Abstract
Following injection challenge of rainbow trout with the Gram‐positive pathogen Renibacterium salmoninarum, serum nitrate levels increased indicative of NO production. The timing and amount of nitrate produced varied with the virulence of the bacterial strain used, with the highest levels seen in fish challenged with the most virulent (autoaggregating) strain. Immunization with a killed R. salmoninarum preparation in Freund’s incomplete adjuvant significantly increased nitrate levels after challenge. Inducible nitric oxide synthase (iNOS) transcript expression was detectable in rainbow trout tissues after injection challenge with R. salmoninarum, and its induction in the gills was both quick (between 3 and 6 hr) and relatively prolonged (lasting several days). iNOS expression in the kidney was also seen at a later stage (24 hr) but appeared to switch off relatively rapidly. Bath challenge with R. salmoninarum also induced iNOS expression in gill, and a variable expression in the gut and kidney also occurred. These results highlight the importance of the gills, not only as a point of entry of pathogens but also as a tissue capable of mounting an immune response.
Introduction
The production of nitrogen radicals is a well known antimicrobial mechanism in mammalian macrophages. In this pathway l‐arginine is used to produce nitric oxide (NO) and l‐citrulline by the action of nitric oxide synthase (NOS). NO is subsequently converted to nitrite and nitrate non‐enzymatically in the presence of an aerobic aqueous environment. 1 In mammals, three isoforms of NOS have been identified. Two of these are constitutively expressed, mainly in the brain (nNOS) and endothelial cells (eNOS). These isoforms are Ca2+/calmodulin dependent, allowing a very quick and low level response. The third isoform is inducible (iNOS), and is mainly present in macrophages but may also be found in hepatocytes, chondrocytes, retinal epithelial cells and osteoblasts. This isoform is Ca2+/calmodulin independent and requires the stimulus of an inducing agent to be expressed. In contrast to nNOS and eNOS, this cytokine‐induced isoform is slower to act (because gene expression must be ‘turned on’) but leads to a high concentration of NO. 2 Human iNOS is 1153 amino acids in length and is the shortest of the three isoforms (c. approx. 1430 amino acids for nNOS and 1200 amino acids for eNOS). All three isoforms share consensus binding sites for cofactors such as nicotinamide adenine dinucleotide phosphate (NADPH), flavine adenine dinucleotide (FAD), flavine mononucleotide (FMN) and calmodulin (CaM), 3,4 which make them very similar to cytochrome P‐450 reductase. iNOS is unique in lacking a stretch of 40 amino acids between the CaM and FMN binding sites present in the other two isoforms.
NOS activity has been described in many organisms ranging from single‐celled slime mould 5 to humans. 6 This suggests that the l‐arginine/NO pathway developed early in evolution and underlines the importance of NO production in the biology of these organisms. In fish, the presence of NOS has been shown in nervous tissues of species as diverse as lamprey (Lampetra planeri), lungfish (Neoceratodus forsteri and Protopterus dolloi), carp (Carassius carassius), channel catfish (Ictalurus punctatus), Atlantic salmon (Salmo salar), roach (Rutilus rutilus) and jeju (Hoplerythrinus unitaeniatus). 7–15 The activity of an inducible NOS has been shown in channel catfish (Ictalurus punctatus) leucocytes from fish challenged with Edwardsiella ictaluri16 and in a goldfish macrophage cell line after stimulation with LPS. 17–19 The iNOS gene has been partially sequenced in several species. 20,21In vivo, iNOS expression has been detected by reverse transcription–polymerase chain reaction (RT–PCR) in kidney macrophages and gills of rainbow trout challenged with an attenuated strain of the Gram‐negative pathogen Aeromonas salmonicida.20,21 In the present study, the ability of a Gram‐positive intracellular fish pathogen Renibacterium salmoninarum (the causative agent of bacterial kidney disease (BKD) in salmonid fish) to stimulate NO production in vivo and in vitro has been studied using a variety of autoaggregating and non‐autoaggregating strains. Among R. salmoninarum strains, autoaggregation is known to be correlated with virulence. 22 In mammals, NO production by macrophages can be up‐regulated by cytokines, e.g. interferon‐γ. 23,24 Antigen‐primed T lymphocytes in fish are known to produce a macrophage‐activating factor on exposure to specific antigen in vitro. 25 Experiments were therefore performed to investigate if NO production in response to injection of R. salmoninarum was enhanced by prior immunization of rainbow trout with an R. salmoninarum bacterin. In addition, RT–PCR was used to detect the expression of iNOS in different tissues of rainbow trout at different times after challenge with the BKD bacterium. The results show that fish infected with a virulent strain produced the highest levels of NO, that this response was enhanced in immunized fish and that iNOS expression could be detected in the gill and kidney. The possible role of NO in the clearance of this pathogen during the early stages of infection is discussed.
Materials and methods
Fish
Rainbow trout (Oncorhynchus mykiss), weighing 400–700 g, were obtained from local fish farms free of BKD and maintained in fibreglass tanks in a closed, recirculating freshwater system at 16° for at least 2 weeks prior to use. They were fed commercial trout pellets (EWOS Ltd, Livingston, UK) twice daily.
Bacteria
Renibacterium salmoninarum MT 426, MT 405 and MT 251 (non‐autoaggregating) and MT 1729 (autoaggregating) were obtained from the Marine Laboratory (Aberdeen, UK) culture collection and grown on Mueller–Hinton agar at 16° for 2–4 weeks. Strain MT 426 was formerly considered virulent 26 but had lost its virulence relative to MT 1729 in challenge experiments 27 presumably from routine subculturing. For the challenge, the bacteria were scraped off the agar and suspended in 0·15 m phosphate‐buffered saline pH 7·2 (PBS). For immunization purposes, strain MT 426 was heat‐killed by autoclaving at 115° for 20 min and was subsequently suspended in PBS at a concentration of 1 × 108 cells/ml or 1 × 109 cells/ml.
Immunization
Fish anaesthetized in 25 µg ethyl‐4‐aminobenzoate (Benzocaine, BDH, Poole, UK)/ml water, were injected intraperitoneally (i.p.) with 100 µl of the above killed bacterial suspensions (i.e. 107 or 108 cells per fish) as used in previous studies, 28,29 alone or mixed with an equal volume of Freund’s incomplete adjuvant (FIA). Additionally, groups of fish were injected with FIA or PBS. Four weeks after immunization, 10 fish per group were injection challenged as described below and serum nitrite/nitrate responses compared with unchallenged, control fish.
Challenge
For challenge by injection, rainbow trout were moved to tanks where the water was supplied from the recirculating freshwater system but where the outflow went to waste after hypochlorite treatment. After being anaesthetized as above, fish were injected i.p. with the different bacterial strains at 1 × 108 live bacteria/fish. Lower challenge doses are known to result in a high infection rate but no mortalities. 30 Immunized fish were challenged with strain MT 426 only. Control fish were injected with PBS at the time of challenge. At different times after the injection (3 hr to 5 days), fish were anaesthetized and blood extracted (0·5 ml) from the caudal vein. The blood was allowed to clot, centrifuged and serum collected. Serum samples were stored at –20° until the end of the experiment. The fish were also dissected and tissue samples collected.
For the bath challenge, fish were placed overnight in tanks containing 100 l of static, aerated water. The following day, live bacteria of strain M 426 were added to the water to give a final concentration of 6 × 105 cells/ml, a standard immersion challenge concentration used for many fish bacterial pathogens. 31 Fish were left in the bacterial suspension for 24 hr and then killed and tissues removed for RNA extraction, as described below.
Isolation of rainbow trout macrophages
Macrophages were isolated from rainbow trout head kidney as described by Secombes, 32 24 hr after the fish were challenged by injection. Briefly, under aseptic conditions, the head kidney was removed, pushed through a 100‐µm nylon mesh and suspended in Leibovitz L‐15 medium (L15, Gibco, Paisley, UK) supplemented with 2% fetal calf serum (FCS, Gibco), 100 µg penicillin/ml, 100 µg streptomycin/ml (P/S, Gibco) and 10 units heparin/ml (Sigma, Poole, UK). The suspensions were then loaded onto a 51/34% discontinuous Percoll density gradient and centrifuged at 400 g for 35 min. The band of cells at the interface of the Percoll gradient was collected, counted in a haemocytometer, adjusted to 1 × 107 cells/ml L15, 5% FCS, P/S, and plated in 96‐well plates (Nunc, Paisley, UK) at 100 µl/well. After 24 hr, non‐adherent cells were removed by washing the cultures twice with Hank’s balanced salt solution (HBSS, Gibco) before extraction of RNA.
Nitrite/nitrate measurement
Serum nitrate was assayed using the nitrate reductase method. 33 Serum diluted 1 : 20 in PBS was plated into 96‐well plates (Greiner, Stonehouse, UK), at 100 µl per well. Three µl nitrate reductase mix (containing 1·5 µg nitrate reductase/ml, 3·3 mg NADPH/ml and 0·13 mg flavine adenine dinucleotide/ml distilled water) were added to each well and incubated at 27° for 1 hr. Because the presence of NADPH interferes with the nitrite determination, it was subsequently oxidized by adding 1·5 µl of 25 mg l‐lactic dehydrogenase (type I)/ml and 10 µl 100 mm sodium pyruvate and incubated at 37° for 1 hr. At the end of these reactions, nitrate is reduced to nitrite, which was measured using the Griess reagent (1% sulphanilamide/0·1% naphthylethylene diamine dihydrochloride/2·5% H3PO4). After addition of 100 µl Griess reagent per well the absorbance was read in a microplate reader at 550 nm, using serial dilutions of sodium nitrate treated as above as standards. The data were analysed by one and two way anova as appropriate.
RNA extraction
Total RNA was isolated from a variety of freshly excised tissues (blood, gill, gut, kidney, liver and spleen) or macrophages using RNAzol B (RNA isolation solvent, Biogenesis). Samples of tissues weighing 200–300 mg were sonicated in RNAzol B for approximately 5 min in an ice bath and mixed with 1/10th volume of chloroform. The suspension was then centrifuged at 15 000 g for 15 min, to allow separation into two phases. The clear upper phase containing the RNA was aspirated and placed in a fresh tube. An equal volume of isopropanol was then added followed by centrifugation at 15 000 g for 15 min. The pellet was washed in 75% ethanol, dried, redissolved in diethylpyrocarbonate‐treated water and stored at –70°.
Reverse transcription
Reverse transcription to cDNA was carried out as described by Laing et al. 20 Briefly, 10 µg RNA were incubated with 2 µl oligo (DT) 12–18 (500 µg/ml) for 10 min. Then, 8 µl 5× first strand buffer; 4 µl 0·1 m dithiothreitol (DTT); 2 µl 10 mm dinucleoside triphosphate (dNTP) mix (10 mm each diadenosine triphosphate (dATP), diguanosine triphosphate (dGTP), dicytosine triphosphate (dCTP) and dithymidine triphosphate (dTTP) at neutral pH) (all from Gibco) were added, mixed and incubated for 2 min at 42°. After this incubation period, 2 µl Superscript II reverse transcriptase (Gibco) was added and the mixture incubated at 42° for 50 min. The reaction was terminated by heating to 70° for 15 min. The resulting cDNA was stored at 4°.
PCR
To detect iNOS message in rainbow trout by RT–PCR it is necessary to perform a nested PCR. 21 For the first round of PCR, the primers used to amplify the cDNA were NOS M (forward) (5′‐GTTGGTACATGGGCACTGA‐3′) and NOS N (reverse) (5′‐CACCTACTTCCTGGACATTAC‐3′). NOS P (forward) (5′‐CTTCAACTCCAGGTTGCTCTG‐3′) and NOS Q (reverse) (5′‐GTTCTGACACCTTCCGAGTGA‐3′) were used for the second round of PCR. All primers were used at 10 pm/ml and were purchased from Genosys (Pampisford, UK). Amplification was performed in 25 µl reactions containing 2 µl dNTP, 0·25 µl Taq DNA polymerase (5 µ/µl; Bioline, London, UK), 1 µl Taq buffer (50 mm; Bioline) containing MgCl2, 2·5 µl 10× reaction buffer (Bioline) and 1 µl cDNA (or reaction mix from the first round of PCR). The cycling protocol was as given in Table 1.
Table 1.
Cycling protocol
| Temperature (°C) | Time (min) | No. of cycles |
|---|---|---|
| 94 | 5 | 1 |
| 94 | 1 | 35 |
| 60 | 1 | 35 |
| 72 | 1 | 35 |
| 72 | 10 | 1 |
Primers for β‐actin were used as a positive control for PCR, since the gene encoding β‐actin is highly conserved and expressed constitutively in all tissues. The primers were: actin forward (5′‐ATCGTGGGCGCCCCAGGCACC‐3′) and reverse (5′‐CTCCTTAATGTCACGCACGATTTC‐3′). Reactions were prepared as described above but only one round of PCR was required. PCR products were visualized on a 1% agarose gel stained with ethidium bromide, using an ultraviolet gel imaging system (UVP). A 100‐bp ladder was used as a size marker. On one occasion the products were also cloned using a TOPO cloning kit (Invitrogen, Leek, the Netherlands), plasmid DNA extracted using a plasmid miniprep kit (Qiagen, Crawley, UK) and sequenced using an ABI 377 Automated Sequencer (Applied Biosystems, Warrington, UK) to confirm the correct gene was being amplified.
Results
Nitrite/nitrate levels in serum of rainbow trout after injection challenge with R. salmoninarum
Non‐autoaggregating R. salmoninarum strains elicited a significantly (P < 0·05) higher response compared to controls at day 4, the first sampling time after challenge (Fig. 1). Fish injected with strains MT 405 and MT 251 also showed significantly (P < 0·05) increased nitrate levels at day 12. The autoaggregating strain did not elicit a significant increase in nitrate levels above the control until day 8 (P < 0·001). However, after this time the nitrate levels continued to increase until day 21, the last sampling time before most of the fish in this group died. The nitrate levels in the other groups were the same as the control values from day 16 to the end of the experiment at day 34. In a further experiment designed to define more precisely the early period when nitrate levels were increased postchallenge, fish were sampled every second day. It was found that following injection with strain MT 426, significant increases (P < 0·05) were apparent by day 2 and remained elevated at day 4 and day 6.
Figure 1.
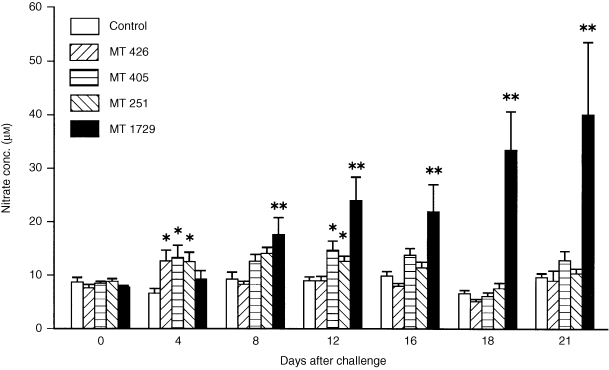
Effect of R. salmoninarum challenge on serum nitrate levels in rainbow trout. Fish were injected intraperitoneally with PBS (control) or with one of four strains of R. salmoninarum at 1 × 108 bacteria/fish. Bars represent the mean + SE of 10 fish. *P < 0·05; **P < 0·001.
Nitrite/nitrate levels in serum of immunized rainbow trout following challenge with R. salmoninarum
Fish injected with killed bacteria at two different doses 1 month prior to challenge gave similar results to each other (Fig. 2). At day 4 post‐challenge, the typical increase in nitrate levels relative to unchallenged fish was observed (although not significantly in the fish previously immunized with killed bacteria in PBS). However, fish immunized with killed bacteria in FIA had significantly increased (P < 0·05) nitrate levels relative to the other challenged groups, and this was particularly apparent in fish immunized with the higher bacterial dose (Fig. 2b). At later times (up to day 32) immunized fish showed no significant increases compared with controls (data not shown).
Figure 2.
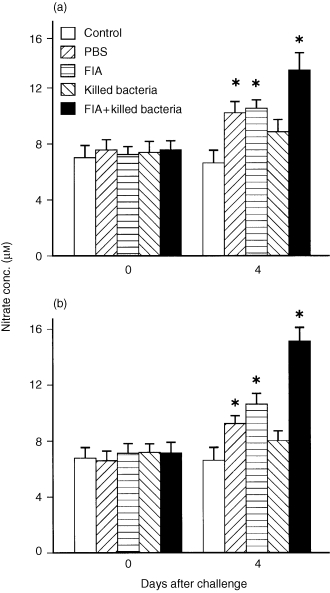
Effect of R. salmoninarum challenge (1 × 108 bacteria/fish of strain MT 426) on serum nitrate levels in rainbow trout immunized 4 weeks earlier with PBS or killed R. salmoninarum preparations at (a) 1 × 107 bacteria/fish or (b) 1 × 108 bacteria/fish, with or without Freund’s incomplete adjuvant (FIA). All groups were injected i.p. with the live bacteria except for the control fish which were injected with PBS. Bars represent the mean+SE of 10 fish. *P < 0·05.
iNOS expression in different tissues of rainbow trout after injection challenge with R. salmoninarum
A range of tissues were sampled 24 hr after injection with R. salmoninarum to examine the sites of iNOS expression, and included blood, gills, gut, kidney, liver and spleen. Only gill and kidney cDNA gave a correct sized (749 bp) product with the iNOS primers (Fig. 3), demonstrating the expression of the iNOS gene. The products were sequenced and confirmed to be trout iNOS. The β‐actin positive controls gave correct sized products (541 bp) in all the tissues used, confirming the presence of an intact cDNA template. Subsequent experiments showed 24 hr was optimal for maximal expression (see below) and so other time points were not studied for all tissues. Control fish that were anaesthetized and injected with saline did not show iNOS expression in gills or kidney 24 hr later.
Figure 3.
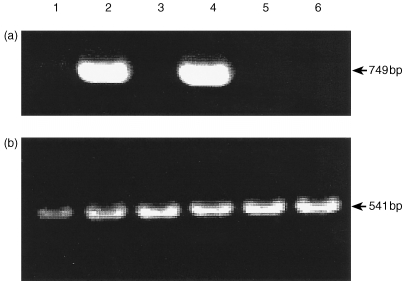
Detection of iNOS (a) and β‐actin (b) expression in various tissues of rainbow trout 24 hr after i.p. challenge with R. salmoninarum (strain MT 426). Lanes: 1, blood; 2, gill; 3, gut; 4, head kidney; 5, liver; 6, spleen. Data are for one fish and are representative of the three fish analysed.
Kinetics of iNOS expression in gills and kidney after injection challenge with R. salmoninarum
Three hours after injection challenge, no iNOS expression was detected in either the gills or kidney (Fig. 4a). At 6 hr and 12 hr, a clear iNOS band was present in gill tissue samples of all three fish. In addition, one fish showed a faint band in kidney samples (Fig. 4b, c). One day after challenge, both kidney and gill were consistently positive for iNOS expression (Fig. 5a). However, after this time iNOS appeared to be switched off in kidney tissue such that at 3 days post‐challenge only one fish showed a faint band (Fig. 5b) and at 5 days post‐injection no iNOS expression was detectable in the kidney (Fig. 5c). At both time points iNOS was positive in the gills and β‐actin controls were positive in both tissues.
Figure 4.
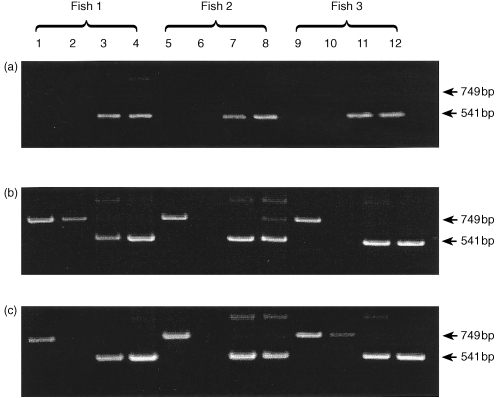
Time course of iNOS mRNA expression in gill and head kidney of three rainbow trout after i.p. challenge with R. salmoninarum (strain MT 426). Lanes 1, 5 and 9, iNOS transcript in gill; lanes 2, 6 and 10, iNOS transcript in head kidney; lanes 3, 7 and 11, β‐actin transcript in gill; lanes 4, 8 and 12, β‐actin transcript in head kidney. (a) 3 hr after i.p. challenge; (b) 6 hr after i.p. challenge; (c) 12 hr after i.p. challenge.
Figure 5.
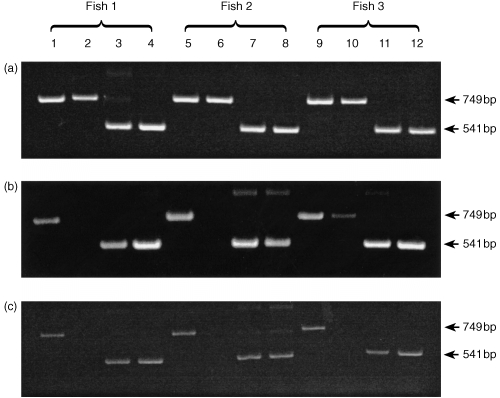
Time course of iNOS mRNA expression in gill and head kidney of three rainbow trout after i.p. challenge with R. salmoninarum (strain MT 426). Lanes 1, 5 and 9, iNOS transcript in gill; lanes 2, 6 and 10, iNOS transcript in head kidney; lanes 3, 7 and 11, β‐actin transcript in gill; lanes 4, 8 and 12, β‐actin transcript in head kidney. (a) One day after i.p. challenge; (b) 3 days after i.p. challenge; (c) 5 days after i.p. challenge.
iNOS expression in head kidney macrophages after injection challenge with R. salmoninarum
Head kidney macrophages were isolated from fish 24 hr after injection challenge with R. salmoninarum and were cultured overnight before RNA extraction to ensure good purity of the cells. The samples were negative for iNOS in RT–PCR although the β‐actin bands were clearly seen (data not shown), demonstrating that the cDNA template was intact and PCR conditions were adequate.
iNOS expression in tissues after immersion challenge with R. salmoninarum
Kidney, gill and gut were investigated for iNOS expression 24 hr after bath challenge. A total of six fish were analysed in this experiment and variation between individuals was apparent. In all fish examined iNOS expression was detectable in the gill (Fig. 6). In four of six fish examined iNOS expression was detectable in the kidney after immersion challenge and in half of the fish the gut samples were also positive. Products corresponding to β‐actin (positive controls) were visualized for all the tissue samples.
Figure 6.
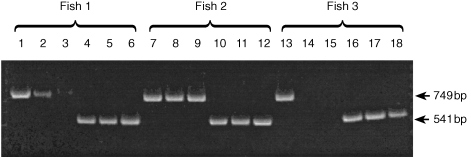
iNOS expression in tissues of three rainbow trout (out of six analysed) 24 hr after immersion challenge with R. salmoninarum (strain MT 426). Lanes 1, 7 and 13, iNOS transcript in gill; lanes 2, 8 and 14, iNOS transcript in gut; lanes 3, 9 and 15, iNOS transcript in head kidney. Lanes 4, 10 and 16, β‐actin transcript in gill; lanes 5, 11 and 17, β‐actin transcript in gut; lanes 6, 12 and 18, β‐actin transcript in head kidney.
Discussion
In mammals, cells from different tissues are known to produce NO in response to stimulation by pathogens or their products, and this is suggested to be an important antimicrobial mechanism. 2 In fish, recent sequence evidence has demonstrated the presence of iNOS at this level of phylogeny. The partial rainbow trout and goldfish iNOS sequences 20,21 show about 70% homology with mammalian iNOS and 85% homology to each other. The regions corresponding to several iNOS cofactor‐binding domains were sequenced and shown to be highly conserved. This sequence information has allowed the present study to be performed, where RT–PCR has been used to examine the kinetics and sites of iNOS expression in rainbow trout following challenge with the Gram‐positive salmonid pathogen R. salmoninarum. Because iNOS expression is controlled primarily at the transcriptional rather than post‐transcriptional level, 34 RT–PCR should be a reliable method to detect whether NO production has been switched on by targeting the expression of mRNA for the enzyme that regulates its release.
After i.p. injection of R. salmoninarum, iNOS expression was detected only in the gill and head kidney. These trout tissues were previously shown to express iNOS 48 hr after injection challenge with an attenuated (ΔaroA) strain of the Gram‐negative bacterium Aeromonas salmonicida but not in unchallenged fish. 21 Interestingly, in the present study blood leucocytes did not show iNOS expression, eliminating the possibility that the signal originated from contaminating blood cells and indicating that the gill contains cell types important in mounting this immune reaction. Nevertheless, injected R. salmoninarum may be taken up by circulating phagocytes that subsequently concentrate in lymphoid tissues such as the gill and kidney. Indeed, tissues such as kidney and spleen are known to have a role in the trapping and processing of blood‐borne compounds in teleost fish, 35 and macrophages associated with venous sinusoids in the kidney are known to trap bacteria introduced into the blood. 36 Thus, it is surprising that the spleen was negative for iNOS expression in the present study. However, in rainbow trout injected with R. salmoninarum the kidney showed signs of infection before the spleen, 37 so the possibility of iNOS expression in spleen at a later time cannot be ruled out.
The present results show that there is a relatively quick induction of iNOS expression in gills, between 3 and 6 hr post‐injection with R. salmoninarum. Sasaki et al. 38 detected expression of iNOS mRNA in spleen and kidney of mice 3 hr after intravenous infection with Staphylococcus aureus. Lamarque et al. 39 showed iNOS activity in the duodenum and duodenal cells of rats 4 hr after administration of an extract of Helicobacter pylori. Why iNOS expression in gills occurs faster than in kidney is not clear. It may be that bacteria‐containing phagocytes or extracellular bacteria in the blood travel faster to a well vascularized tissue such as the gills. Another possibility is that, because of their exposed location, the gills are better equipped to deal with invading pathogens and possess more responsive cells, which is supported by the fact that the response in gills lasted longer than in kidney. Moore et al. 40 have shown that gill tissues are able to take up latex microspheres from the environment and retain them for as long as 24 days after exposure, primarily within epithelial cells and underlying phagocytes, demonstrating their important role in local immunity. On the other hand, whilst expression of iNOS in the kidney is slower to appear than in the gill, it is possible that once induced the infection is controlled more quickly allowing rapid down‐regulation of iNOS transcription. These experiments were performed with an avirulent strain of R. salmoninarum which may not have survived in vivo for many days.
Bath challenge was also effective at inducing iNOS mRNA expression in the tissues sampled. R. salmoninarum is known to infect salmonids via contaminated food, 41 cohabitation with infected fish 42 and via the fecal–oral route, 43 so this route may be more representative of a natural challenge. The way the bacterium crosses the epithelial layer is unknown but the bacterium seems capable of promoting its intake by non‐professional phagocytic cells 44,45 and infections can be established by bath challenge. 42 The present results show strong and consistent iNOS expression in the gill 24 hr following immersion exposure. The individual differences seen in expression in gut and kidney after bath challenge may reflect individual susceptibility to invasion by R. salmoninarum because of genetic factors or past history.
An ultrastructural study of kidney of rainbow trout challenged with R. salmoninarum has shown that the bacterium is taken up by a range of cells (sinusoidal cells, macrophages, reticular and barrier cells) that may act as reservoirs of the infection. 37 On the other hand, an in vitro study of gill tissue 46 showed that infection with R. salmoninarum produced an increased number of monocytes/macrophages and eosinophilic granular cells. This suggests that cell differentiation occurs in gill, probably mediated by cytokines, and underlines the importance of the gill in the process of mounting immune reactions. Invading pathogens may be taken to these tissues, physically contained there by leucocytes and leucocyte‐like cells as they mount a response, involving the release of reactive oxygen and nitrogen species and maybe antibodies. 47,48 Physical containment has been suggested as a strategy to kill invading pathogens while reducing tissue damage to a minimum. 49
Isolated head kidney macrophages from challenged trout did not express iNOS in contrast to results with a goldfish macrophage cell line which can be stimulated to express iNOS and produce NO. 19,20 More recently, Wiegertjes et al50 reported NO production in vitro by carp macrophages stimulated with LPS and with the parasite Trypanoplasma borreli. As the trout cells were cultured for 24 hr before use, in order to obtain high purity cultures, it is possible that essential factors (cytokines?) required for iNOS expression were lacking and resulted in a rapid down‐regulation of expression. Indeed, kidney total leucocyte suspensions cultured in parallel during this period also lost detectable iNOS expression by 24 hr (unpublished observation).
In addition to iNOS expression, an increase in serum nitrate levels relative to control samples was detected in rainbow trout after injection with R. salmoninarum and supports the contention that a functional iNOS enzyme was induced. Control serum levels were high relative to mammals, presumably reflecting differences in nitrogen metabolism, where fish produce ammonia as the main end‐product which is excreted across the gills. In chinook salmon naturally infected (clinically evident) with R. salmoninarum an increase in iNOS activity (citrulline formation) in the head kidney has also been reported, compared to clinically normal fish. 16 In the present work, the fact that the avirulent strains elicited increased nitrate levels relatively early post‐challenge and that no mortalities were seen, suggest that this small but significant production may be sufficient to control the disease at this stage. This early production could be increased by prior immunization of fish with killed bacteria in FIA, suggesting that immune responses can up‐regulate pathways involved in NO production as seen in mammals with cytokines released from antigen stimulated T cells. 51 NO release in vivo has been shown to correlate well with clearance of pathogens in mammals, as seen with Staphylococcus aureus, 38Leishmania amazonensis, 52Listeria monocytogenes 53 and Trypanosoma cruzi. 54 In contrast, the virulent strain did not elevate nitrate levels until 8 days after challenge. One explanation for this could be the presence in the bacterial capsule of an inhibiting factor, as with Leishmania which possesses phospholipids that prevent NO synthesis. 55 Indeed, a soluble extracellular protein produced by R. salmoninarum is known to inhibit the respiratory burst of trout phagocytes. 56 Another explanation may be the rapid escape of virulent bacteria from phagosomes into the cytoplasm, and their release a few days later, when they may spread throughout the host tissues. 57 The slow growth of strain MT 1729 could also be a factor, particularly if NO release is stimulated by a product of live, metabolizing bacteria. Despite the highest serum nitrate levels being induced by MT 1729, most fish died, suggesting that this virulent strain is resistant to NO or ultimately, that NO contributes towards the success of the pathogen. Precedents for this exist in the mammalian literature, as reported by Doi et al. 58 who showed that the intracellular pathogen Mycobacterium intracellulare induced the release of NO by macrophages in vitro but is resistant to its effects. Moreover, the NO so produced inhibited phagocytosis, favouring the extracellular spread of the bacteria.
In conclusion, these results demonstrate that nitrate levels increase in rainbow trout during infection with the Gram‐positive pathogen R. salmoninarum, indicative of NO production, although the timing and amount of nitrate produced varies with the virulence of the strain used. iNOS expression is detectable in rainbow trout tissues after injection and bath challenge with R. salmoninarum, and its induction in the gills is both quick and relatively prolonged. These results highlight the importance of the gills, not only as a point of entry of pathogens but also as a tissue capable of mounting an immune response.
Acknowledgments
This work was supported by a studentship to JJCP from the National Council for Science and Technology (CONACyT) Mexico and a grant from The Leverhulme Trust (F/152/Q). Thanks go to Mr I. Stewart for technical assistance.
References
- 1.Marletta MA, Yoon PS, Iyengar R, Leaf CD, Wishnok JS. Macrophage oxidation of l‐arginine to nitrite and nitrate: nitric oxide is an intermediate. Biochemistry. 1988;27:8706. doi: 10.1021/bi00424a003. [DOI] [PubMed] [Google Scholar]
- 2.Clark IA, Rockett KA. Nitric oxide and parasitic disease. Adv Parasitol. 1996;37:1. doi: 10.1016/s0065-308x(08)60218-3. [DOI] [PubMed] [Google Scholar]
- 3.Bredt DS, Hwang PM, Glatt CE, Lowenstein C, Reed RR, Snyder SH. Cloned and expressed nitric oxide synthase structurally resembles cytochrome p‐450 reductase. Nature. 1991;351:714. doi: 10.1038/351714a0. [DOI] [PubMed] [Google Scholar]
- 4.Bredt DS, Ferris CD, Snyder SH. Nitric oxide synthase regulatory sites: phosphorylation by cyclic AMP‐dependent protein kinase C, and calcium/calmodulin protein kinase; identification of flavin and calmodulin binding sites. J Biol Chem. 1992;267:10976. [PubMed] [Google Scholar]
- 5.Werner‐felmeyer G, Golderer G, Werner ER, Grobner P, Wachter H. Pteridine biosynthesis and nitric oxide synthase in Physarium polycephalum. Biochem J. 1994;304:105. doi: 10.1042/bj3040105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nozaki Y, Hasegawa Y, Ichiyama S, Nakashima I, Shimokata K. Mechanism of nitric oxide‐dependent killing of Mycobacterium bovis BCG in human alveolar macrophages. Infect Immun. 1997;65:3644. doi: 10.1128/iai.65.9.3644-3647.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Greenstreet EH, Djamgoz MBA. Nitric oxide induces light‐adaptive morphological changes in retinal neurons. Neuroreport. 1994;6:109. doi: 10.1097/00001756-199412300-00029. [DOI] [PubMed] [Google Scholar]
- 8.Huque T, Brand JG. Nitric oxide synthase activity of the taste organ of the channel catfish, Ictalurus punctatus. Comp Biochem Physiol. 1994;108B:481. doi: 10.1016/0305-0491(92)90088-9. [DOI] [PubMed] [Google Scholar]
- 9.Hylland P, Nilsson GE. Evidence that acetylcholine mediates increased cerebral blood flow velocity in crucian carp through a nitric oxide‐dependent mechanism. J Cerebral Blood Flow Metab. 1995;15:519. doi: 10.1038/jcbfm.1995.64. [DOI] [PubMed] [Google Scholar]
- 10.Li ZS, Furness JB. Nitric oxide synthase in the enteric nervous system of the rainbow trout, Salmo gairdneri. Arch Histol Cytol. 1993;56:185. doi: 10.1679/aohc.56.185. [DOI] [PubMed] [Google Scholar]
- 11.Ostholm T, Holmqvist BI, Alm P, Ekstrom P. Nitric oxide in the CNS of the Atlantic salmon. Neurosci Lett. 1994;168:233. doi: 10.1016/0304-3940(94)90458-8. [DOI] [PubMed] [Google Scholar]
- 12.Schober A, Malz CR, Schober W, Meyer DL. NADPH‐diaphorase in the central nervous system of the larval lamprey (Lampetra planeri) J Comp Neurol. 1994;345:94. doi: 10.1002/cne.903450107. [DOI] [PubMed] [Google Scholar]
- 13.Schober A, Meyer DL, Von Bartheld CS. Central projections of the nervus terminalis and the nervus preaeopticus in the lungfish brain revealed by nitric oxide synthase. J Comp Neurol. 1994;349:1. doi: 10.1002/cne.903490102. [DOI] [PubMed] [Google Scholar]
- 14.Soderstrom V, Hylland P, Nilsson GE. Nitric oxide synthase inhibitor blocks acetylcoline induced increase in brain flow in rainbow trout. Neurosci Lett. 1995;197:191. doi: 10.1016/0304-3940(95)11927-o. [DOI] [PubMed] [Google Scholar]
- 15.Staples JF, Zapol WM, Bloch KD, Kawai N, Val VMF, Hochachka PW. Nitric oxide responses of air‐breathing and water‐breathing fish. Am J Physiol. 1995;268:R816. doi: 10.1152/ajpregu.1995.268.3.R816. [DOI] [PubMed] [Google Scholar]
- 16.Schoor WP, Plumb JA. Induction of nitric oxide synthase in channel catfish Ictalurus punctatus by Edwardsiella ictaluri. Dis Aquat Org. 1994;19:153. [Google Scholar]
- 17.Neumann NF, Fagan D, Belosevic M. Macrophage activating factor (s) secreted by mitogen stimulated goldfish kidney leukocytes synergize with bacterial lipopolysaccharide to induce nitric oxide production in teleost macrophages. Dev Comp Immunol. 1995;19:473. doi: 10.1016/0145-305x(95)00032-o. [DOI] [PubMed] [Google Scholar]
- 18.Neumann NF, Belosevic M. Deactivation of primed respiratory burst response of goldfish macrophages by leukocyte‐derived macrophage activating factor (s) Dev Comp Immunol. 1996;20:427. doi: 10.1016/s0145-305x(96)00029-8. [DOI] [PubMed] [Google Scholar]
- 19.Wang R, Neumann NF, Shen Q, Belosevic M. Establishment and characterisation of a macrophage cell line from the goldfish. Fish Shellfish Immunol. 1995;5:329. [Google Scholar]
- 20.Laing KJ, Grabowski PS, Belosevic M, Secombes CJ. A partial sequence for nitric oxide synthase from a goldfish (Carassius auratus) macrophage cell line. Immunol Cell Biol. 1996;74:374. doi: 10.1038/icb.1996.65. [DOI] [PubMed] [Google Scholar]
- 21.Laing KJ, Hardie LJ, Aartsen W, Grabowski PS, Secombes CJ. Expression of an inducible nitric oxide synthase gene in rainbow trout Oncorhynchus mykiss. Dev Comp Immunol. 1999;23:71. doi: 10.1016/s0145-305x(98)00036-6. [DOI] [PubMed] [Google Scholar]
- 22.Bruno DW. Presence of a saline extractable protein associated with virulent strains of the fish pathogen Renibacterium salmoninarum. Bull Eur Ass Fish Pathol. 1990;10:8. [Google Scholar]
- 23.Lorsbach RB, Russell SW. A specific sequence of stimulation is required to induce synthesis of the antimicrobial molecule nitric oxide by mouse macrophages. Infect Immun. 1992;60:2133. doi: 10.1128/iai.60.5.2133-2135.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park J, Rikihisa Y. l‐Arginine dependent killing of intracellular Ehrlichia risticii by macrophages treated with gamma interferon. Infect Immun. 1992;60:3504. doi: 10.1128/iai.60.9.3504-3508.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marsden MJ, Cox D, Secombes CJ. Antigen‐induced release of macrophage activating factor from rainbow trout Oncorhynchus mykiss leucocytes. Vet Immunol Immunopathol. 1994;42:199. doi: 10.1016/0165-2427(94)90008-6. [DOI] [PubMed] [Google Scholar]
- 26.Hardie LJ, Ellis AE, Secombes CJ. In vitro activation of rainbow trout macrophages stimulates killing of Renibacterium salmoninarum concomitant with augmented generation of respiratory burst products. Dis Aquat Org. 1996;25:175. [Google Scholar]
- 27.Campos‐perez JJ. Aberdeen, UK: University of Aberdeen; The role of reactive oxygen and nitrogen species in the immune response of rainbow trout to Renibacterium salmoninarum. PhD Thesis. [Google Scholar]
- 28.Munro ALS, Bruno DW. Vaccination against bacterial kidney disease. In: Ellis AE, editor. Fish Vaccination. London: Academic Press.; 1988. p. p.124. [Google Scholar]
- 29.Evenden AJ, Grayson TH, Gilpin ML, Munn CB. Renibacterium salmoninarum and bacterial kidney disease – the unfinished jigsaw. Ann Rev Fish Dis. 1993;3:87. [Google Scholar]
- 30.McCarthy DH, Croy TR, Amend DF. Immunization of rainbow trout, Salmo gairdneri Richardson, against bacterial kidney disease: Preliminary efficacy evaluation. J Fish Dis. 1984;7:65. [Google Scholar]
- 31.Thompson I, Fletcher TC, Houlihan DF, Secombes CJ. The effect of dietary vitamin A on the immunocompetence of Atlantic salmon (Salmo salar L.) Fish Physiol Biochem. 1994;12:513. doi: 10.1007/BF00004453. [DOI] [PubMed] [Google Scholar]
- 32.Secombes CJ. Isolation of salmonid macrophages and analysis of their killing activity. In: Stolen JS, Fletcher TC, Anderson DP, Robertson BS, Van Muiswinkel WB, editors. Techniques in Fish Immunology. I. New Jersey: SOS Publications, Fair Haven,; 1990. p. 137. [Google Scholar]
- 33.Schmidt HHHW, Seifert R, Bohme EJN. Formation and release of nitric‐oxide from human‐neutrophils and HL‐60 cells induced by a chemotactic peptide, platelet activating factor and leukotriene‐B4. FEBS Lett. 1989;244:357. doi: 10.1016/0014-5793(89)80562-9. [DOI] [PubMed] [Google Scholar]
- 34.Morris SM, Billiar TR. New insights into the regulation of inducible nitric oxide synthase. Am J Physiol. 1994;266:E829. doi: 10.1152/ajpendo.1994.266.6.E829. [DOI] [PubMed] [Google Scholar]
- 35.Zapata AG, Chiba A, Varas A. Cells and tissues of the immune system of fish. In: Iwama G, Nakanishi T, editors. The Fish Immune System: Organism, Pathogen and Environment. London: Academic Press; 1996. p. p.1. [Google Scholar]
- 36.Ferguson HW. Renal portal phagocytosis of bacteria in rainbow trout (Salmo gairdneri Richardson): ultrastructural observations. Can J Zool. 1984;62:2505. [Google Scholar]
- 37.Flaño E, Lopez‐fierro P, Razquin B, Kaattari SL, Villena A. Histopathology of the renal and splenic haemopoietic tissues of coho salmon Oncorhynchus kisutch experimentally infected with Renibacterium salmoninarum. Dis Aquat Org. 1996;24:107. [Google Scholar]
- 38.Sasaki S, Miura T, Nishkawa S, Yamada K, Hirasue M, Nakane A. Protective role of nitric oxide in Staphylococcus aureus infection in mice. Infect Immun. 1998;66:1017. doi: 10.1128/iai.66.3.1017-1022.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lamarque D, Kiss J, Tankovic J, Flejou JF, Delchier JC, Whittle BJ. Induction of nitric oxide synthase in vivo and cell injury in rat duodenal epithelium by a water soluble extract of Helicobacter pylori. Brit J Pharmacol. 1998;123:1073. doi: 10.1038/sj.bjp.0701706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moore JD, Ototake M, Nakanishi T. Particulate antigen uptake during immersion immunisation of fish: the effectiveness of prolonged exposure and the roles of skin and gill. Fish Shellfish Immunol. 1998;8:393. [Google Scholar]
- 41.Wood JW, Wallis J. Research Briefs 6, Portland, Oregon: Fisheries Commission; 1955. Kidney disease in adult chinook salmon and its transmission by feeding to young chinook salmon; p. 32. [Google Scholar]
- 42.Murray CB, Evelyn TPT, Beacham TD, Barner LW, Ketcheson JE, Prosperi‐porta L. Experimental induction of bacterial kidney disease in chinook salmon by immersion and cohabitation challenges. Dis Aquat Org. 1992;12:91. [Google Scholar]
- 43.Balfry SK, Albright LJ, Evelyn TPT. Horizontal transfer of Renibacterium salmoninarum among farmed salmonids via the fecal‐oral route. Dis Aquat Org. 1996;25:63. [Google Scholar]
- 44.Evelyn TPT. Infection and disease. In: Iwama G, Nakanishi T, editors. The Fish Immune Sytem: Organism, Pathogen and Environment. London: Academic Press; 1996. p. p.339. [Google Scholar]
- 45.McIntosh D, Meaden PG, Austin B. A simplified PCR‐based method for the detection of Renibacterium salmoninarum utilizing preparations of rainbow trout (Oncorhynchus mykiss) lymphocytes. App Environ Microbiol. 1997;62:3929. doi: 10.1128/aem.62.11.3929-3932.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Flaño E, Lopez‐fierro P, Razquin BE, Villena AJ. In vitro differentiation of eosinophilic granular cells in Renibacterium salmoninarum‐infected gill cultures from rainbow trout. Fish Shellfish Immunol. 1996;6:173. [Google Scholar]
- 47.Davidson GA, Lin SH, Secombes CJ, Ellis AE. Detection of specific and ‘constitutive’ antibody secreting cells in the gills, head kidney and peripheral blood leucocytes of dab (Limanda limanda) Vet Immunol Immunopathol. 1997;58:363. doi: 10.1016/s0165-2427(97)00017-2. [DOI] [PubMed] [Google Scholar]
- 48.Lin S‐H, Ellis AE, Davidson GA, Secombes CJ. Migratory, respiratory burst and mitogenic responses of leucocytes isolated from the gills of rainbow trout (Oncorhynchus mykiss) Fish Shellfish Immunol. 1999;9:211. [Google Scholar]
- 49.Akaike T, Suga M, Maeda H. Free radicals in viral pathogenesis: molecular mechanisms involving superoxide and No. Proc Soc Exp Biol Med. 1998;217:64. doi: 10.3181/00379727-217-44206. [DOI] [PubMed] [Google Scholar]
- 50.Wiegertjes GF, Saeij JPJ, Mossnik MH, Regeer RR, Stet RJM. The 4th Nordic Symposium on Fish Immunology Abstracts. Denmark: Hirtshals; 1998. Immunogenetic resistance to the parasitic protozoan Trypanoplasma borreli in carp (Cyprinus carpio L.) [Google Scholar]
- 51.Migliorini P, Corradin G, Corradin SB. Macrophage production as a sensitive and rapid assay for the quantitation of murine IFN‐γ. J Immunol Methods. 1991;139:107. doi: 10.1016/0022-1759(91)90357-l. [DOI] [PubMed] [Google Scholar]
- 52.Giorgio S, Linares E, Ischiropoulos H, Von Zuben FJ, Yamada A, Augusto O. In vivo formation of electron paramagnetic resonance‐detectable nitric oxide and nitrotyrosine is not impaired during murine leishmaniasis. Infect Immun. 1998;66:807. doi: 10.1128/iai.66.2.807-814.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Boockvar KS, Granger DL, Poston RM, et al. Nitric oxide produced during murine listeriosis is protective. Infect Immun. 1994;62:1089. doi: 10.1128/iai.62.3.1089-1100.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vespa GNR, Cunha FQ, Silva JS. Nitric oxide is involved in control of Trypanosoma cruzi‐induced parasitemia and directly kills the parasite in vitro. Infect Immun. 1994;62:5177. doi: 10.1128/iai.62.11.5177-5182.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Proudfoot L, O'donnell CA, Liew FY. Glycoinositolphospholipids of Leishmania major inhibit nitric oxide synthesis and reduce leishmanicidal activity in murine macrophages. Eur J Immunol. 1995;25:745. doi: 10.1002/eji.1830250318. [DOI] [PubMed] [Google Scholar]
- 56.Densmore CL, Smith SA, Holladay SD. In vitro effects of the extracellular protein of Renibacterium salmoninarum on phagocyte function in brook trout. Vet Immunol Immunopathol. 1998;62:349. doi: 10.1016/s0165-2427(98)00101-9. [DOI] [PubMed] [Google Scholar]
- 57.Gutemberger SK, Duimstra JR, Rohovec JS, Fryer JL. Intracellular survival of Renibacterium salmoninarum in trout mononuclear phagocytes. Dis Aquat Org. 1997;28:93. [Google Scholar]
- 58.Doi T, Ando M, Akaike T, Suga M, Sato K, Maeda H. Resistance to nitric oxide in Mycobacterium avium complex and its implication in pathogenesis. Infect Immun. 1993;61:1980. doi: 10.1128/iai.61.5.1980-1989.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]


