | 2 | – | 3 | 2 | 2 | – | 2 | 1 | – | – |
| Wheat flour | 3 | 3 | – | 2 | 3 | – | – | 2 | 1 | – | – |
| Total IgE (kU/l) | 1129 | 1383 | 186 | 2000 | 1288 | 82·5 | 158 | 471 | 506 | 203 | <2·0 |
Values shown for total immunoglobulin E (IgE) levels are given in kU/l.
Immunoglobulin E (IgE) levels to environmental allergen sources are expressed as radioallergosorbent test (RAST) classes: RAST class 0, < 0·35 kUA/l (dash); RAST class 1, 0·36–0·7 kUA/l; RAST class 2, 0·8–3·6 kUA/l; RAST class 3, 3·7–17 kUA/l; RAST class 4, 18–50 kUA/l; RAST class 5, 51–99 kUA/l; and RAST class 6, >100 kUA/l.
IgE immunoblotting
The human epithelial cell line A431, which has been established from an epidermoid vulvar carcinoma, was purchased from the American Type Culture Collection (ATCC; Rockville, MD). The cells were cultured in RPMI-1640 medium supplemented with 2 mm l-glutamine, 50 mmβ-mercaptoethanol (β-MET), 100 U/ml of penicillin, 100 µg/ml of streptomycin (Gibco, Gaithersburg, MD) and 10% fetal calf serum (FCS) (Sera Lab, Crawley Down, Sussex, UK). Outdated platelet concentrates from normal human donors were obtained from the local blood bank. A431 cells as well as platelets were washed twice in phosphate-buffered saline (PBS) and then homogenized in sodium dodecyl sulphate (SDS) lysis buffer.25 Extracts were separated by denaturing polyacrylamide electrophoresis25 and then blotted onto nitrocellulose (Schleicher & Schuell, Dassel, Germany), as described previously.26 Nitrocellulose-blotted proteins were exposed to patients' sera diluted 1 : 10 in buffer A (50 mm sodium phosphate, pH 7·5, 0·5% w/v Tween, 0·5% w/v bovine serum albumin, 0·05% w/v NaN3). Bound human IgE antibodies were detected with 125I-labelled anti-human IgE antibodies (radioallergosorbent test [RAST]; Pharmacia, Uppsala, Sweden) diluted 1 : 10.8
Immunoabsorption
Preparation of allergen extracts
Five grams of raw material (birch pollen, timothy grass pollen, mugwort pollen, wheat flour, peanut, guinea-pig-, dog-, cat-, horse-hair dander, pig, cod fish-muscle, Cladosporium herbarum, Aspergillus fumigatus and house dust mites; Allergon, AB, Välinge, Sweden) were homogenized with an ultraturrax in 50 ml of water containing 5 mm phenylmethylsulphonyl fluoride (PMSF) and further extracted under continuous agitation at 4° for 20 min. Supernatants obtained after centrifugation at 4°, 40 000 g, 30 min were frozen and lyophilized. Protein concentrations were determined in the extracts using the MICRO-BCA protein assay (Pierce, Rockford, IL).
Coupling of allergen extracts to cyanogen bromide (CNBr)-activated sepharose immunoabsorption
Allergen extracts were coupled to CNBr-activated sepharose 4B according to the manufacturer's instructions (Pharmacia). Five millilitres of serum from three atopic persons was incubated with 1 ml of allergen-coupled gel by end-over-end rotation overnight at 4°. Serum was recovered by centrifugation (4°, 5 min, 5000 g) and was then incubated with the next sepharose-coupled allergen extract. Specific and total IgE levels were determined after each depletion step by CAP-RAST measurements (Pharmacia) to monitor the degree and specificity of the IgE depletion.
Analysis of the IgE VH-gene usage
RNA isolation
PBMC (1–5 × 107 cells) from 10 atopic patients and the non-allergic control were isolated by Ficoll–Paque density-gradient centrifugation at the time of serum collection. Total cellular RNA was isolated using the guanidine isothiocyanate method and CsCl gradient centrifugation.27
cDNA synthesis and PCR
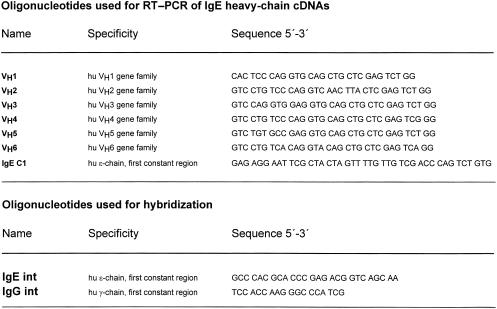
First-strand cDNA was synthesized with an oligonucleotide primer specific for the first exon of the ε constant region, IgE C1 (Fig. 1) using total RNA as template and an RNA PCR kit (Perkin Elmer-Cetus, Norwalk, CT). Equal aliquots of the RT reaction were individually PCR amplified for each VH primer using 25 pm of the VH primers (VH1–VH6, Fig. 1) as described previously.28 The quality of the RNA was checked by performing RT–PCR reactions with primers for human kappa light chains, as described previously.28
Figure 1.
List of oligonucleotides used for reverse transcription–polymerase chain reaction (RT–PCR) amplification of the immunoglobulin E (IgE) heavy chain-encoding cDNAs and for hybridization.
Southern blot hybridization
Equal aliquots (0·1 of the final volume) of each PCR reaction were separated in a 1·5% agarose gel and transferred to nitrocellulose. The identity of the PCR products was confirmed by hybridization with a 32P-labelled oligonucleotide specific for the Cε1 cDNA (IgE int,Fig. 1), as described previously.29 The same membrane was stripped and hybridized with a 32P-labelled oligonucleotide specific for the Cγ1 cDNA (IgG int,Fig. 1), as described previously.29
Results
Characterization of atopic patients
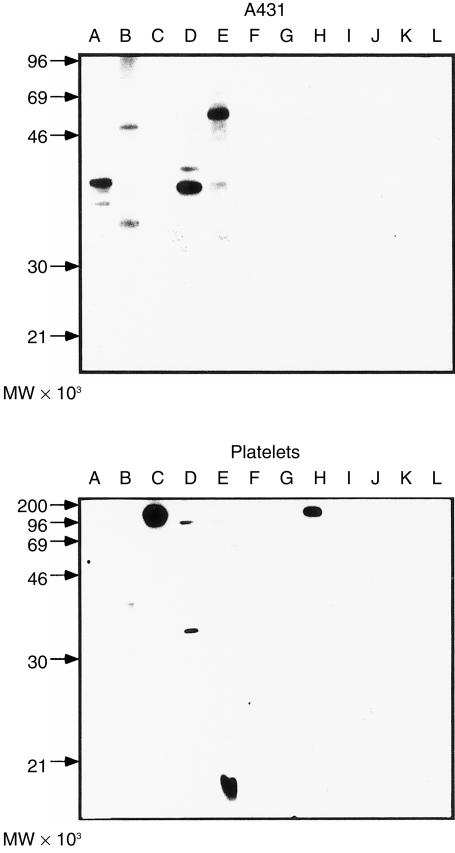
In this study we included 10 atopic individuals (five women, five men; mean age 38·4 years) and one non-atopic female (34 years of age) (Table 1). Patients were selected to cover different forms and severity of atopy. Six of the atopic patients suffered from AD with additional symptoms of mucosal atopy (rhinoconjunctivitis, food allergy, asthma) (A–E, H, Table 1). Four atopic individuals suffered from mucosal forms of atopy (rhinitis, conjunctivitis, allergic asthma, food allergy) but not from AD (F, G, I, J,Table 1). Determination of total serum IgE levels (Table 2) showed that four AD patients (A, B, D, E) had highly elevated total serum IgE levels (average 1450 kU IgE/l), whereas AD patients C and H, as well as the patients with mucosal atopy (F, G, I, J), had lower IgE levels, ranging from 82·5 to 506 kU/l (Table 2). AD patients A, B, D and E, with high levels of total serum IgE, suffered from various forms of mucosal atopy and were sensitized against a broad variety of environmental allergens (plant pollens, animal hair dander, moulds, mites, food) (Table 2). Patients suffering only from mucosal forms of atopy were sensitized against fewer environmental allergens (Table 2). When we analysed the sera of the atopic individuals for IgE antibodies to human proteins (by immunoblotting) we found that AD patients, but not patients with mucosal forms of atopy or the non-atopic individual, reacted with a variety of autoallergens in nitrocellulose-blotted human epithelial extracts as well as platelet extracts (Fig. 2). In AD sera A, B, D and E, IgE autoantibodies were present against 25 000–100 000-molecular weight (MW) A431 proteins (Fig. 2, upper panel), and AD sera B, C, D, E and H displayed IgE autoreactivity against platelet-derived proteins of 15 000–150 000 MW (Fig. 2, lower panel). In summary, we included patients in our study who suffered from skin and several mucosal manifestations of atopy with a broad sensitization to environmental and autoallergens (e.g. patients A, B, C, D, E and H) as well as atopic patients with fewer mucosal manifestations who were sensitized against fewer allergens and lacked IgE autoantibodies (e.g. patients F, G, I and J).
Figure 2.
Sera from severe atopic individuals contain immunoglobulin E (IgE) autoantibodies. Nitrocellulose-blotted proteins from a human epithelial cell line (A431) (a) and from human platelets (b) were exposed to sera from atopic patients suffering from symptoms of atopy (lanes A–J) (see Tables 1 and 2) and, for control purposes, with serum from a non-atopic individual (lane K) or with buffer alone (lane L). Bound IgE was detected with 125I-labelled anti-human IgE antibodies. Molecular weights (× 10−3) are displayed on the left of the figure.
Allergen-specific IgE antibodies account for most of the total serum IgE levels in atopic patients
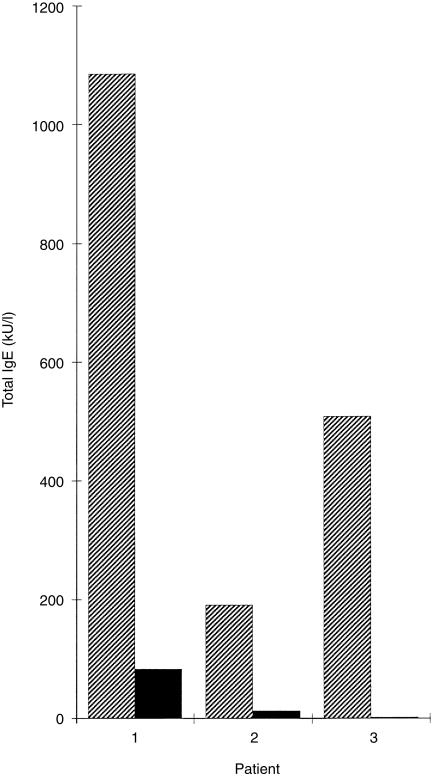
It has been suggested that highly elevated serum IgE levels in atopic individuals may result from a polyclonal IL-4-driven secretion of IgE without defined antigen specificity.12 In order to investigate the contribution of allergen-specific IgE to total serum IgE levels in the atopic individuals investigated in our study, we determined first the presence of allergen-specific IgE antibodies by RAST measurements for each individual. For the RAST measurements we selected representative allergen sources that contained cross-reactive epitopes (e.g. timothy grass pollen contains most of the IgE epitopes present in other grass and corn species).24 Similarly to the principle described by Gleich & Jacob,30 we preabsorbed the sera to sepharose-coupled allergen extracts according to the patient's sensitization pattern, e.g. total serum IgE levels of two AD patients (1 = patient A; 2 = patient C; Fig. 3, Tables 1 and 2), also containing IgE antibodies to autoallergens, were reduced to 8% and 7% after preabsorption with environmental allergens (1 = A: timothy grass pollen, birch pollen, mugwort pollen, cat, dog, horse, guinea-pig, mite, cod fish, pork, peanut, wheat flour; 2 = C: timothy grass pollen, birch pollen, cat, horse, mite, Aspergillus). The remaining low levels of total IgE in the sera of AD patients are probably directed against autoallergens because both preabsorbed sera failed to react with exogenous allergens. Preabsorption of serum from a patient with mucosal atopy lacking IgE autoantibodies (3 = patient I; Fig. 3, Tables 1 and 2) with environmental allergens (timothy grass pollen, birch pollen, mugwort pollen, mite, peanut, wheat flour) removed all IgE from the serum. Serum aliquots taken after adsorption to a given allergen extract contained the same levels as untreated serum of IgE specific for a not yet adsorbed extract, thus confirming the specificity of the immunoadsorption experiments (data not shown). In summary, our results demonstrate that most, if not all, IgE present in the sera of the atopic individuals investigated is directed against specific allergens and does not result from polyclonal production of IgE without antigen specificity.
Figure 3.
Estimation of the percentage of total immunoglobulin E (IgE) directed against environmental allergen sources. Total serum IgE levels (y-axis) were determined in sera from two atopic dermatitis (AD) patients with IgE autoantibodies (patients 1 and 2) and in serum from a rhinoconjunctivitis (RC) patient without IgE autoantibodies (patient 3) before (cross-hatched bars), and after (black bars) absorption with environmental allergens.
IgE antibodies of atopic individuals exhibit a broad usage of VH families
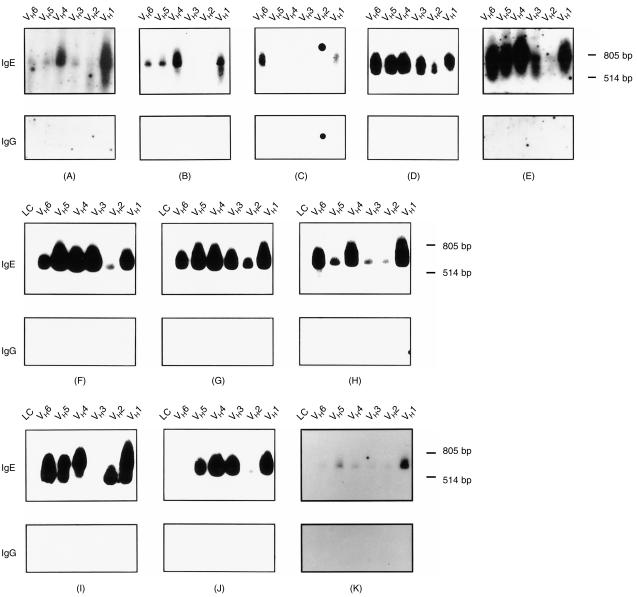
When we hybridized IgE heavy chain-encoding cDNAs (obtained by RT–PCR amplification) from PBMC of the 10 atopic individuals and a non-atopic person with an IgE-specific oligonucleotide probe we found that:
Certain atopic individuals (e.g. A, D, E, F, G, H; Fig. 4) and the non-atopic person used all six VH-family genes to a varying degree.
Certain atopics (e.g. B, I, J; Fig. 4) used all but one or two VH-family genes.
Only one atopic person (C; Fig. 4) exhibited a preferential usage of two VH-families (VH1, VH6).
Figure 4.
Analysis of the immunoglobulin E (IgE) VH-gene usage in patients suffering from various grades of atopy and in a non-atopic individual. cDNAs coding for IgE heavy chain fragments were generated from the peripheral blood mononuclear cells (PBMCs) of atopic persons (A–J) and of a non-allergic individual (K) by reverse transcription–polymerase chain reaction (RT–PCR) using different VH-gene primers (lanes VH1–VH6; Fig. 2). Nitrocellulose-blotted IgE heavy chain-encoding cDNAs and, for control purposes, light chain-encoding cDNAs (F–K: lanes LC) were hybridized with 32P-labelled IgE- or IgG-specific oligonucleotide probes (Fig. 1). The position of the 805-bp and 514-bp molecular size marker bands are indicated on the right of each blot.
The PCR products hybridized with the IgE-specific oligonucleotide probe between the 514- and 804-bp DNA marker bands, which is in agreement with the expected size of the PCR product of ≈ 650 bp. The specificity of the hybridization results is supported by two observations:
The IgE-specific oligonucleotide probe failed to hybridize with light chain-encoding cDNA products present on the same nitrocellulose membranes (Fig. 4: panels F–K, lanes LC).
An immunoglobulin G (IgG)-specific oligonucleotide probe failed to hybridize with the IgE-encoding heavy chain cDNAs (Fig. 4: panels A–K: IgG).
Because of the close relationship of the VH1- and VH7-gene families,31 we have not attempted to amplify VH7-gene products in our experiments. In summary, our results demonstrate that IgE antibodies from atopic individuals exhibit no preferential VH usage.
Neither type and severity of atopic symptoms nor recognition of certain allergens are associated with a particular VH-usage pattern in atopic patients
The comparison of type (i.e. skin or mucosal manifestations) and severity (i.e. number of manifestations) with the usage of certain VH families showed that patients suffering from severe forms of atopy (e.g. B, D) used only certain (B: VH1, VH4–6) or all (D: VH1–6) VH families (Tables 1 and 2; Fig. 4). Thus, type or severity of atopy were not associated with a certain VH-gene usage. Likewise, we observed no association between the number of allergens recognized and the usage of certain VH genes. We found patients who were sensitized against a broad panel of environmental allergens and autoallergens with a broad (A, D, H) or limited (B) VH-gene usage (Fig. 4). Patients who reacted with few environmental allergens exhibited a broad (G) or limited (J) VH usage (Fig. 4). IgE heavy chain cDNAs using all VH-family genes could be PCR amplified, even from the PBMC of a non-atopic individual whose total and allergen-specific IgE levels were below the detection levels.
Discussion
A preferential usage of certain VH genes in the generation of disease-specific/eliciting/protective antibodies has been reported for several immunologically mediated diseases.13–18 Recent evidence for a limited VH usage in atopy came from the observations that ε transcripts obtained from the PBMC of three atopic dermatitis patients19 and spleen-derived lymphocytes of an asthmatic individual20 preferentially used the VH5 family. In addition, molecular and structural analyses of allergens indicates that many atopic individuals are sensitized against a limited panel of cross-reactive allergens and epitopes.5–6 A preferential usage of certain VH genes in the formation of allergen-specific IgE antibodies would thus be not unexpected and perhaps be of relevance as it might allow the development of therapeutic strategies for atopic disorders, which aim at the deletion of certain IgE-producing B cells.
In this study we analysed the VH-gene usage in ε transcripts from PBMC of 10 patients suffering from various degrees of atopy who reacted with a varying spectrum of environmental allergens and autoallergens.8–10 In order to avoid a bias towards a certain VH family caused by varying subcloning efficacies of IgE-encoding PCR products, we chose a hybridization-based screening method. IgE VH1–6 heavy chain cDNAs were independently obtained from PBMC by RT–PCR using primers specific for VH1–6 in conjunction with a Cε1-specific oligonucleotide. RT–PCR experiments were performed using 1–5 × 107 PBMC from each donor. Given the assumption that 20% of the PBMC are B cells and that ≈ 0·79% of those express IgE32 we calculated that the cell numbers used should allow us to obtain from each VH family, by RT–PCR, sufficient and representative IgE-encoding PCR products. The results obtained showed that it was indeed possible to obtain IgE-encoding PCR products with each of the VH-family primers. Using internal IgE-specific, as well as IgG-specific, oligonucleotides for hybridization we were able to confirm that the VH-family specific PCR products obtained coded for IgE. We found that IgE transcripts of atopic individuals exhibited a broad usage of all VH families with no bias to a certain VH family. Neither the type and manifestations of atopy nor the number of allergens/allergen sources recognized were associated with a certain pattern of VH usage. The assumption that the IgE transcripts analysed in our study probably code for allergen-specific IgE antibodies is supported by the following arguments:
Immunoabsorption experiments demonstrated that most, if not all, IgE could be depleted from the sera of atopic individuals by absorption to sepharose-coupled allergens.
It was shown previously that PBMC from allergic patients represent a suitable source for allergen-specific IgE antibodies.29,33
Support for the finding that allergen-specific IgE antibodies use various VH families comes also from the molecular analysis of recombinant human IgE Fabs with specificity for grass pollen allergens. These IgE Fabs were obtained from a combinatorial IgE library that was constructed from PBMC of a grass pollen allergic patient suffering from allergic rhinoconjunctivitis only.28 IgE Fabs with specificity for the major timothy grass pollen allergen, Phl p 5, used the VH3 family, whereas IgE Fabs with specificity for the major timothy grass pollen allergens, Phl p 1 and Phl p 2, utilized the VH3 and VH4 families, respectively.28,34 A similar broad usage of VH families was reported for human IgG antibodies (BAB1–5), which exhibited specificity for the major birch pollen allergen, Bet v 1.35–36 BAB1 and BAB2 used the VH3 family, BAB3 and BAB4 the VH2 family, and BAB5 the VH5 family. The latter data are of particular interest inasmuch as they demonstrate that different VH families can be engaged in the formation of antibodies against a single allergen.
In this study we did not analyse the sequences of the IgE variable regions. However the sequence analysis of IgE antibodies with specificity for the major timothy grass pollen allergens, Phl p 1 and Phl p 2, has revealed that even IgE antibodies with specificity for a single allergen can utilize different complementarity-determining region (CDR) sequences.34 Taken together, it seems that sensitization to allergens activates a great variety of different B-cell clones, which can utilize a broad spectrum of VH-family genes. This fact must be borne in mind if therapeutic concepts for atopic diseases are considered that aim at the deletion of IgE-secreting allergen-specific B-cell clones.
Acknowledgments
This work was supported by grant F0506 of the Austrian Science Fund.
References
- 1.Kay AB, editor. Allergy and Allergic Diseases. Oxford: Blackwell Science; 1997. [Google Scholar]
- 2.Casolaro V, Georas SN, Song Z, Ono SJ. Biology and genetics of atopic disease. Curr Opin Immunol. 1996;9:788. doi: 10.1016/s0952-7915(96)80007-0. [DOI] [PubMed] [Google Scholar]
- 3.Segal DM, Taurog JD, Metzger H. Dimeric immunoglobulin E serves as a unit signal for mast cell degranulation. Proc Natl Acad Sci USA. 1977;41:457. doi: 10.1073/pnas.74.7.2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stingl G, Maurer D. IgE-mediated allergen presentation via Fc epsilon RI on antigen-presenting cells. Int Arch Allergy Immunol. 1997;113:24. doi: 10.1159/000237499. [DOI] [PubMed] [Google Scholar]
- 5.Valenta R, Kraft D. Recombinant allergens for diagnosis and therapy of allergic diseases. Curr Opin Immunol. 1995;7:751. doi: 10.1016/0952-7915(95)80043-3. [DOI] [PubMed] [Google Scholar]
- 6.Valenta R, Vrtala S, Laffer S, Spitzauer S, Kraft D. Recombinant allergens. Allergy. 1998;53:552. doi: 10.1111/j.1398-9995.1998.tb03930.x. [DOI] [PubMed] [Google Scholar]
- 7.Valenta R, Steinberger P, Duchêne M, Kraft D. Immunological similarities among allergens: prerequisite for a specific and component based therapy of allergy. Immunol Cell Biol. 1996;74:187. doi: 10.1038/icb.1996.26. [DOI] [PubMed] [Google Scholar]
- 8.Valenta R, Maurer D, Steiner R, et al. Immunoglobulin E response to human proteins in atopic patients. J Invest Dermatol. 1996;107:203. doi: 10.1111/1523-1747.ep12329617. [DOI] [PubMed] [Google Scholar]
- 9.Natter S, Seiberler S, Hufnagl P, et al. Isolation of cDNA clones coding for IgE autoantigens with serum IgE from atopic dermatitis patients. FASEB J. 1998;12:1559. doi: 10.1096/fasebj.12.14.1559. [DOI] [PubMed] [Google Scholar]
- 10.Valenta R, Natter S, Seiberler S, et al. Molecular characterization of an autoallergen, Hom s 1, identified by serum IgE from atopic dermatitis patients. J Invest Dermatol. 1998;111:1178. doi: 10.1046/j.1523-1747.1998.00413.x. [DOI] [PubMed] [Google Scholar]
- 11.Juhlin L, Johansson SGO, Bennich H, Högman C, Thyresson N. Immunoglobulin E in dermatoses. Levels in atopic dermatitis and urticaria. Arch Dermatol. 1969;100:12. doi: 10.1001/archderm.100.1.12. [DOI] [PubMed] [Google Scholar]
- 12.Poulsen L. Differential IgE regulation in atopic dermatitis and inhalant allergy. ACI International. 1998;10 [Google Scholar]
- 13.Adderson EE, Shackelford PG, Quinn A, Wilson PM, Cunningham MW, Insel RA. Restricted immunoglobulin VH usage and VDJ combinations in the human response to Haemophilus influenzae type b capsular polysaccharide. Nucleotide sequences of monospecific anti-Haemophilus antibodies and polyspecific antibodies cross-reacting with self antigens. J Clin Invest. 1993;91:2734. doi: 10.1172/JCI116514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ikematsu H, Harindranat N, Euki Y, Notkins AL, Casali P. Clonal analysis of a human antibody response. II. Sequences of the VH genes of human IgM, IgG and IgA to rabies virus reveal preferential utilization of VHIII segments and somatic hypermutation. J Immunol. 1993;150:1325. [PMC free article] [PubMed] [Google Scholar]
- 15.Pascual V, Capra JD. VH4–21, a human gene segment overrepresented in the autoimmune repertoire. Arthritis Rheum. 1992;35:11. doi: 10.1002/art.1780350103. [DOI] [PubMed] [Google Scholar]
- 16.Andris JS, Abraham SR, Pascual V, Pistillo MP, Mantero S, Ferrara GB, Capra D. The human antibody repertoire: heavy and light chain variable region gene usage in six alloantibodies specific for human HLA class I and class II alloantigens. Mol Immunol. 1995;32:1105. doi: 10.1016/0161-5890(95)00071-2. 10.1016/0161-5890(95)00071-2. [DOI] [PubMed] [Google Scholar]
- 17.Newkirk MM, Rauch J, Mageed RAK, Jefferis R, Posnett DN, Silverman GJ. Restricted immunoglobulin variable region gene usage by hybridoma rheumatoid factors from patients with systemic lupus erythematosus and rheumatoid arthritis. Mol Immunol. 1993;30:255. doi: 10.1016/0161-5890(93)90054-f. [DOI] [PubMed] [Google Scholar]
- 18.Yang YY, Fischer P, Leu SJC, Olee T, Carson DA, Chen PP. IgG rheumatoid factors isolated by the surface displaying phage library technique. Immunogenetics. 1997;45:301. doi: 10.1007/s002510050209. 10.1007/s002510050209. [DOI] [PubMed] [Google Scholar]
- 19.van Der Stoep N, Van Der Linden J, Logtenberg T. Molecular evolution of the human immunoglobulin E response: high incidence of shared mutations and clonal relatedness among ε VH5 transcripts from three unrelated patients with atopic dermatitis. J Exp Med. 1993;177:99. doi: 10.1084/jem.177.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Snow RE, Chapman CJ, Frew AJ, Holgate ST, Stevenson FK. Analysis of Ig VH region genes encoding IgE antibodies in splenic B lymphocytes of a patient with asthma. J Immunol. 1995;154:5576. [PubMed] [Google Scholar]
- 21.Tilgner J, Golembowsky S, Kersten B, Sterry W, Jahn S. VH genes expressed in peripheral blood IgE-producing B cells from patients with atopic dermatitis. Clin Exp Immunol. 1997;107:528. doi: 10.1046/j.1365-2249.1997.d01-960.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Janezic A, Chapman CJ, Snow RE, Hourihane JO, Warner JO, Stevenson FK. Immunogenetic analysis of the heavy chain variable regions of IgE from patients allergic to peanuts. J Allergy Clin Immunol. 1998;101:391. doi: 10.1016/S0091-6749(98)70253-2. [DOI] [PubMed] [Google Scholar]
- 23.Hanifin JM, Rajka G. Diagnostic features of atopic dermatitis. Acta Dermatol Venereol Suppl. 1980;92:44. [Google Scholar]
- 24.Niederberger V, Laffer S, Fröschl R, Kraft D, Rumpold H, Kapiotis S, Valenta R, Spitzauer S. IgE antibodies to recombinant pollen allergens (Phl p 1, Phl p 2, Phl p 5, and Bet v 2) account for a high percentage of grass pollen-specific IgE. J Allergy Clin Immunol. 1998;101:258. doi: 10.1016/s0091-6749(98)70391-4. [DOI] [PubMed] [Google Scholar]
- 25.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 26.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets. Procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davis LG, Dibner MD, Battey JF. Basic Methods in Molecular Biology. New York: Elsevier; 1986. [Google Scholar]
- 28.Steinberger P, Kraft D, Valenta R. Construction of a combinatorial IgE library from an allergic patient: isolation and characterization of human IgE Fabs with specificity for the major timothy grass pollen allergen, Phl p 5. J Biol Chem. 1996;271:10967. doi: 10.1074/jbc.271.18.10967. 10.1074/jbc.271.18.10967. [DOI] [PubMed] [Google Scholar]
- 29.Steinberger P, Bohle B, Di Padova F, Wrann M, Liehl E, Scheiner O, Kraft D, Valenta R. Allergen-specific IgE production of committed B cells from allergic patients in vitro. J Allergy Clin Immunol. 1995;96:209. doi: 10.1016/s0091-6749(95)70010-2. [DOI] [PubMed] [Google Scholar]
- 30.Gleich GJ, Jacob GL. Immunoglobulin E antibodies to pollen allergens account for high percentage of total immunoglobulin E protein. Science. 1975;190:1106. doi: 10.1126/science.1188389. [DOI] [PubMed] [Google Scholar]
- 31.van Dijk KW, Mortari F, Kirkham PM, Schroeder HW, Milner EC. VH7 gene family consists of a small, polymorphic group of six to eight gene segments dispersed throughout the VH locus. Eur J Immunol. 1993;23:832. doi: 10.1002/eji.1830230410. [DOI] [PubMed] [Google Scholar]
- 32.Kasaian MT, Meyer CH, Nault AK, Bond JF. An increased frequency of IgE-producing B cell precursors contributes to the elevated levels of plasma IgE in atopic subjects. Clin Exp Allergy. 1995;25:749. doi: 10.1111/j.1365-2222.1995.tb00013.x. [DOI] [PubMed] [Google Scholar]
- 33.Dolecek C, Steinberger P, Susani M, Kraft D, Valenta R, Boltz-Nitulescu G. Effects of IL-4 and IL-13 on total and allergen specific IgE production by cultured PBMC from allergic patients determined with recombinant allergens. Clin Exp Allergy. 1995;25:879. doi: 10.1111/j.1365-2222.1995.tb00031.x. [DOI] [PubMed] [Google Scholar]
- 34.Flicker S, Steinberger P, Laffer S, Denepoux S, Lebecque S, Kraft D, Valenta R. Expression in Escherichia coli and purification of antibody fragments (Fabs) with specificity for major pollen allergens. Allergy (abstr) 1997;52:11. [Google Scholar]
- 35.Visco V, Dolecek C, Denépoux S, et al. Human IgG monoclonal antibodies that modulate the binding of specific IgE to birch pollen Bet v 1. J Immunol. 1996;157:956. [PubMed] [Google Scholar]
- 36.Denepoux S, Eibensteiner PB, Steinberger P, et al. Molecular characterization of human IgG monoclonal antibodies specific for the major birch pollen allergen Bet v 1. Anti-allergen IgG can enhance the anaphylactic reaction. FEBS Lett. 2000;465:39. doi: 10.1016/s0014-5793(99)01703-2. 10.1016/s0014-5793(99)01703-2. [DOI] [PubMed] [Google Scholar]