Introduction
The generation of B lymphocytes from committed progenitor cells is a complex process involving the transit of cells through several critical stages of development. Throughout their transit, developing B cells are subject to choices between survival, proliferation or death; fates that are dictated by combinations of intrinsic and extrinsic signals. Thus, failure to express certain signalling molecules on their surface at particular stages of development results in the cell failing to receive a viability and/or proliferative signal. Likewise, failure to link surface receptor expression to survival machinery will result in cell death. During the early stages of B-cell development, one such receptor molecule is the pre-B-cell receptor (pre-BCR). The pre-BCR is a heterodimer composed of an immunoglobulin (Ig) heavy chain molecule (IgH) covalently associated with an immunoglobulin light chain-like molecule called the surrogate light chain (SL).1–3 The SL itself is made up of two non-covalently associated proteins called lambda-5 (λ5) and VpreB, which together form a molecule having structural homology with conventional light (IgL) chains.4,5 Thus, in the SL, λ5 replaces a light chain constant region and VpreB the variable part. It is likely that the membrane pre-BCR complex is composed of two IgH molecules associated with two SLs.1 However, SL can reach the surface in the absence of IgH but in association with other molecules, for example with a glycoprotein of approximately 130 000 MW, so-called gp130.6,7 In this case, the surface SL-containing complex is referred to as a pro-BCR.8 The detailed description of SL structure must await crystallographic analysis, but recently Melchers has proposed a model of SL chain assembly.9 In this review we will discuss the molecular structure and gene organization of pre-BCR components and attempt to outline how surface expression of the pre-BCR impacts on cell survival, differentiation, proliferation and repertoire selection of developing B cells.
General scheme of B lymphocyte development
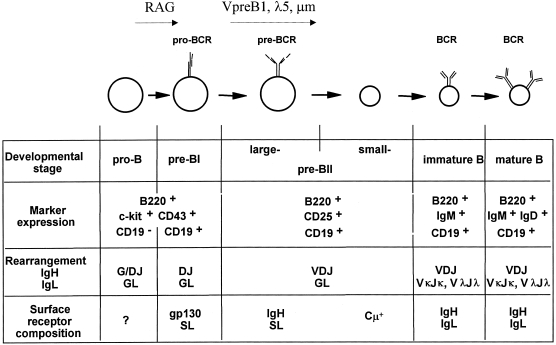
Different stages of B-cell development can be defined by the use of a combination of cell-surface markers and the status of immunglobulin gene rearrangements10,11 (Fig. 1, B-lymphocyte development in adult bone marrow). Following commitment to the B-cell lineage, an event which is still poorly understood, the first stage of the B-cell lineage is called a pro-B cell and is defined phenotypically as a B220+CD19− c-kit+CD43+CD25−IgM−IgD− cell in which there does not have to be any immunoglobulin gene rearrangement.12,13 It should be noted though that some pro-B cells have DJH rearrangements but only on one allele.13 Thus pro-B cells are found in mice in which the recombinase-activating genes (RAG) have been deleted,14,15 and in such mice, they are CD19+. However, pro-B cells do contain RNA transcripts for components of the SL but it is unclear at the present time whether they contain SL protein either in their cytoplasm or at the cell surface.13,16 It is possible that SL might reach the cell surface in association with molecules other than IgH, for example gp130.6,7
Figure 1.
B-lymphocyte development in the adult mouse bone marrow.
The next stage of B-cell development is the pre-BI cell which is phenotypically similar to a pro-B cell except for the expression of CD19 (B220+CD19+c-kit+CD43+CD25− IgM−IgD−) but which genotypically can be distinguished by having completed the first step in immunoglobulin gene rearrangement, namely rearrangement of DH to JH segments. As shown by single-cell analysis, most pre-BI cells have rearranged DH to JH on both alleles.17 It should be noted, however, that DH to JH rearrangement is not a marker per se of B-lymphocyte commitment. Thus, T-cell lines, clones and hybridomas can show DJ rearrangements and as recently shown by Rolink et al., pre-B-cell clones from Pax-5 KO mice containing DJ rearrangements at the immunoglobulin locus and incapable of further B-cell development,18,19 could upon in vivo transfer, reconstitute other hemopoietic lineages, including that of T cells.20 In pre-BI cells, the second step in immunuglobulin rearrangement, VH to DJH rearrangement takes place and this step is unique to committed B cells. Cells with successful VDJH rearrangements contain IgH protein in their cytoplasm (cµ+ cells) and become large pre-BII cells which are phenotypically B220+CD19+c-kit−CD43−CD25+IgM−IgD−. It is at this pre-BI to pre-BII transition and at the large pre-BII stage that SL plays its major role.
As with all immunoglobulin gene rearrangements,21 VD and DJ junctions are generated randomly. The success of VDJH rearrangements is only judged when the protein product thus created is expressed at the cell surface. Because of the three base-pair rule and in-built stop codons in DJH genes, the majority of VDJH rearrangements will not be successful in that the DNA sequence generated will not encode a protein. However, each cell is given multiple chances of rearranging immunoglobulin genes successfully and generating an IgH protein. Even if the VDJH rearrangement that has occurred encodes a protein, the developing B-cell must test the quality of the IgH protein product. The major criterion of quality is whether the IgH protein can successfully pair with SL and be expressed at the cell surface as a pre-BCR molecule.22 Cells that fail to express a pre-BCR molecule run the gauntlet of impending cell death. Cell viability at this point in B-cell development is dependent upon extrinsic factors including IL-7. It is known that signals from the IL-7 receptor (IL-7R) impinge on cell survival genes such as bcl-2, and over-expression of these survival genes at this point in development may prevent some of the cell death that would otherwise occur.23 However, it is unclear whether signals from the pre-BCR influence IL-7R expression or whether the number of IL-7R molecules per pre-BI cell varies. Careful analysis reveals that there is considerable cell loss in the transition from pre-BI to pre-BII cells presumably because failure to successfully express a pre-BCR results in cell death.
In theory, VH to DJH rearrangement can occur on both alleles. However, single-cell analysis reveals that this occurs in only half the pre-BII and more mature B cells and that the other half have rearranged only one allele and have the other allele in the DJ configuration.17 The explanation for this failure to rearrange the second allele in many pre-BII cells is that signalling via the pre-BCR, composed of the IgH protein from the first rearranged allele, prevents or inhibits rearrangement of the second allele, a phenomenon called allelic exclusion. Allelic exclusion, which at the DNA level is clearly not 100% efficient, is probably aided by the fact that there is a dramatic drop in functional RAG-1 and RAG-2 proteins in the transition from pre-BI to pre-BII cells.24 Regulation of functional RAG protein availability is a complex process involving regulation of transcription,25–27 message stability and protein phosphorylation, the latter linked to the proliferative status of the cell.28 Nevertheless, approximately half of the pre-BII cells have rearranged both IgH alleles and sequence analysis reveals that in these cells most, but not all, are rearranged successfully on one allele but unsuccessfully on the other. Transfection experiments reveal that in the few pre-BII cells that successfully rearrange both IgH alleles (i.e. potentially double IgH expressing cells) only one of the two IgH proteins is capable of pairing with SL and reaching the cell surface.22 Thus, IgH allelic exclusion is partially maintained at the level of protein pairing.
At present, it is unclear whether pre-BCR molecules have ligand-recognition function. Even though pre-BCR molecules consisting of IgH and SL associate with other components of the BCR signalling complex and can be shown to deliver signals such as calcium fluxes within pre-B-cell lines,29 it is unclear whether engagement of pre-BCR molecules by ligand influences B-cell development. Certainly using fetal liver organ cultures and anti-immunoglobulin reagents, recognizing IgH molecules in both BCR and pre-BCR complexes, engagement of the BCR but not the pre-BCR resulted in the arrest of B-cell development.30
At the large pre-BII cell stage, cells undergo several rounds of division and DNA analysis indicates that 60–70% of the cells are in S, G2 or M phases of the cell cycle.31 After proliferation, they differentiate into small, resting pre-BII cells, which have the same cell-surface phenotype as their larger precursors. The ratio of large to small pre-BII cells is 1:3 or 1:4 in normal mice but how this ratio is maintained is unclear.32 The signal for pre-BII cell proliferation is initiated by surface expression of the pre-BCR and is probably maintained by the availability of interleukin-7 (IL-7) in the local microenvironment. It is unclear if all pre-BII cell clones proliferate equally. It is distinctly possible that, depending upon the number of pre-BCR molecules expressed, the affinity/avidity of the interaction between IgH and SL, the number of IL-7R molecules and the availability of IL-7, different clones of pre-BII cells may undergo different numbers of cell divisions. Why a large pre-BII cell should stop dividing is also unclear. Certainly, the availability of SL protein drops dramatically in pre-BII cells31 and as recently proposed by Melchers et al. it might be that limiting SL protein availability may dictate the pre-BII cell division number.33 The reduction in SL protein availability in pre-BII cells may in part explain their decreased level of surface pre-BCR expression but with continued expression of IgH molecules in the cytoplasm.31 Molecular analysis indicates that the repertoire of IgH molecules expressed at the cµ+ pre-BI stage can differ greatly from that seen at the large pre-BII cell stage and in peripheral B cells.34 This substantiates the idea that at the clonal level, cell division within the pre-BII compartment varies considerably and suggests that differential proliferative expansion in the pre-BII compartment may have profound influences on subsequent immunoglobulin repertoire generation.
Large pre-BII cells produce sterile Igκ, and some rearranged Igκ transcripts, indicating that these cells are just about to rearrange Igκ. However, sterile and rearranged Igλ transcripts are only detected in small pre-BII cells.35 Thus, IgL rearranges in the pre-BII cell compartment and as soon as a functional IgL protein is formed, the cells differentiate into immature B cells characterized phenotypically as B220+CD19+c-kit−CD43−CD25−IgM+IgD−. Differentiation to immature IgM+ cells is dependent upon the quality of the IgL chains produced. Each IgL chain must be capable of pairing with the IgH already expressed, thereby reaching the cell surface as a BCR. Whether the rules governing IgH/IgL pairing are similar to those governing IgH and SL pairing are unknown. It may well be that the ‘fixed’ IgH protein in individual pre-BII clones selects a certain repertoire of IgL proteins. In addition, the IgH/IgL dimer must not form a surface BCR capable of recognizing self-antigen. If autoantigen is recognized, then further development is arrested. Thus, the transition from small pre-BII to immature B cells marks the stage at which negative selection of the immunoglobulin repertoire by external ligands begins. This arrest of B-cell development can result in either the induction of apoptosis and cell loss or the induction of additional immunoglobulin rearrangements to generate a modified, non-autoreactive BCR.36,37 This latter process has been extensively studied in mice expressing potentially auto-reactive transgenic BCR and has been called receptor editing. However, the impact of receptor editing on the generation of the B-cell repertoire in normal mice is unclear. Once a non-auto-reactive BCR molecule is expressed, cells can leave the bone marrow and mature into B220+CD19+c-kit−CD43−CD25−IgM+IgD+ cells. Both immature and mature B cells are resting cells. From the above account, it is clear that the pre-BCR plays a crucial role in B-cell development.
How do modifications of the pre-BCR affect B-lymphocyte development?
Several of the pre-BCR components necessary for the generation of a complete pre-BCR have been deleted in the mouse germline by homologous recombination and a brief description of the phenotypes of the respective mice is given below.
In mice lacking either RAG-1 or RAG-2 genes, B-cell development is arrested at the pro-B-cell stage and all further B-cell (and T-cell) development is blocked.14,15 Mice deficient in the transmembrane portion of Ig µH (µmT) chain fail to express cell-surface pre-BCR molecules and show a block in B-cell development.38 In addition, the cells that have rearranged VHDJH are allelicly included at the IgH loci.39 In contrast, mice lacking the constant region of µH, but synthesising δH, show a normal phenotype40 suggesting that IgD can substitute for IgM. Thus, the transmembrane portion of IgH is necessary for pre-B lymphocytes to differentiate, to proliferate, and for IgH allelic exclusion. Mice lacking the λ5 gene (λ5T) also show a block in B-cell development.41,42 However, in contrast to the µmT mice, the block is not complete: small numbers of immature/mature B lymphocytes do develop, demonstrating that differentiation can occur in the absence of λ5.
Detailed analysis of cell numbers reveals that in RAG KO, µmT and λ5T mice, the number of pro/pre-BI cells is about twofold higher than that of normal mice.32 It is still unclear how cell viability is maintained in the absence of normal pre-BCR molecules. Interleukin-7 may play a role in that increasing IL-7 availability, by the introduction of an IL-7 transgene, resulted in a further modest (twofold) increase in pro/pre-BI cell numbers.43 That this increase was not greater may reflect the fact that the size of the microenvironment capable of supporting pro- and pre-B-cell survival is limited and that cells must move to other microenvironments to continue their development. This hypothesis is supported by the presence of normal or reduced numbers of pro/pre-BI cells in mice lacking the IL-7Rα,44,45 which are blocked at the same developmental stage as seen in µmT and λ5T mice.
The most prominent effect in λ5T mice is a lack of proliferation of large pre-BII cells, indicated by the ratio of large to small cells in the B220+CD25+ population and further demonstrated in fetal liver organ cultures and in vitro cultures.32,46,47 Interestingly, IgH allelic exclusion is still operating in these mice, which may be due to the expression of a pre-BCR composed of IgH and VpreB alone. The block in λ5T animals affects both the B-1 and B-2 B-cell compartments in young mice. The B-1 compartment reaches normal levels with age, most likely because of their capacity for self-renewal, but this is apparently not the case with B-2 B cells.41 Importantly, the analysis of λ5T mice indicates that the signals emanating from the pre-BCR, which are responsible for pre-BII cell proliferation and allelic exclusion, are generated independently.
In the mouse, in contrast to humans, there are two VpreB genes, which are 97% identical at the nucleotide and amino acid levels, resulting in differences at four out of the total of 142 amino acids in the two proteins.5 At the single-cell level, bone marrow pre-B cells that express λ5 also make VpreB1 mRNA, while ∼30% of these cells express VpreB2 as well.48 Currently, it is not known if the appeared differential expression of VpreB1 and VpreB2 is genuine or owing to a difference in the sensitivity of the detection assays. The VpreB1 and VpreB2 proteins are both recognized by the VP245 monoclonal antobody (mAb) but there is currently no mAb available that distinguishes the two proteins and it is therefore not known if both proteins are present in one and the same cell.6,48 If a cell does make both proteins, and since the pre-BCR is a mixed heterodimer of two IgH and two SL molecules, this implies that a cell can express three different pre-BCRs; a pair of IgH and λ5 molecules associated with either two VpreB1, two VpreB2 or one of each. If this is the case, differential availability of VpreB1 and VpreB2 might affect pairing with IgH and thereby IgH repertoire selection.
Recently, mice lacking the VpreB1 gene only (VpreB1−/−), the VpreB2 gene only (VpreB2−/−) or both genes (VpreB−/−) have been established. The VpreB1−/− mice have been analysed and they show a partial block in B-lymphocyte development at the same stage as that observed in λ5T mice, but the effect is much less marked.49 In the bone marrow, this block results in an increase in the number of pre-BI cells (to the same extent as in RAG KO, µmT and λ5T mice) and a slight decrease in pre-BII and immature B cells (80% of control levels). The ratio of large to small pre-BII cells is the same as in wild-type mice and hence proliferation appears to be normal. The cells express VpreB2 and λ5 both as RNA and protein suggesting that a pre-BCR containing VpreB2 is fully competent. Furthermore, IgH allelic exclusion is also in place. The slight decrease in numbers of large pre-BII cells may be due to a difference between VpreB1 and VpreB2 in their capacity to pair with µH and form a pre-BCR. This demonstrates that VpreB2 on its own is sufficient although not as efficient as both VpreB gene products together. The VpreB2−/− and VpreB−/− mice are currently being analysed and we should soon know what effect the lack of these molecules has, if any, on B-cell differentiation, proliferation, allelic exclusion and repertoire selection.
Regulation of SL gene expression
From the above it is obvious that the control of SL component synthesis and expression is tightly regulated during B-cell development. VpreB and λ5 are expressed only in the B lineage, and furthermore, only at the earlier stages of development, i.e. in pro-B, pre-BI and large pre-BII cells.24,48 This cell lineage and stage-specific expression is regulated at the level of transcription.50–53 Measurements made using semiquantitative reverse transcriptase–polymerase chain reaction (RT–PCR) indicate that RNA steady-state levels of the SL components are relatively similar in the early stages of B-cell development, but undergo a dramatic (100-fold) drop in small pre-BII cells.24,48
The VpreB1 gene is located 4–5 kb upstream of, and in the same transcriptional orientation as, the λ5 gene, while the distance to VpreB2, also located on chromosome 16, is unknown.5,54 Searches for regulatory regions have demonstrated that VpreB1, VpreB2 and λ5 are all transcribed as independent genes and hence have their respective promoter.50,51 These promoters belong to the initiator family in that they lack a TATA-box and initiate transcription at multiple sites.55 The region immediately upstream of each respective gene contains, in addition to the promoter, an enhancer that is preferentially active in pre-B cell lines.51,53,56 The in vivo activity of the λ5 5′ region (promoter plus enhancer) has been studied in transgenic mice where these elements have been used to control the expression of a reporter gene encoding the human CD25 (huTac) molecule.16 In such mice, the highest level of human CD25 transgene expression was found in pre-BI cells, while later stages expressed decreasing levels, a pattern of transgene expression similar to that of the endogenous λ5 gene. Transgenic mice expressing a reporter gene under the control of the VpreB1 enhancer have recently been established and these are currently being analysed to determine whether the VpreB1 enhancer is also active in vivo and, if so, whether it shows stage-specific expression similar to that of the λ5 enhancer. The region 3′ of λ5, together with the λ5 enhancer and genomic region, are required in order to achieve expression from a single-copy transgene.57 This same region may also be important for the expression of a single-copy VpreB1 gene57 but it is not clear how it would influence VpreB2 gene expression as this gene may be located far from the λ5 3′ region.
Transcription factors important for B-cell development and SL gene expression
Several transcription factors have been shown as crucial for B-cell development and some of these have been implied as regulators of VpreB and λ5 gene expression. Since the two genes are always expressed in parallel, the hypothesis would be that they are regulated by the same factors. So far, four transcription factors have been suggested to regulate the transcription of both VpreB and λ5.
The first is early B-cell factor (EBF), which was originally demonstrated to regulate the activity of the mb-1 (Igα) promoter.58,59 EBF was subsequently found to control the activity of the endogenous SL genes, possibly through the VpreB and λ5 enhancers.53,56,60,61 These findings were further supported by the observation that mice lacking EBF, where B-cell development is blocked at the pro-B cell (B220+CD43+) stage, did not express VpreB or λ5, nor mb-1.62
The second factor is E2A, which binds to E-box motifs present in many cell-type-specific genes.63 Recent studies have shown that this factor is important for the activation of endogenous VpreB and λ5 gene expression, possibly through the respective enhancer.60,61 This factor has also been disrupted and mice lacking the E2A gene products (E12 and E47) are blocked at a slightly earlier developmental stage than EBF-deficient mice. As expected, the cells do not express λ5.64,65 Interestingly, mice heterozygous for both EBF and E2A deficiency (EBF+/−E2A+/−) are also blocked in B-cell development and the effect is more pronounced than in heterozygous mice deficient in the individual factors. However, this phenotype is not as strong as in the respective homozygous mice, suggesting a co-ordinated regulation of B-cell development.61 Furthermore, the EBF+/−E2A+/− mice express low levels of VpreB owing to a decreased frequency of VpreB-expressing cells, rather than an overall decrease in expression levels, implying mono-allelic expression of EBF (E2A is bi-allelicly expressed in mature B cells).65
A third factor, Pax-5, has also been implied in VpreB and λ5 gene expression. However both VpreB and λ5 are expressed at seemingly normal levels in mice lacking Pax-5, suggesting that this factor is not required for the activation of these genes.18,66 Pax-5 is important for the control of the B-cell specific CD19 gene67,68 and, as mentioned above, these animals have a block in B-cell development at the pre-BI stage.
A fourth factor implicated as a regulator of the VpreB and λ5 genes is Ikaros, which exists in several forms generated by differential splicing. This transcription factor was first cloned for its capacity to activate the CD3δ enhancer and the terminal deoxynucleotidyl transferase (TdT) promoter.69,70 As shown by gene deletion, Ikaros is also important for the development of B cells as well as other cell types. However, the block is very early in development and it is not possible to analyse its effects on SL gene expression.71 Ikaros has been implied as a suppressor of λ5 gene activity: the λ5 gene was found to be co-localized with Ikaros in transcriptionally inactive hetero-chromatin, but only in cells that did not express the gene, i.e. in a mature B- but not in a pre-B-cell line.72 Thus, EBF and E2A seem to control the activation of VpreB and λ5, while Ikaros, and possibly Pax5, may be more significant in switching off the genes. Hence, a combination of all these factors will ensure that the VpreB and λ5 genes are transcribed in cells at the right time.
Conclusion
As outlined above, the pre-BCR controls a critical stage of B-cell development and pre-B cells failing to express a fully functional pre-BCR are compromised in further development. The pre-BCR is crucial for differentiation, but also for IgH allelic exclusion, proliferation and IgH repertoire selection. However, the lack of individual components seem to affect different signalling pathways, making it possible to study these signalling events separately. Numerous important questions remain unanswered. How do signals generated via the pre-BCR regulate cell proliferation and how is proliferation arrested? How are pre-BCR signals linked to those from the IL-7R? Does the pre-BCR have a ligand? How do the lack of VpreB and the lack of the complete SL affect all these events? Can the transgenic mice that carry particular reporter genes driven by the λ5 and VpreB enhancers tell us anything about the pre-BCR? Finally, how do the multiple transcription factors involved combine to regulate SL gene expression?
Acknowledgments
We appreciate the constructive comments of our colleagues. I.-L.M. is supported by the BBSRC.
References
- 1.Pillai S, Baltimore D. Formation of disulfide-linked µ2ω2 tetramers in pre-B cells by the 18k ω-immunoglobulin light chain. Nature. 1987;329:435–441. doi: 10.1038/329172a0. [DOI] [PubMed] [Google Scholar]
- 2.Karasuyama H, Kudo A, Melchers F. The proteins encoded by the VpreB and lambda 5 pre-B cell-specific genes can associate with each other and with mu heavy chain. J Exp Med. 1990;172:969–72. doi: 10.1084/jem.172.3.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsubata T, Reth M. The products of pre-B cell-specific genes (lambda 5 and VpreB) and the immunoglobulin mu chain form a complex that is transported onto the cell surface. J Exp Med. 1990;172:973–6. doi: 10.1084/jem.172.3.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sakaguchi N, Melchers F. Lambda 5, a new light-chain-related locus selectively expressed in pre-B lymphocytes. Nature. 1986;324:579–82. doi: 10.1038/324579a0. [DOI] [PubMed] [Google Scholar]
- 5.Kudo A, Melchers F. A second gene, VpreB in the lambda 5 locus of the mouse, which appears to be selectively expressed in pre-B lymphocytes. EMBO J. 1987;6:2267–72. doi: 10.1002/j.1460-2075.1987.tb02500.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karasuyama H, Rolink A, Melchers F. A complex of glycoproteins is associated with VpreB/lambda 5 surrogate light chain on the surface of mu heavy chain-negative early precursor B cell lines. J Exp Med. 1993;178:469–78. doi: 10.1084/jem.178.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shinjo F, Hardy RR, Jongstra J. Monoclonal anti-lambda 5 antibody FS1 identifies a 130 kDa protein associated with lambda 5 and Vpre-B on the surface of early pre-B cell lines. Int Immunol. 1994;6:393–9. doi: 10.1093/intimm/6.3.393. [DOI] [PubMed] [Google Scholar]
- 8.Winkler T, Melchers F. Structure and Function of the Pro- and Pre-B-Cell Receptors on B-Lymphoid Lineage Precursor Cells. In: Monroe JAR, editor. Molecular Biology of B-Cell and T-Cell Development. Totowa, NJ: Humana Press Inc; 1999. [Google Scholar]
- 9.Melchers F. Fit for life in the immune system? Surrogate L chain tests H chains that tests L chains (Commentary) Proc Natl Sci USA. 1999;96:2571–3. doi: 10.1073/pnas.96.6.2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hardy R, Carmack C, Shinton S, Kemp J, Hayakawa K. Resolution and characterisation of pro-B and pre-pro-B cell stages in normal mouse bone marrow. J Exp Med. 1991;173:1213–25. doi: 10.1084/jem.173.5.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Osmond DG, Rolink A, Melchers F. Murine B lymphopoiesis: towards a unified model. Immunol Today. 1998;19:65–8. doi: 10.1016/s0167-5699(97)01203-6. 10.1016/s0167-5699(97)01203-6. [DOI] [PubMed] [Google Scholar]
- 12.Li YS, Wasserman R, Hayakawa K, Hardy RR. Identification of the earliest B lineage stage in mouse bone marrow. Immunity. 1996;5:527–35. doi: 10.1016/s1074-7613(00)80268-x. [DOI] [PubMed] [Google Scholar]
- 13.Ogawa M, ten Boekel E, Melchers F. Identification of CD19− B220+c-kit+Flt3/Flk-2+ cells as early B lymphoid precursors before pre-B-I cells in juvenile mouse bone marrow. Int Immunol. 2000;12:313–24. doi: 10.1093/intimm/12.3.313. 10.1093/intimm/12.3.313. [DOI] [PubMed] [Google Scholar]
- 14.Mombaerts P, Iacomini J, Johnson RS, Herrup K, Tonegawa S, Papaioannou VE. RAG-1 deficient mice have no mature B and T lymphocytes. Cell. 1992;68:869–77. doi: 10.1016/0092-8674(92)90030-g. [DOI] [PubMed] [Google Scholar]
- 15.Shinkai Y, Rathbun G, Lam K-P, et al. RAG-2 deficient mice lack mature lymphocytes owing to inability to initiate V (D) J rearrangement. Cell. 1992;68:855–67. doi: 10.1016/0092-8674(92)90029-c. [DOI] [PubMed] [Google Scholar]
- 16.Mårtensson I-L, Melchers F, Winkler TH. A transgenic marker for mouse B lymphoid precursors. J Exp Med. 1997;185:653–61. doi: 10.1084/jem.185.4.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.ten Boekel E, Melchers F, Rolink A. The status of Ig loci rearrangements in single cells from different stages of B cell development. Int Immunol. 1995;7:1013–9. doi: 10.1093/intimm/7.6.1013. [DOI] [PubMed] [Google Scholar]
- 18.Urbanek P, Wang ZQ, Fetka I, Wagner EF, Busslinger M. Complete block of early B cell differentiation and altered patterning of the posterior midbrain in mice lacking Pax5/BSAP. Cell. 1994;79:901–12. doi: 10.1016/0092-8674(94)90079-5. [DOI] [PubMed] [Google Scholar]
- 19.Nutt SL, Urbanek P, Rolink A, Busslinger M. Essential functions of Pax5 (BSAP) in pro-B cell development: difference between fetal and adult B lymphopoiesis and reduced V-to-DJ recombination at the IgH locus. Genes Dev. 1997;11:476–91. doi: 10.1101/gad.11.4.476. [DOI] [PubMed] [Google Scholar]
- 20.Rolink AG, Nutt SL, Melchers F, Busslinger M. Long-term in vivo reconstitution of T-cell development by Pax5-deficient B-cell progenitors. Nature. 1999;401:603–6. doi: 10.1038/44164. [DOI] [PubMed] [Google Scholar]
- 21.Tonegawa S. Somatic generation of antibody diversity. Nature. 1983;302:575–81. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- 22.ten Boekel E, Melchers F, Rolink AG. Precursor B cells showing H chain allelic inclusion display allelic exclusion at the level of pre-B cell receptor surface expression. Immunity. 1998;8:199–207. doi: 10.1016/s1074-7613(00)80472-0. [DOI] [PubMed] [Google Scholar]
- 23.Lu L, Chaudhury P, Osmond DG. Regulation of cell survival during B lymphopoiesis: apoptosis and Bcl-2/Bax content of precursor B cells in bone marrow of mice with altered expression of IL-7 and recombinase-activating gene-2. J Immunol. 1999;162:1931–40. [PubMed] [Google Scholar]
- 24.Grawunder U, Leu TM, Schatz DG, Werner A, Rolink AG, Melchers F, Winkler TH. Down-regulation of RAG1 and RAG2 gene expression in preB cells after functional immunoglobulin heavy chain rearrangement. Immunity. 1995;3:601–8. doi: 10.1016/1074-7613(95)90131-0. [DOI] [PubMed] [Google Scholar]
- 25.Monroe RJ, Chen F, Ferrini R, Davidson L, Alt FW. RAG2 is regulated differentially in B and T cells by elements 5′ of the promoter. Proc Natl Acad Sci USA. 1999;96:12713–8. doi: 10.1073/pnas.96.22.12713. 10.1073/pnas.96.22.12713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Monroe RJ, Seidl KJ, Gaertner F, et al. RAG2: GFP knockin mice reveal novel aspects of RAG2 expression in primary and peripheral lymphoid tissues. Immunity. 1999;11:201–12. doi: 10.1016/s1074-7613(00)80095-3. [DOI] [PubMed] [Google Scholar]
- 27.Yu W, Nagaoka H, Jankovic M, et al. Continued RAG expression in late stages of B cell development and no apparent re-induction after immunization [see comments] Nature. 1999;400:682–7. doi: 10.1038/23287. [DOI] [PubMed] [Google Scholar]
- 28.Li Z, Dordai DI, Lee J, Desiderio S. A conserved degradation signal regulates RAG-2 accumulation during cell division and links V (D) J recombination to the cell cycle. Immunity. 1996;5:575–89. doi: 10.1016/s1074-7613(00)80272-1. [DOI] [PubMed] [Google Scholar]
- 29.Misener V, Downey GP, Jongstra J. The immunoglobulin light chain related protein lambda 5 is expressed on the surface of mouse pre-B cell lines and can function as a signal transducing molecule. Int Immunol. 1991;3:1129–36. doi: 10.1093/intimm/3.11.1129. [DOI] [PubMed] [Google Scholar]
- 30.Ceredig R, Rolink AG, Melchers F, Andersson J. The B cell receptor, but not the pre-B cell receptor, mediates arrest of B cell differentiation. Eur J Immunol. 2000;30:759–67. doi: 10.1002/1521-4141(200003)30:3<759::AID-IMMU759>3.0.CO;2-M. 10.1002/(sici)1521-4141(200003)30:03<759::aid-immu759>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 31.Karasuyama H, Rolink A, Shinkai Y, Young F, Alt FW, Melchers F. The expression of VpreB/λ5 surrogate light chain in early bone marrow precursor B cells of normal and B cell deficient mutant mice. Cell. 1994;77:133–43. doi: 10.1016/0092-8674(94)90241-0. [DOI] [PubMed] [Google Scholar]
- 32.Rolink A, Grawunder U, Winkler TH, Karasuyama H, Melchers F. IL-2 receptor alpha chain (CD25, TAC) expression defines a crucial stage in pre-B cell development. Int Immunol. 1994;6:1257–64. doi: 10.1093/intimm/6.8.1257. [DOI] [PubMed] [Google Scholar]
- 33.Melchers F, ten Boekel E, Yamagami T, Andersson J, Rolink A. The roles of preB and B cell receptors in the stepwise allelic exclusion of mouse IgH and L chain gene loci. Semin Immunol. 1999;11:307–17. doi: 10.1006/smim.1999.0187. 10.1006/smim.1999.0187. [DOI] [PubMed] [Google Scholar]
- 34.ten Boekel E, Melchers F, Rolink AG. Changes in the V (H) gene repertoire of developing precursor B lymphocytes in mouse bone marrow mediated by the pre-B cell receptor. Immunity. 1997;7:357–68. doi: 10.1016/s1074-7613(00)80357-x. [DOI] [PubMed] [Google Scholar]
- 35.Engel H, Rolink A, Weiss S. B cells are programmed to activate kappa and lambda for rearrangement at consecutive developmental stages. Eur J Immunol. 1999;29:2167–76. doi: 10.1002/(SICI)1521-4141(199907)29:07<2167::AID-IMMU2167>3.0.CO;2-H. 10.1002/(sici)1521-4141(199907)29:07<2167::aid-immu2167>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 36.Goodnow CC, Crosbie J, Jorgensen H, Brink RA, Basten A. Induction of self-tolerance in mature peripheral B lymphocytes. Nature. 1989;342:385–91. doi: 10.1038/342385a0. [DOI] [PubMed] [Google Scholar]
- 37.Nemazee DA, Bürki K. Clonal deletion of B lymphocytes in a transgenic mouse bearing anti-MHC class I antibody genes. Nature. 1989;337:562–6. doi: 10.1038/337562a0. [DOI] [PubMed] [Google Scholar]
- 38.Kitamura D, Roes J, Kuhn R, Rajewsky K. A B cell-deficient mouse by targeted disruption of the membrane exon of the immunoglobulin mu chain gene. Nature. 1991;350:423–6. doi: 10.1038/350423a0. [DOI] [PubMed] [Google Scholar]
- 39.Kitamura D, Rajewsky K. Targeted Disruption of m chain membrane exon causes loss of heavy-chain allelic exclusion. Nature. 1992;356:154–6. doi: 10.1038/356154a0. [DOI] [PubMed] [Google Scholar]
- 40.Lutz C, Ledermann B, Kosco-Vilbois MH, Ochsenbein AF, Zinkernagel RM, Köhler G, Brombacher F. IgD can largely substitute for loss of IgM function in B cells. Nature. 1998;393:797–801. doi: 10.1038/31716. [DOI] [PubMed] [Google Scholar]
- 41.Kitamura D, Kudo A, Schaal S, Muller W, Melchers F, Rajewsky K. A critical role of λ5 protein in B cell development. Cell. 1992;69:823–31. doi: 10.1016/0092-8674(92)90293-l. [DOI] [PubMed] [Google Scholar]
- 42.Rolink A, Karasuyama H, Grawunder U, Haasner D, Kudo A, Melchers F. B cell development in mice with a defective lambda 5 gene. Eur J Immunol. 1993;23:1284–8. doi: 10.1002/eji.1830230614. [DOI] [PubMed] [Google Scholar]
- 43.Ceredig R, Andersson J, Melchers F, Rolink A. Effect of deregulated IL-7 transgene expression on B lymphocyte development in mice expressing mutated pre-B cell receptors. Eur J Immunol. 1999;29:2797–807. doi: 10.1002/(SICI)1521-4141(199909)29:09<2797::AID-IMMU2797>3.0.CO;2-8. 10.1002/(sici)1521-4141(199909)29:09<2797::aid-immu2797>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 44.Peschon JJ, Morrisey PJ, Grqbstein KH, et al. Early lymphocyte expansion is severely impaired in interleukin 7 receptor-deficient mice. J Exp Med. 1994;180:1955–60. doi: 10.1084/jem.180.5.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Corcoran AE, Riddell A, Krooshoop D, Venkitaraman AR. Impaired immunoglobulin gene rearrangement in mice lacking the IL-7 receptor. Nature. 1998;391:904–7. doi: 10.1038/36122. [DOI] [PubMed] [Google Scholar]
- 46.Ceredig R, ten Boekel E, Rolink A, Melchers F, Andersson J. Fetal liver organ cultures allow the proliferative expansion of pre-B receptor-expressing pre-B-II cells and the differentiation of immature and mature B cells in vitro. Int Immunol. 1998;10:49–59. doi: 10.1093/intimm/10.1.49. 10.1093/intimm/10.1.49. [DOI] [PubMed] [Google Scholar]
- 47.Rolink AG, Winkler T, Melchers F, Andersson J. Precursor B cell receptor-dependent B cell proliferation and differentiation does not require the bone marrow or fetal liver environment. J Exp Med. 2000;191:23–32. doi: 10.1084/jem.191.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dul JL, Argon Y, Winkler T, ten Boekel E, Melchers F, Mårtensson IL. The murine VpreB1 and VpreB2 genes both encode a protein of the surrogate light chain and are co-expressed during B cell development. Eur J Immunol. 1996;26:906–13. doi: 10.1002/eji.1830260428. [DOI] [PubMed] [Google Scholar]
- 49.Mårtensson A, Argon Y, Melchers F, Dul J, Mårtensson I-L. Partial block in B lymphocyte development at the transition into the pre-B cell receptor stage in VpreB1 deficient mice. Int Immunol. 1999;11:453–60. doi: 10.1093/intimm/11.3.453. 10.1093/intimm/11.3.453. [DOI] [PubMed] [Google Scholar]
- 50.Okabe T, Bauer SR, Kudo A. Pre-B lymphocyte-specific transcriptional control of the mouse VpreB gene. Eur J Immunol. 1992;22:31–6. doi: 10.1002/eji.1830220106. [DOI] [PubMed] [Google Scholar]
- 51.Mårtensson I-L, Melchers F. PreB cell-specific λ5 gene expression due to suppression in non preB cells. Int Immunol. 1994;6:863–72. doi: 10.1093/intimm/6.6.863. [DOI] [PubMed] [Google Scholar]
- 52.Yang J, Glozak MA, Blomberg BB. Identification and localization of a developmental stage-specific promoter activity from the murine lambda 5 gene. J Immunol. 1995;155:2498–514. [PubMed] [Google Scholar]
- 53.Persson C, Mårtensson A, Mårtensson IL. Identification of a tissue- and differentiation stage-specific enhancer of the VpreB1 gene. Eur J Immunol. 1998;28:787–98. doi: 10.1002/(SICI)1521-4141(199803)28:03<787::AID-IMMU787>3.0.CO;2-H. 10.1002/(sici)1521-4141(199803)28:03<787::aid-immu787>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 54.Kudo A, Sakaguchi N, Melchers F. Organization of the murine Ig-related lambda 5 gene transcribed selectively in pre-B lymphocytes. EMBO J. 1987;6:103–7. doi: 10.1002/j.1460-2075.1987.tb04725.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mai S, Mårtensson IL. The c-myc protein represses the lambda 5 and TdT initiators. Nucleic Acids Res. 1995;23:1–9. doi: 10.1093/nar/23.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mårtensson A, Mårtensson I-L. Early B cell factor binds to a site critical for lambda5 core enhancer activity. Eur J Immunol. 1997;27:315–20. doi: 10.1002/eji.1830270145. [DOI] [PubMed] [Google Scholar]
- 57.Sabbattini P, Georgiou A, Sinclair C, Dillon N. Analysis of mice with single and multiple copies of transgenes reveals a novel arrangement for the lambda5-VpreB1 locus control region. Mol Cell Biol. 1999;19:671–9. doi: 10.1128/mcb.19.1.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hagman J, Travis A, Grosschedl R. A novel lineage-specific nuclear factor regulates mb-1 gene transcription at the early stages of B cell differentiation. EMBO J. 1991;10:3409–17. doi: 10.1002/j.1460-2075.1991.tb04905.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hagman J, Belanger C, Travis A, Turck CW, Grosschedl R. Cloning and functional characterization of early B-cell factor, a regulator of lymphocyte-specific gene expression. Genes Dev. 1993;7:760–73. doi: 10.1101/gad.7.5.760. [DOI] [PubMed] [Google Scholar]
- 60.Sigvardsson M, O'riordan M, Grosschedl R. EBF and E47 collaborate to induce expression of the endogenous immunoglobulin surrogate light chain genes. Immunity. 1997;7:25–36. doi: 10.1016/s1074-7613(00)80507-5. [DOI] [PubMed] [Google Scholar]
- 61.O'riordan M, Grosschedl R. Coordinate regulation of B cell differentiation by the transcription factors EBF and E2A. Immunity. 1999;11:21–31. doi: 10.1016/s1074-7613(00)80078-3. [DOI] [PubMed] [Google Scholar]
- 62.Lin H, Grosschedl R. Failure of B-cell differentiation in mice lacking the transcription factor EBF. Nature. 1995;376:263–7. doi: 10.1038/376263a0. [DOI] [PubMed] [Google Scholar]
- 63.Murre C, McCaw PS, Baltimore D. A new DNA Binding and Dimerization Motif in Immunoglobulin Enhancer Binding, daughterless, MyoD, and myc Proteins. Cell. 1989;56:773–83. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- 64.Bain G, Maandag EC, Izon DJ, et al. E2A proteins are required for proper B cell development and initiation of immunoglobulin gene rearrangements. Cell. 1994;79:885–92. doi: 10.1016/0092-8674(94)90077-9. [DOI] [PubMed] [Google Scholar]
- 65.Zhuang Y, Soriano P, Weintraub H. The helix-loop-helix gene E2A is required for B cell formation. Cell. 1994;79:875–84. doi: 10.1016/0092-8674(94)90076-0. [DOI] [PubMed] [Google Scholar]
- 66.Nutt SL, Thevenin C, Busslinger M. Essential functions of Pax-5 (BSAP) in pro-B cell development. Immunobiology. 1997;198:227–35. doi: 10.1016/S0171-2985(97)80043-5. [DOI] [PubMed] [Google Scholar]
- 67.Kozmik Z, Wang S, Dorfler P, Adams B, Busslinger M. The promoter of the CD19 gene is a target for the B-cell-specific transcription factor BSAP. Mol Cell Biol. 1992;12:2662–72. doi: 10.1128/mcb.12.6.2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Adams B, Dörfler P, Aguzzi A, Kozmik Z, Urbánek P, Maurer-Fogy I, Busslinger M. Pax-5 encodes the transcription factor BSAP and is expressed in B lymphocytes, the developing CNS, and adult testis. Genes Devel. 1992;6:1589–607. doi: 10.1101/gad.6.9.1589. [DOI] [PubMed] [Google Scholar]
- 69.Georgopoulos K, Moore DD, Derfler B. Ikaros, an early lymphoid-specific transcription factor and a putative mediator for T cell commitment. Science. 1992;258:808–12. doi: 10.1126/science.1439790. [DOI] [PubMed] [Google Scholar]
- 70.Hahm K, Ernst P, Lo K, Kim GS, Turck C, Smale ST. The lymphoid transcription factor LyF-1 is encoded by specific, alternatively spliced mRNAs derived from the Ikaros gene. Mol Cell Biol. 1994;14:7111–23. doi: 10.1128/mcb.14.11.7111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Georgopoulos K, Bigby M, Wang JH, Molnar A, Wu P, Winandy S, Sharpe A. The Ikaros gene is required for the development of all lymphoid lineages. Cell. 1994;79:143–56. doi: 10.1016/0092-8674(94)90407-3. [DOI] [PubMed] [Google Scholar]
- 72.Brown KE, Guest SS, Smale ST, Hahm K, Merkenschlager M, Fisher AG. Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell. 1997;91:845–54. doi: 10.1016/s0092-8674(00)80472-9. [DOI] [PubMed] [Google Scholar]