Abstract
Differential expression of ovine CD1 was examined using a combination of reverse transcription–polymerase chain reaction (RT–PCR) with sequence-specific primers and Northern and in situ hybridization techniques. The aim of the study was to establish the patterns of CD1 expression at the molecular level and address questions posed by previous studies in other species regarding expression patterns of CD1. A ‘pan-CD1’ probe based on the exon 4 (alpha 3) region was used in addition to isotype-specific probes for SCD1B (the exon 3 region of clone SCD1B42) and SCD1D (the exon 3 region of clone SCD1D). Widespread expression of CD1 (including thymus, peripheral blood lymphocytes, lung and intestine) was identified using both the exon 4 and SCD1D probe. SCD1B expression was more restricted, being identified in equivalent levels only in the thymus and in scattered populations of dendritic cells. These results highlight the difference in expression patterns between group 1 and group 2 CD1 family members and establish SCD1D as the CD1 family member with the widest pattern of expression, consistent with a differential role for the different CD1 family members.
Introduction
CD1 molecules are a family of cell-surface-associated glycoproteins, now recognized as having a role in antigen presentation, with some similarities to classical major histocompatibility complex (MHC) classes I and II. The CD1 family can be broadly divided into two groups – the CD1b group (group 1) and the CD1d group (group 2). Sequencing of CD1e (only identified in humans and guinea pigs to date) indicates that this is an intermediate group with similarities to both group 1 and group 2.1,2 Rodents only possess members of the CD1d group whereas humans, sheep, cattle and rabbits possess members of both groups. Recent studies in the guinea pig have demonstrated group 1 CD1 and a single CD1e homologue.2 The evolutionary conservation of CD1d antigens by the majority of species is consistent with a key role for these molecules in the immune response (recently reviewed in ref. 3).
Analysis of tissue-specific expression of CD1 molecules in the sheep has previously been carried out using a range of monoclonal antibodies (mAbs).4,5 These antibodies have been clustered, according to their reactivity on bovine tissues, as BoCD1w1, BoCD1w2 and BoCD1w3.6 BoCD1w2 mAbs recognize antigens with a staining pattern consistent with CD1b expression (i.e. cortical thymocytes and dendritic cells). That these mAbs recognize CD1b family antigens was further demonstrated by the amino-terminal sequencing of the antigen recognized by the mAb CC14. This antigen has 100% amino-terminal homology with the predicted sequence of the sheep CD1 gene SCD1B-42.7 The mAbs BoCD1w1 and BoCD1w3 recognize molecules with a wider cellular distribution – in addition to thymocytes and dendritic cells, they recognize antigen on macrophages, B cells and monocytes,5 consistent with a ‘CD1d-like’ pattern of expression.
At the molecular level, several distinct CD1B-like genes (SCD1B42, SCD1B52, SCD1T10 and SCD1A25) and a CD1D-like gene (SCD1D) have been identified in the sheep.7,8 The aim of this study was to establish the patterns of CD1 expression at the molecular level and address questions posed by previous studies in other species regarding expression patterns of the two major CD1 groups. To obtain this data, a combination of Northern hybridization, reverse transcription–polymerase chain reaction (RT–PCR) and in situ hybridization was carried out on a range of tissues.
Materials and methods
Northern hybridization
Total RNA was isolated from various tissues using TRIreagent (Sigma Chemical Co., Poole, Dorset, UK). mRNA was then purified on oligodT cellulose and electrophoresed in 1% agarose gels containing 1 × MOPS (20 mm 3-[N-morpholino] propanesulfonic acid, pH 7), 10 mm EDTA and 2% formaldehyde. The running buffer was 1 × MOPS. mRNA was then transferred to nylon membrane (Hybond-XL; Amersham Pharmacia Biotech, Buckinghamshire, UK). Membranes were prehybridized (for 30 min at 68°) in Ultrahyb (Ambion, Austin, TX) prior to hybridization (for 26 hr at 68°) with 32P-labelled exon 4 probe in fresh hybridization solution. Posthybridization washes comprised 2 × 5-min washes in 2 × SSC (saline-sodium citrate), 0·1% sodium dodecyl sulphate (SDS) at 68° followed by 2 × 15-min washes in 0·1 × SSC, 0·1% SDS at 68°. Washed blots were then exposed to Kodak X-OMAT AR film (Kodak, Hertforshire, UK) at −80°.
RT–PCR
cDNA synthesis was carried out using random hexanucleotide primers, according to standard protocols. PCR was performed with Taq DNA polymerase using the following sense/antisense primer pairs: ATPase: 5′-GCTGACTTGGTCATCTGC-3′, 3′-CAGGTAGGTTTGAGGGGATAC-5′; exon 4/alpha 3: 5′-TGAAGCCTGGCTGTCCAGT-3′, 3′-CCAGTACAGGATGATATCC-5′; SCDID exon 3: 5′-CAGGGCACGTTCAGCGACC-3′, 3′-CATGGAGGAGCCAGTGCACCG-5′; SCD1B52 exon 3: 5′-GTCAACAGCACATGGGCTC-3′, 3′-TGTGGGGTGTCCTTGCCTCTG-5′.
The basic components of each reaction were 50 mm KCl, 10 mm Tris-HCl (pH 8·3), 1·5 mm MgCl2 100 µmol of each dNTP (Pharmacia), DNA template, 25–50 pmoles of each primer and 2 U Taq polymerase. PCR products were separated on 1% w/v agarose gels (Type I: low electroendosmosis EEO; Sigma).
In situ hybridization
Tissue preparation and pretreatment
Tissues were obtained immediately post-mortem from lambs killed at the Royal (Dick) School of Veterinary Studies and fixed in 10% neutral-buffered formalin for a maximum of 48 hr. Tissues were routinely processed, and 4-µm sections were cut onto slides pretreated with Biobond tissue section adhesive (British Biocell International, Cardiff, UK). Slides were dewaxed in xylene, rehydrated through graded ethanol to phosphate-buffered saline (PBS), then treated for 15 min in PBS containing 0·3% Triton-X-100. Sections were permeabilized for 20–40 min at 37° in 0·1 m Tris-EDTA buffer containing proteinase K (10–20 µg/ml). (Proteinase K digestion concentration and time were optimized for each individual tissue type). Following digestion, sections were postfixed at 4° in PBS containing 4% paraformaldehyde and rinsed in PBS. Prehybridization was carried out for 1 hr at 42° in 4 × SSC containing 50% v/v deionized formamide.
Probe generation
Probes were generated from cDNA clones and primer sequences selected to allow generation of ≈ 300-bp sequences. The following probes were generated: a ‘pan’ CD1 probe (exon 4 derived), a SCD1B-specific probe (clone SCD1B42, exon 3/alpha 2 region) and a SCD1D-specific probe (clone SCD1D, exon 3/alpha 2 region). The percentage homology of the SCD1B42 probe to the other SCD1B sequences was as follows: SCD1B52, 92%; SCD1A25, 80%; SCD1T10, 76%; SCD1D, 54%. As such, this probe would be expected to hybridize to all the sheep CD1B-like sequences under the hybridization conditions used, but not to SCD1D. The SCD1D probe, whilst being 100% homologous to SCD1D, was only 50–56% homologous to the SCD1B sequences and would therefore not be expected to hybridize to these sequences. It is possible, however, that hybridization to other, as yet unknown, CD1D-like sequences could occur in a similar manner to that described for the CD1B-like sequences. Amplified cDNA was cloned using the TOPO TA Cloning kit (Invitrogen, Groningen, the Netherlands) to generate sense and antisense riboprobes using T7 and Sp6 RNA polymerase, according to standard protocols. Probes were labelled with digoxigenin by incorporating digoxigenin labelling mix (Boehringer Mannheim, Sussex, UK) into the synthesis reaction. Synthesized RNA probes were pelleted, washed twice in 75% ethanol, resuspended in diethyl pyrocarbonate (DEPC)-treated water and stored at −70°.
Hybridization
Probes were diluted in hybridization buffer (40% deionized formamide, 10% dextran sulphate, 1 × Denhardt's solution, 4 × SSC, 10 mm dithiothreitol, 1 mg/ml of yeast transfer RNA) and hybridization was carried out at 42° for 16 hr.
After hybridization, sections were washed at 37° as follows: 2 × 15-min washes in 2 × SSC; 2 × 15-min washes in 1 × SSC; and 2 × 30-min washes in 0·1 × SSC. Slides were then washed at room temperature in 100 mm Tris-HCl (pH 7·5), 150 mm NaCl and incubated for 30 min at room temperature in 100 mm Tris-HCl (pH 7·5), 150 mm NaCl, 0·1% Triton-X-100, 2% normal sheep serum (Scottish Antibody Production Unit [SAPU], Lanarkshire, UK).
Visualization
Slides were incubated for 2 hr in 100 mm Tris-HCl (pH 7·5) containing 150 mm NaCl, 0·1% Triton-X-100, 2% normal sheep serum and 25 U/ml antidigoxigenin alkaline phosphatase (Boehringer Mannheim), washed for 20 min in 100 mm Tris-HCl (pH 7·5), 150 mm NaCl and for 10 min in 100 mm Tris-HCl (pH 9·5), 100 mm NaCl, 50 mm MgCl2. Slides were then developed for 2–6 hr in nitro blue tetrazolium/5-Bromo-4-chloro-3-indolyl phosphate (NBT/BCIP) solution in 100 mm Tris-HCl (pH 9·5), 100 mm NaCl and 50 mm MgCl2 containing 5 mm levamisole. The reaction was stopped by incubation in 10 mm Tris-HCl (pH 7·5) containing 1 mm EDTA. Sections were lightly counterstained in 0·1% fast green FCF for 1–2 min then washed in tap water for 10 min and mounted using aqueous mounting medium (Immu-mount; Shandon, Pittsburgh, PA).
Results
Northern hybridization
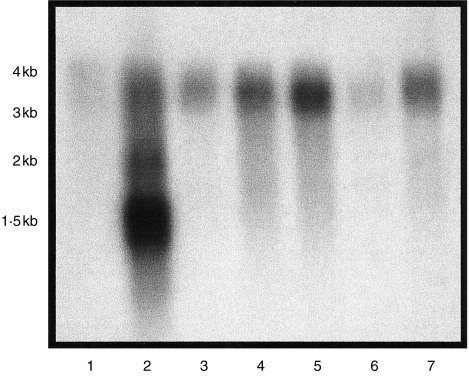
Northern hybridization using an exon 4/alpha 3 probe identified abundant and multiple RNA transcripts in the thymus RNA population (Fig. 1). A single 3·5-kb transcript was present in all other populations examined.
Figure 1.
Northern blot of mRNA probed with conserved exon 4 sequence. Lane 1, lymphocyte; lane 2, thymus; lane 3, small intestine; lane 4, spleen; lane 5, lung; lane 6, liver; lane 7, peripheral lymph node.
RT–PCR
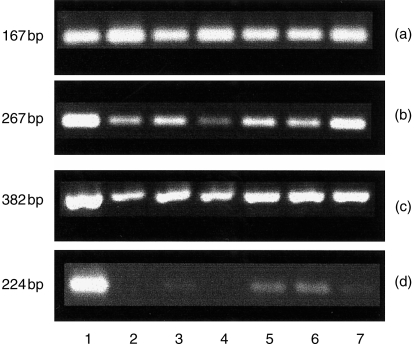
RT–PCR was used to further examine the differential expression of ovine CD1 genes (Fig. 2). Consistent with the results of Northern hybridization, probes specific for the exon 4 region showed expression in all sites examined (thymus, small intestine, lymphocytes, liver, lymph node, lung and spleen) (Fig. 2b). Similar widespread expression was evident using primers specific for SCD1D (Fig. 2c). In contrast, SCD1B42-specific primers showed a more restricted expression pattern, being detectable by this means in thymus, lymph node and (strong expression) and spleen and lymphocyte populations (weak expression) (Fig. 2d).
Figure 2.
Reverse transcription–polymerase chain reaction (RT–PCR) analysis of: lane 1, thymus; lane 2, small intestine; lane 3, lymphocyte; lane 4, liver; lane 5, peripheral lymph node; lane 6, lung; lane 7, spleen. (a) ATPase; (b) exon 4; (c) SCD1D; (d) SCD1B42.
The specificity of these PCR reactions had previously been confirmed by Southern hybridization and probing of products with specific internal oligonucleotides (data not shown).
In situ hybridization
A range of tissues were examined by in situ hybridization, including thymus, lymph node, spleen, intestine (including ileal Peyer's patch), spinal cord and lung.
In view of strong expression of CD1 by cortical thymocytes, this tissue was used as a positive control for all probes in the experiment. Table 1 summarizes the results of in situ hybridization (using the exon 4, SCD1D and SCD1B42 probes) in the tissues listed above.
Table 1.
Summary of in situ hybridization data
| Tissue | Exon 4, SCD1D | SCD1B42 |
|---|---|---|
| Thymus | Cortex | Cortex |
| Lymph node | Follicles, paracortical DCs | Paracortical DCs |
| Spleen | WP lymphocytes, RP macrophages | DCs |
| Lung | BALT lymphocytes | – |
| Intestine | Macrophages, Peyer's patches | – |
| CNS | Glial cells, neurons | – |
BALT, bronchial/bronchiolar-associated lymphoid tissue; CNS, central nervous system; DCs, dendritic cells; RP, red pulp; WP, white pulp.
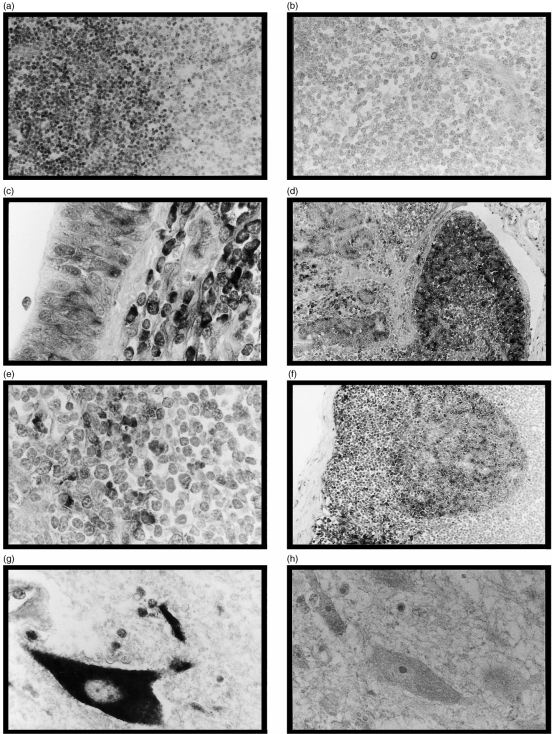
All antisense probes gave similar results when hybridized to thymus sections. Strong staining of cortical thymocytes was identified with scattered, more intensely stained cells in the outer cortex close to the capsule (Fig. 3a). Minimal staining of medullary thymocytes was evident with scattered dendritic cells staining positively within the medullary compartment. In all other areas, the pattern of staining with the exon 4 and SCD1D probes was identical whereas SCD1B42 showed a more restricted distribution.
Figure 3.
In situ hybridization of various tissues with digoxigenin-labelled riboprobes developed using an alkaline phosphatase system. The counterstain used was 0·1% fast green FCF. (a) Thymus; exon 4 antisense; magnification × 250. (b) Thymus; exon 4 sense; magnification × 250. (c) Lung; SCD1D antisense; magnification × 400. (d) Distal ileum; SCD1D antisense; magnification × 100. (e) Lymph node; SCD1B42 antisense; magnification × 400. (f) Lymph node; SCD1D antisense; magnification × 100. (g) Central nervous system (CNS) (spinal cord); SCD1D antisense; magnification × 400. (h) CNS (spinal cord); SCD1D sense; magnification × 400.
Within the lung, the exon 4 and SCD1D probes showed strong staining of cells in the peribronchiolar/peribronchial regions (lymphocytes) (Fig. 3c). No significant staining was identified with the SCD1B42 probe.
Within the intestine, lamina propria macrophages, Peyer's patch lymphocytes, and crypt and villous epithelial cells showed positive reactivity with exon 4 and SCD1D probes (Fig. 3d). No staining was observed with the SCD1B42 probe.
Within lymphoid tissue, dendritic cells of splenic red pulp and paracortical lymph node dendritic cells stained with all three probes (Fig. 3e). In addition, the exon 4 and SCD1D probes also stained follicular B cells (Fig. 3f).
Central nervous system (CNS) tissue (cervical spinal cord) showed no reactivity with the SCD1B42 probe. The exon 4 and SCD1D probes showed staining of microglial cells and neuronal cell bodies (Fig. 3g). No staining was evident with either sense riboprobe (Fig. 3h).
Discussion
The CD1 family is now established as representing an important third lineage of antigen-presenting molecules.9,10 The major difference between CD1 molecules and the classical antigen-presenting molecules (MHC classes I and II) is that the antigens presented by CD1 molecules are predominantly lipids and glycolipids. Consistent with this role in antigen presentation, cells expressing CD1 include ‘professional’ antigen-presenting cells such as dendritic cells and Langerhans' cells. On the basis of differential expression and sequence analysis, CD1 molecules can be divided into two groups: group 1 (CD1a, CD1b and CD1c) and group 2 (CD1d).
The Northern hybridization results are consistent with alternative splicing of ovine CD1 within the thymus. This phenomenon has been previously described in humans where all the CD1 genes, except CD1D, demonstrated alternative splicing with specific cryptic splice sites present in the cDNA of CD1B and CD1E.11 Further evidence of alternative splicing was reported by Woolfson & Milstein12 who provided evidence for exon 4 of the CD1 genes being under alternative splicing control.
Constitutive expression of the group 1 CD1 molecules is largely confined to cortical thymocytes and dendritic cells. Data concerning expression of group 2 (CD1d) antigens is, however, less straightforward. In situ hybridization studies on mouse intestine using a probe based on the exon 4 region of mouse CD1D showed expression at the base of the crypts in Paneth cells but not on villous epithelium.13 This result was surprising in that immunohistochemistry had previously shown expression of murine CD1 on gastrointestinal epithelium.14 More recent studies using mAbs showed that the principal site of CD1 expression in the mouse was on haemopoietic cells, including constitutive expression on nearly all T and B cells, macrophages and dendritic cells. Interestingly it was not detectable on the great majority of intestinal epithelial cells.15 Studies on human CD1d, in contrast, have localized expression of CD1 antigen to the apical and lateral regions of small and large intestinal epithelial cells in both glycosylated and non-glycosylated forms.16
These results indicate that there are interspecies differences in the expression of group 2 CD1 genes and antigens. The recent identification of a group 2/CD1D gene in sheep,7 which shares some but not all of the group 2 ‘class specific’ residues, suggests that group 2 CD1 antigens may comprise a heterogeneous family in a similar way to the CD1a–c molecules that form the group 1 family.
In addition to the differences in expression, it is clear from recent studies that the antigen-recognition properties of group 1 and group 2 CD1 molecules also differ in several ways. Differences include the microbial antigens which can be presented and the nature of the TCR receptors capable of recognizing the presented antigen (reviewed in ref. 17). Recently, mouse CD1 molecules (group 2) have been shown to present lipoglygan antigens from Plasmodium falciparum and Trypanosoma brucei to NK1.1 helper T cells.18 It is perhaps not surprising then that there are marked differences in the expression patterns of group 1 and group 2 family members given the apparent functional differences which exist between the two groups.
The results described in this work identify a similar dichotomy of CD1 expression in the sheep with SCD1D expression identified in a range of cells including cortical thymocytes, B cells and dendritic cells. In all tissues examined, the pattern of staining generated with the exon 4 probe was identical to that obtained with the SCD1D probe, establishing that this isotype has the widest pattern of expression in the sheep. Of particular note is the staining identified within the CNS where strong expression in neuroglial cells and, most interestingly, in neurones was identified. It is well recognized that the CNS is an ‘immune privileged’ site with low levels of expression of classical MHC molecules. Previous studies of neural tissue from human patients with various immune-mediated nervous system diseases have shown up-regulation of CD1b on hypertrophic astrocytes in chronic active lesions of multiple sclerosis19 and on endoneurial macrophages and myelinated nerve fibres in acute inflammatory demyelinating polyradiculoneuropathy.20 Expression of CD1 by ovine microglial cell cultures has also been described.21 The results described here confirm constitutive group 2 CD1 expression within the CNS of sheep. Recent experiments using CD1 knockout mice have shown that CD1-reactive T cells are required for the development of systemic tolerance to antigen introduced to the immune-privileged site of the eye.22 It is plausible that constitutive group 2 CD1 expression within the CNS is consistent with a role for SCD1D in immune regulation in this site.
In conclusion, we have extended previous investigations, which used mAbs to examine expression of ovine CD1. The combination of RT–PCR and generation of isotype-specific probes for hybridization studies has confirmed the marked differences in expression of group 1 and group 2 CD1, establishing widespread expression of CD1D distinct from the more restricted expression of group 1 family members. These data provide a valuable baseline for future investigations on modulation of CD1 expression in various disease states.
Acknowledgments
This work was funded by BBSRC grant 15/S11255.
References
- 1.Calabi F, Jarvis JM, Martin L, Milstein C. Two classes of CD1 genes. Eur J Immunol. 1989;19:285–92. doi: 10.1002/eji.1830190211. [DOI] [PubMed] [Google Scholar]
- 2.Dascher CC, Hiromatsu K, Naylor JW, et al. Conservation of a CD1 multigene family in the guinea pig. J Immunol. 1999;163:5478–88. [PubMed] [Google Scholar]
- 3.Brossay L, Kronenberg M. Highly conserved antigen-presenting function of CD1d molecules. Immunogenetics. 1999;50:146–51. doi: 10.1007/s002510050590. [DOI] [PubMed] [Google Scholar]
- 4.Dutia BM, Hopkins J. Analysis of the CD1 cluster in sheep. Vet Immunol Immunopathol. 1991;27:189–94. doi: 10.1016/0165-2427(91)90099-x. [DOI] [PubMed] [Google Scholar]
- 5.Rhind SM, Dutia BM, Howard CJ, Hopkins J. Discrimination of 2 subsets of CD1 molecules in the sheep. Vet Immunol Immunopathol. 1996;52:265–70. doi: 10.1016/0165-2427(96)05576-6. 10.1016/0165-2427(96)05576-6. [DOI] [PubMed] [Google Scholar]
- 6.Howard CJ, Naessens J. Summary of workshop findings for cattle. Vet Immunol Immunopathol. 1993;39:25–48. doi: 10.1016/0165-2427(93)90161-v. [DOI] [PubMed] [Google Scholar]
- 7.Rhind SM, Hopkins J, Dutia BM. Amino-terminal sequencing of sheep CD1 antigens and identification of a sheep CD1D gene. Immunogenetics. 1999;49:225–30. doi: 10.1007/s002510050483. 10.1007/s002510050483. [DOI] [PubMed] [Google Scholar]
- 8.Ferguson ED, Dutia BM, Hein WR, Hopkins J. The sheep CD1 gene family contains at least 4 CD1B homologues. Immunogenetics. 1996;44:86–96. 10.1007/s002510050094. [PubMed] [Google Scholar]
- 9.Porcelli SA, Modlin RL. The CD1 system: antigen-presenting molecules for T cell recognition of lipids and glycolipids. Annu Rev Immunol. 1999;17:297–329. doi: 10.1146/annurev.immunol.17.1.297. [DOI] [PubMed] [Google Scholar]
- 10.Porcelli SA, Segelke BW, Sugita M, Wilson IA, Brenner MB. The CD1 family of lipid antigen-presenting molecules. Immunol Today. 1998;19:362–8. doi: 10.1016/s0167-5699(98)01289-4. 10.1016/s0167-5699(98)01289-4. [DOI] [PubMed] [Google Scholar]
- 11.Calabi F, Bilsland CAG, Yu CY, Bradbury A, Terta Belt K, Martin LH, Milstein C. Recent progress in the molecular study of CD1. In: Knapp W, editor. Leukocyte Typing IV. Oxford: Oxford University Press; 1989. pp. 254–8. [Google Scholar]
- 12.Woolfson A, Milstein C. Alternative splicing generates secretory isoforms of human CD1. Proc Natl Acad Sci USA. 1994;91:6683–7. doi: 10.1073/pnas.91.14.6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lacasse J, Martin LH. Detection of CD1 messenger-RNA in paneth cells of the mouse intestine by in situ hybridization. J Histochem Cytochem. 1992;40:1527–153. doi: 10.1177/40.10.1382091. [DOI] [PubMed] [Google Scholar]
- 14.Bleicher PA, Balk SP, Hagen SJ, Blumberg RS, Flotte TJ, Terhorst C. Expression of murine CD1 on gastrointestinal epithelium. Science. 1990;250:679–82. doi: 10.1126/science.1700477. [DOI] [PubMed] [Google Scholar]
- 15.Brossay L, Jullien D, Cardell S, Sydora BC, Burdin N, Modlin RL, Kronenberg M. Mouse CD1 is mainly expressed on hemopoietic-derived cells. J Immunol. 1997;159:1216–24. [PubMed] [Google Scholar]
- 16.SomnayWadgaonkar K, Nusrat A, Kim HS, Canchis WP, Balk SP, Colgan BRS. Immunolocalization of CD1d in human intestinal epithelial cells and identification of a beta(2)-microglobulin-associated form. Int Immunol. 1999;11:383–92. doi: 10.1093/intimm/11.3.383. 10.1093/intimm/11.3.383. [DOI] [PubMed] [Google Scholar]
- 17.Burdin N, Kronenberg M. CD1-mediated immune responses to glycolipids. Curr Opin Immunol. 1999;11:326–31. doi: 10.1016/s0952-7915(99)80052-1. 10.1016/s0952-7915(99)80052-1. [DOI] [PubMed] [Google Scholar]
- 18.Schofield L, McConville MJ, Hansen D, Campbell AS, FraserReid B, Grusby MJ, Tachado SD. CD1d-restricted immunoglobulin G formation to GPI-anchored antigens mediated by NKT cells. Science. 1999;283:225–9. doi: 10.1126/science.283.5399.225. 10.1126/science.283.5399.225. [DOI] [PubMed] [Google Scholar]
- 19.Battistini L, Fischer FR, Raine CS, Brosnan CF. CD1b is expressed in multiple sclerosis lesions. J Neuroimmunol. 1996;67:145–51. doi: 10.1016/0165-5728(96)00045-8. 10.1016/s0165-5728(96)00045-8. [DOI] [PubMed] [Google Scholar]
- 20.KhaliliShirazi A, Gregson NA, Londei M, Summers L, Hughes RAC. The distribution of CD1 molecules in inflammatory neuropathy. J Neurol Sci. 1998;158:154–63. doi: 10.1016/s0022-510x(98)00121-x. 10.1016/s0022-510x(98)00121-x. [DOI] [PubMed] [Google Scholar]
- 21.Ebrahimi B, Allsopp TE, Fazakerley JK, Harkiss GD. Phenotypic characterisation and infection of ovine microglial cells with Maedi-Visna Virus. J Neurovirology. 2000;6:320–8. doi: 10.3109/13550280009030758. [DOI] [PubMed] [Google Scholar]
- 22.Sonoda KH, Exley M, Snapper S, Balk SP, SteinStreilein J. CD1-reactive natural killer T cells are required for development of systemic tolerance through an immune-privileged site. J Exp Med. 1999;190:1215–25. doi: 10.1084/jem.190.9.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]