Abstract
The type III effector protein encoded by avirulence gene B (AvrB) is delivered into plant cells by pathogenic strains of Pseudomonas syringae. There, it localizes to the plasma membrane and triggers immunity mediated by the Arabidopsis coiled-coil (CC)-nucleotide binding (NB)-leucine-rich repeat (LRR) disease resistance protein RPM1. The sequence unrelated type III effector avirulence protein encoded by avirulence gene Rpm1 (AvrRpm1) also activates RPM1. AvrB contributes to virulence after delivery from P. syringae in leaves of susceptible soybean plants, and AvrRpm1 does the same in Arabidopsis rpm1 plants. Conditional overexpression of AvrB in rpm1 plants results in leaf chlorosis. In a genetic screen for mutants that lack AvrB-dependent chlorosis in an rpm1 background, we isolated TAO1 (target of AvrB operation), which encodes a Toll-IL-1 receptor (TIR)-NB-LRR disease resistance protein. In rpm1 plants, TAO1 function results in the expression of the pathogenesis-related protein 1 (PR-1) gene, suggestive of a defense response. In RPM1 plants, TAO1 contributes to disease resistance in response to Pto (P. syringae pathovars tomato) DC3000(avrB), but not against Pto DC3000(avrRpm1). The tao1–5 mutant allele, a stop mutation in the LRR domain of TAO1, posttranscriptionally suppresses RPM1 accumulation. These data provide evidence of genetically separable disease resistance responses to AvrB and AvrRpm1 in Arabidopsis. AvrB activates both RPM1, a CC-NB-LRR protein, and TAO1, a TIR-NB-LRR protein. These NB-LRR proteins then act additively to generate a full disease resistance response to P. syringae expressing this type III effector.
Exposure to pathogens has driven the evolution of a sophis-ticated plant immune system that controls complex defense mechanisms for limiting infection (1). Plants express disease resistance proteins whose activation results in the restriction of pathogen growth and, typically, a localized cell death termed the hypersensitive response (HR). The most common class of disease resistance proteins consists of a central nucleotide binding (NB) site and a C-terminal leucine-rich repeat (LRR). NB-LRR proteins can be further subdivided based on the presence of a coiled-coil (CC) or a Drosophila Toll-IL-1 receptor (TIR) domain at the N terminus. In response to infection by Gram-negative bacterial phytopathogens, specific NB-LRR proteins are activated via recognition of a pathogen-encoded type III effector protein. This recognition can occur in an indirect manner, whereby type III effectors are perceived via their modification of host target proteins. Hence, NB-LRR proteins can survey the integrity of intracellular host proteins targeted by type III effectors acting as virulence factors. When a particular type III effector activates an NB-LRR protein, that type III effector is defined as an “avirulence protein.” This interaction is specific, in that a single allele of a NB-LRR gene is typically responsible for mediating resistance to a pathogen isolate expressing a particular type III effector (1).
One exception to this rule is provided by the Pseudomonas syringae type III effector proteins avirulence gene B (AvrB) and avirulence gene RPM1 (AvrRpm1); each can elicit disease resistance mediated by the Arabidopsis CC-NB-LRR protein RPM1 (2, 3). After their delivery into host cells by the type III secretion pilus, AvrB and AvrRpm1 undergo N-terminal myristoylation that facilitates association with the plant plasma membrane (4). Here, these type III effectors associate with the membrane-bound host target protein, RIN4 (RPM1-interacting protein 4) (5). AvrB-RIN4 or AvrRpm1-RIN4 interactions are correlated with the phosphorylation of RIN4 and activation of RPM1 (6). Interestingly, in Glycine max (soybean) resistance to pathogen strains expressing AvrB or AvrRpm1 can be encoded by two different CC-NB-LRR genes, Rpg1-b and Rpg1-r, respectively (7, 8). Genetic data confirms that AvrB and AvrRpm1 have multiple host targets in Arabidopsis rpm1 plants (9), suggesting that different NB-LRR receptors may have evolved to monitor the integrity of independent targets of these two type III effectors. Yet, the ability of AvrRpm1 and AvrB to trigger disease resistance in Arabidopsis has never been separated.
Transgenic in planta conditional expression of AvrB in rpm1 plants leads to leaf chlorosis (4). AvrB-induced chlorosis is RIN4-independent (9), but requires the host protein RAR1 (required for Mla-resistance 1) (10). RAR1 is part of a chaperone complex that functions to positively regulate steady-state accumulation of numerous NB-LRR proteins (9, 11–13). Additionally, RAR1 is a positive regulator of basal disease resistance against virulent pathogens (13). Because AvrB can coimmunoprecipitate RAR1, the suggestion has recently arisen that RAR1 might be a virulence target of AvrB (10).
We performed a genetic screen to isolate mutants compromised for AvrB-induced chlorosis. This screen was performed in Mt-0, an ecotype (inbred genetic line) of Arabidopsis from which RPM1 is naturally deleted (14), but which responds to AvrB with leaf chlorosis (4). We defined and isolated TAO1 (target of AvrB operation), a gene encoding a disease resistance protein of the TIR-NB-LRR class. TAO1 activation correlates with defense gene expression. A specific mutant allele of TAO1 posttranscriptionally suppresses the steady-state accumulation of RPM1. In the Col-0 (RPM1) accession, TAO1 is required for a full resistance response to P. syringae pathovars tomato (Pto) DC3000(avrB) but not to Pto DC3000(avrRpm1), showing that recognition of these two type III effectors is genetically separable in Arabidopsis. TAO1-dependent phenotypes, like disease resistance mediated by all TIR-NB-LRR proteins, requires phytoalexin deficient 4 (PAD4). Our data provide further evidence that CC and TIR subclasses of NB-LRR proteins can function additively in disease resistance responses against biotrophic pathogens.
Results
TAO1 Encodes a TIR-NB-LRR Protein Required for AvrB-Induced Chlorosis in rpm1 Host Plants.
We designed a conditional genetic screen to isolate loci required for AvrB-induced chlorosis in Mt-0 (rpm1). A stable single insertion transgenic line carrying a dexamethasone (DEX)-inducible avrB expression system [DEX:avrB-HA (15)] was established in Mt-0 (see Methods). Seed from this line was mutagenized, and ≈400,000 M2 plants were screened for loss of AvrB-induced chlorosis after DEX treatment (Fig. 1A). DEX-inducible AvrB-HA expression was confirmed in mutant (fully green) M3 progeny by Western blot analysis (Fig. 1B). We used transient Agrobacterium transformation of DEX:avrB-HA on several M3 progeny from each putative M2 mutant plant to eliminate mutations in the DEX:avrB-HA expression system. In this assay, plants mutated in the DEX:avrB-HA transgene were complemented by the transient expression of DEX:avrB-HA and display chlorosis after DEX treatment. These lines were not studied further.
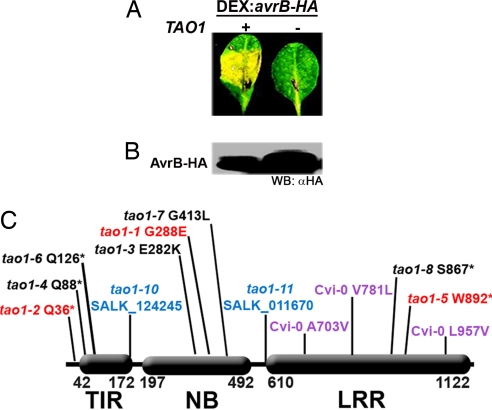
Fig. 1.
TAO1 is a TIR-NB-LRR R protein required for AvrB-induced chlorosis in rpm1 host plants. (A) Mt-0 leaves were inoculated with Agrobacterium containing T-DNA with a DEX:avrB-HA transgene (4). Leaves were treated with DEX 48 h after inoculation. The picture was taken 72 h after inoculation. (B) Western blot showing accumulation of AvrB-HA at 8 h after DEX treatment. (C) Deduced structure of TAO1 alleles recovered in Mt-0 (red and black), Col-0 Salk T-DNA insertion lines (blue), and as polymorphisms in Cvi-0 (purple). For all missense mutations, the wild-type Mt-0 residue is listed first. TIR, amino acids 42–172. NB, amino acids 197–492. LRR, amino acids 610-1122. All TAO1 alleles in Mt-0 were generated by EMS mutagenesis except tao1–8, which is a fast neutron deletion of 1 bp in codon S667. Red tao1 alleles represent alleles that were out-crossed away from the DEX:avrB-HA transgene and used for further analyses. The tao1–10 (Salk_124245) insertion begins at amino acid 168. The tao1–11 (Salk_011670) insertion begins at amino acid 597. The only amino acid difference between Mt-0 and Col-0 is V489M. Genomic TAO1 sequences for Mt-0 (EU031442) and Cvi-0 (EU031443) have been deposited in GenBank.
Genetic complementation crosses indicated eight allelic, recessive tao1 mutants that were also allelic to the naturally occurring tao1 phenotype of Cvi-0 (4). Transgenic DEX:avrB-HA rpm1 tao1–2 (Mt-0) and DEX:avrB-HA rpm1 tao1–5 (Mt-0) lines were crossed independently to rpm1–3 TAO1 (Col-0) for map-based cloning of TAO1. After DEX induction, F1 plants exhibited host cell chlorosis, confirming that both tao1–2 and tao1–5 were recessive (data not shown). We isolated nonchlorotic plants in the F2 generation. Two genotypes were present among these nonchlorotic F2 plants: (i) those that lacked the DEX:avrB-HA transgene and (ii) those that contained the DEX:avrB-HA transgene and were homozygous tao1. We performed PCR with avrB-specific primers to distinguish between these classes; plants lacking the DEX:avrB-HA transgene were discarded. From the tao1–5 cross, ≈1,000 informative F2 plants showed that TAO1 was tightly linked to the bottom of chromosome 5. In a smaller F2 population, the tao1–2 mutant allele also mapped to the same location, which was in agreement with data previously presented for the Cvi-0 loss of function allele of TAO1 (4). To avoid mis-scoring caused by transgene silencing, we used transient assays with Agrobacterium tumefaciens (DEX:avrB-HA) of F3 progeny to confirm the tao1 genotypes of F2 individuals. This approach, combined with the sequence identification of mutations in eight tao1 alleles (designated tao1–1 to tao1–8; Fig. 1C), identified TAO1 as the Mt-0 allele of the Col-0 gene At5g44510.
TAO1 encodes a TIR-NB-LRR protein of the TNL-G clade (16). There are no other NB-LRR genes at the TAO1 locus in the reference Col-0 genome sequence. The deduced TAO1 protein is only 26% similar to RPM1 and 25% similar to RPG1-B (soybean). For further analyses, we chose three alleles: tao1–2, an early stop mutation after Q36 and a likely null; tao1–1, a missense mutation at G238E in the NB domain; and tao1–5, a nonsense mutation at W892 that results in the deletion of the last five LRRs (13 through 17) (Fig. 1C). Sequencing of the naturally occurring tao1 allele in Cvi-0 revealed three amino acid changes compared with the Mt-0 TAO1 sequence at 703, 781, and 957 (Fig. 1C). Col-0 TAO1 shares these amino acids with Mt-0, while also having a polymorphism at amino acid 489.
TAO1 Activation Results in Expression of Pathogenesis-Related Protein-1 (PR-1).
We next evaluated whether TAO1 activation resulted in expression of a common defense response gene, PR-1. Five days after induction with DEX, DEX:avrB-HA rpm1 tao1–2 plants were nonchlorotic, whereas the DEX:avrB-HA rpm1 TAO1 plants displayed strong leaf chlorosis (Fig. 2A). TAO1-mediated chlorosis correlated with an increase in PR-1 expression in leaf tissue (Fig. 2A). Thus, TAO1 function results in the induction of a common defense response marker. Similar results were found for DEX:avrB-HA rpm1 tao1–5 (data not shown).
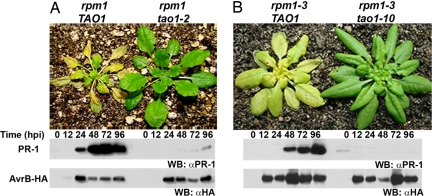
Fig. 2.
TAO1 activation correlates with defense gene expression in both Mt-0 and Col-0. Five-week-old plants were sprayed with 20 μM Dex to induce expression of AvrB-HA. (A) Mt-0 and tao1–2. (B) Col-0(rpm1–3) and rpm1–3 tao1–10. (Top) Photograph was taken 5 days after DEX treatment. (Middle) Western blot showing PR-1 protein levels. (Bottom) Western blot analysis showing AvrB-HA protein levels. Equivalent amounts of total proteins were loaded in all lanes (data not shown). The experiment is representative of three independent replicates.
To analyze TAO1 function in Col-0, a better-studied genetic background for disease resistance studies than Mt-0, we used two mutant alleles designated tao1–10 (SALK_124245), and tao1–11 (SALK_011670) that carry exon insertions in At5g44510 (Fig. 1D). Both are TAO1 transcript nulls [supporting information (SI) Fig. S1]. These lines were crossed to an isogenic DEX:avrB-HA rpm1–3 TAO1 line, and DEX:avrB-HA rpm1–3 tao1–10 and DEX:avrB-HA rpm1–3 tao1–11 lines were selected from the resulting progeny. Five days after induction with DEX, the DEX:avrB-HA rpm1–3 TAO1 plants were chlorotic, whereas the DEX:avrB-HA rpm1–3 tao1–10 lines were nonresponsive (Fig. 2B). Furthermore, TAO1 activation correlated with an increase in PR-1 expression (Fig. 2B). Similar results were found for DEX:avrB-HA rpm1–3 tao1–11 (data not shown). Hence, tao1–10 and tao1–11 lose TAO1 function in response to AvrB expression in planta.
RPM1 Function Is Lost Specifically in tao1–5.
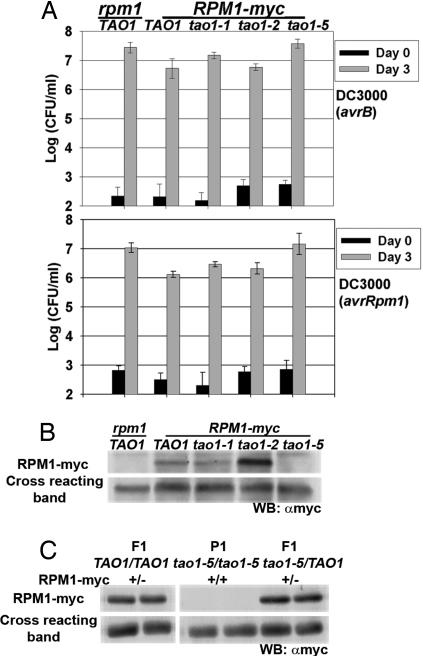
We next addressed whether TAO1 contributes to RPM1-mediated disease resistance. For this, we introduced an RPM1 transgene containing a C-terminal myc epitope tag, expressed from the native RPM1 promoter (17), into Mt-0 (rpm1 TAO1). A line carrying a single copy of the RPM1-myc transgene was selected and then crossed into the tao1–1, tao1–2, and tao1–5 mutant backgrounds (previously back-crossed and selected for loss of the DEX:avrB-HA transgene) (see Methods). Stable RPM1-myc TAO1, RPM1-myc tao1–1, RPM1-myc tao1–2, and RPM1-myc tao1–5 lines were selected, and RPM1 function was assessed. After hand infiltration with Pto DC3000(avrB) or Pto DC3000(avrRpm1), RPM1 function resulted in only a 5- and 10-fold growth restriction of bacteria, respectively (Fig. 3A), which is much less bacterial growth restriction than observed in Col-0 (RPM1) (Fig. 4A). This weak RPM1-mediated growth restriction is likely caused by lower accumulation of RPM1-myc in Mt-0 relative to Col-0 (Fig. S2). Lower RPM1-myc accumulation may be the result of genetic background differences between Mt-0 and Col-0 that control RPM1 accumulation or transgene positional effects. In RPM1-myc tao1–1 and RPM1-myc tao1–2, there was essentially no effect on the weak RPM1-mediated growth restriction of Pto DC3000(avrRpm1) or Pto DC3000(avrB) (Fig. 3A). Interestingly, RPM1 function was lost in the RPM1-myc tao1–5 background in response to either Pto DC3000(avrB) or Pto DC3000(avrRpm1) (Fig. 3A).
Fig. 3.
Weak RPM1 function in Mt-0 is suppressed posttranscriptionally by tao1–5. (A) Four-week old plants were hand-infiltrated with Pto DC3000(avrB) (Upper) or Pto DC3000(avrRpm1) (Lower) at 105 cfu/ml. Error bars represent the SD among four samples. This experiment is indicative of three independent replicates. Similar results were seen with dip infections (data not shown). (B and C) Western blot analysis showing RPM1-myc accumulation in plant lines with the noted genotypes. Cross-reacting band indicates equivalent amounts of total protein loading. Samples from hybrid (F1) and parental (P1) lines are noted above their respective genotypes. These experiments are indicative of four (B) and two (C) independent replicates. The higher than wild-type level of RPM-myc accumulation in the tao1–2 background shown in B was not reproducible.
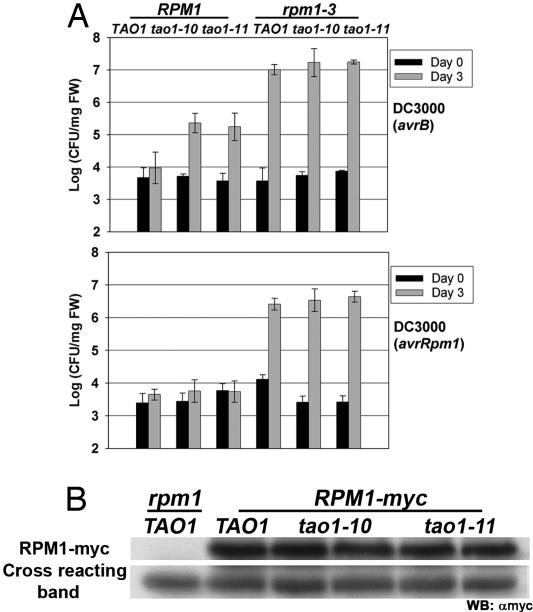
Fig. 4.
TAO1 contributes to disease resistance against Pto DC3000(avrB) but not Pto DC3000(avrRpm1). (A) Two- to 3-week-old plants were dip-infiltrated with Pto DC3000(avrB) (Upper) or Pto DC3000(avrRpm1) (Lower) at a bacterial concentration of 2.5 × 107 cfu/ml. Error bars represent the SD among four samples. This experiment is representative of three independent replicates. (B) Western blot analysis showing RPM1-myc accumulation in plant lines with the noted genotypes. The cross-reacting band serves as an indicator of equivalent amounts of total protein loaded. This experiment is representative of two independent replicates.
RPM1 Does Not Accumulate in the tao1–5 Background.
We postulated two possible reasons why RPM1 function is lost in the tao1–5 background. The tao1–5 mutant protein could either prevent RPM1 signaling, or the tao1–5 mutant protein might negatively regulate RPM1 steady-state accumulation. To distinguish between these possibilities, the RPM1-myc TAO1, RPM1-myc tao1–1, RPM1-myc tao1–2, and RPM1-myc tao1–5 lines were subjected to Western blot analysis for RPM1-myc. Despite some experimental variation, RPM1 accumulated normally in RPM1-myc tao1–1 and RPM1-myc tao1–2 (Fig. 3B). By contrast, RPM1 accumulation was abolished in RPM1-myc tao1–5 (Fig. 3B). RT-PCR analysis revealed that this effect on RPM1 is posttranscriptional (Fig. S3). Interestingly, the effect of tao1–5 on RPM1 accumulation was recessive (Fig. 3C).
TAO1 Is Required for Full Disease Resistance Against Pto DC3000(avrB) in Col-0.
We introgressed the tao1–10 and tao1–11 null alleles into rpm1–3. Homozygous rpm1–3 tao1–10 and rpm1–3 tao1–11 lines were selected to evaluate TAO1 function in both the presence and absence of RPM1. Interestingly, after dip inoculation in a bacterial suspension of Pto DC3000(avrB), both the RPM1 tao1–10 and RPM1 tao1–11 lines displayed ≈10-fold less disease resistance than RPM1 TAO1 (Fig. 4A). This effect of TAO1 was not seen with hand infiltrations (data not shown), suggesting that TAO1 may act early in pathogen infection. Hence, TAO1 contributes ≈1–5% of the full resistance to Pto DC3000(avrB) in the presence of RPM1. We observed no enhanced susceptibility to Pto DC3000(avrB) in the rpm1–3 tao1–10 and rpm1–3 tao1–11 lines, indicating that the weak TAO1 contribution to disease resistance is too low to measure in the absence of RPM1 (Fig. 4A). We observed full RPM1-mediated growth restriction in the RPM1 tao1–10 and RPM1 tao1–11 lines after dip inoculation of Pto DC3000(avrRpm1) (Fig. 4B). Hence, TAO1 contributes specifically to disease resistance triggered by Pto DC3000(avrB) and does not contribute to RPM1-mediated disease resistance in response to Pto DC3000(avrRpm1). As with Pto DC3000(avrB), we observed no enhanced susceptibility of the rpm1–3 tao1–10 and rpm1–3 tao1–11 lines to Pto DC3000(avrRpm1) (Fig. 4A).
Although RPM1-mediated resistance to Pto DC3000(avrRpm1) was unaffected in the tao1–10 and tao1–11 lines, the possibility existed that TAO1 affects RPM1 steady-state accumulation. This scenario would indicate that different accumulation levels of RPM1 are required for resistance to Pto DC3000(avrB) versus Pto DC3000(avrRpm1). To address this possibility, the rpm1–3 tao1–10 and rpm1–3 tao1–11 lines were crossed to a rpm1–3 RPM1-myc TAO1 line (17). Stable rpm1–3 RPM1-myc tao1–10 and rpm1–3 RPM1-myc tao1–11 lines were selected. Western blot analysis revealed that RPM1 accumulation was equivalent in the tao1–10 and tao1–11 lines relative to the TAO1 parental line (Fig. 4B). Hence, loss of TAO1 does not affect RPM1 steady-state accumulation. Therefore, the contribution of TAO1 to disease resistance against Pto DC3000(avrB) in Col-0 is likely to be at the signaling level.
TAO1 Requires PAD4 for Function.
The Arabidopsis PAD4 protein functions in both basal defense and TIR-NB-LRR-mediated disease resistance (18, 19). To assess whether PAD4 is required for TAO1 function, we crossed the DEX:avrB-HA rpm1–3 line to the pad4–1 mutant line (20) and selected homozygous F2 rpm1–3 pad4–1 lines that contained the DEX:avrB-HA transgene. After inducing AvrB expression in planta, we observed that both TAO1-mediated chlorosis and PR-1 expression were lost in the rpm1–3 pad4–1 lines (Fig. S4A). Furthermore, inoculation with Pto DC3000(avrB) and Pto DC3000(avrRpm1) on pad4–1 TAO1 RPM1 lines revealed that the TAO1 defense function against Pto DC3000(avrB) is also lost in a pad4–1 mutant (Fig. S4B). Thus, these results demonstrate that TAO1 requires PAD4 for function.
Discussion
AvrB is a bacterial type III effector protein that causes modification of the Arabidopsis host protein RIN4, subsequently activating the NB-LRR protein RPM1 (5, 6, 21) or contributing to pathogen virulence in susceptible plants (7). The sequence-independent type III effector AvrRpm1 acts similarly (22). In the absence of RPM1, in planta expression of AvrB produces a host cell chlorosis originally speculated to be indicative of an AvrB virulence function (4). Because AvrB-mediated chlorosis is RIN4 independent, AvrB must have additional targets within the Arabidopsis cell (9). Our aims were to (i) identify host proteins required for AvrB-induced chlorosis, (ii) assess whether and how these host components affect RPM1 function triggered by either AvrB or AvrRpm1, and (iii) clarify whether these host factors are specific for the responses to AvrB. We identified only one gene in our mutant screen, and it encodes TAO1, a TIR-NB-LRR disease resistance protein.
Our data demonstrate that TAO1 activation contributes to disease resistance in response to Pto DC3000(avrB), but not in response to Pto DC3000(avrRpm1) (Fig. 4A). To explain the functional relationship between AvrB, TAO1, and RPM1, we propose the following model (Fig. S5). In rpm1 plants, TAO1 activation in response to AvrB is insufficient to cause a HR (data not shown) or significantly restrict Pto DC3000(avrB) growth (Fig. 4A). However, chlorosis and expression of a common defense protein, PR-1, are induced (Fig. 2; see also Fig. S5 Left). These data demonstrate that TAO1 is a weak disease resistance protein that cannot create sufficient defense response amplitude to restrict Pto DC3000(avrB) growth in the absence of RPM1. In tao1 plants, RPM1 activation in response to AvrB results in a HR in addition to a significant restriction of Pto DC3000(avrB) growth (Fig. 4A) (Fig. S5 Center). Finally, the additive functions of RPM1 and TAO1 facilitate full disease resistance to Pto DC3000(avrB) (Fig. 4A) (Fig. S5 Right).
Because TAO1 function is RIN4 independent (9) and RIN4 is required for RPM1 function (5), TAO1 perception of AvrB might occur at a cellular site that lacks RIN4 and RPM1. Recently, two studies (6, 21) identified AvrB mutants that are unable to activate RPM1, but still activate TAO1. For example, the AvrB T125A mutation abolished both the RIN4–AvrB interaction and the ability to trigger RPM1, but retained the ability to activate TAO1 (6, 21). Additionally, the AvrB D297A mutation retained interaction with RIN4 and lost RPM1 activation but still elicited TAO1 (6, 21). These data suggest that there are likely to be structural differences in how AvrB targets host proteins and consequently triggers either RPM1 or TAO1. Despite the potentially separate mechanisms of TAO1 and RPM1 activation, both require AvrB membrane localization via myristoylation (4). These data may indicate that like RPM1 and RIN4, a second putative protein(s), targeted by AvrB and associated with TAO1, is also present at the plasma membrane.
Interestingly, the steady-state levels of RPM1 are significantly diminished in tao1–5 (Fig. 3B). Because the other loss-of-function mutant alleles tested, tao1–1 (presumptive P-loop dead) and tao1–2 (presumptive null), do not affect RPM1 accumulation and tao1–5 homozygosity is required for its effects on RPM1, tao1–5 acts as a recessive gain-of-function allele (Fig. 3 B and C). This phenotype is reminiscent of the effects of specific alleles of hsp90.2 on RPM1 accumulation (23, 24). The tao1–5 allele exhibits no other observable phenotypic defects (data not shown). A tao1–5-encoded protein could sequester RPM1 directly or negatively affect RPM1 stability through its effects on an intermediate host protein. Three host proteins shown to be required for RPM1 steady-state accumulation are RIN4 and the general NB-LRR chaperone complex members RAR1 and HSP90.2 (5, 12). Because neither RPS2 function nor RPS5 steady-state accumulation are compromised in the tao1–5 background (data not shown), it is unlikely that the tao1–5 mutant protein sequesters RIN4 or RAR1. Hence, we speculate that HSP90.2 or an as-yet-undiscovered protein that contributes to RPM1 steady-state accumulation is altered by the mutant tao1–5-encoded product.
Recently, Shang et al. (10) demonstrated that RAR1 is required for AvrB-dependent chlorosis in rpm1 plants (10). They also showed that AvrB-induced chlorosis correlated with increased growth of a Pto DC3000(hrpL) mutant that lacks the type III pilus and therefore cannot deliver type III effectors to the plant cell. This relaxation of basal defense responses was RAR1 dependent. Also, when overexpressed, AvrB and RAR1 can be coimmunoprecipitated (10) and also allow reactivation of a split-luciferase reporter (albeit under conditions where the AvrB myristoylation site is buried in the reporter fusion and mislocalization of AvrB is likely) (25). These results led the authors to conclude that RAR1 is a “virulence target” of AvrB, although no direct interaction of AvrB and RAR1 was demonstrated (10). Our data demonstrate that AvrB-induced chlorosis is mediated by the TIR-NB-LRR protein TAO1 and that TAO1 functions as a “weak R gene” (Figs. 2 and 4A). Thus, there is a contradiction: is the observed host cell chlorosis indicative of AvrB virulence targeting of RAR1, or does it represent a TAO1-mediated disease resistance response, which in turn requires RAR1?
There are several possible explanations for the differing interpretations presented in ref. 10 and here regarding what AvrB-mediated chlorosis represents. First, the increase in Pto DC3000(hrpL) growth observed after induced AvrB overexpression in planta was COI1 dependent, whereas AvrB-induced chlorosis was COI1 independent (10). Hence, our observation of AvrB-mediated, TAO1-dependent chlorosis can be genetically separated from AvrB-mediated, COI1-dependent suppression of basal defense (10). Second, our data show that AvrB-mediated chlorosis requires the host protein PAD4 (Fig. S4), a positive regulator of both basal defense and TIR-NB-LRR-mediated disease resistance (18, 19). Third, a very strong regimen of conditional overexpression of AvrB [30 μM DEX spray 2 days before the infection, and also continually throughout the Pto DC3000(hrpL) infection] was used in ref. 10. Using a regimen of two times 10 μM DEX treatment, but only 1 day before infection, we were able to fully induce AvrB-dependent chlorosis in TAO1 plants, but did not observe increased growth of Pto DC3000(hrcC) (data not shown). These data suggest that the increased growth of Pto DC3000(hrpL) shown in ref. 10 is caused by extraordinary levels of AvrB expression in planta, resulting in extensive tissue collapse. Our data demonstrate that TAO1 contributes to the restriction of pathogen growth when AvrB is delivered naturally via a type III pilus from Pto DC3000 (Fig. 4A). Furthermore, we note that using the same experimental conditions as in ref. 10, it was shown that AvrB-mediated chlorosis correlated with a growth reduction of Pto DC3000 (33). This growth reduction was RAR1 dependent, consistent with a role for NB-LRR proteins in basal defense (13), but difficult to reconcile with the data presented in ref. 10. RAR1 regulates the function of many NB-LRR proteins by acting as a positive regulator of NB-LRR stability (9, 11–13). Hence, the simplest scenario to explain the sum of the data presented here and in ref. 10 is that the loss of AvrB-dependent chlorosis in a rar1 background indicates that TAO1 requires RAR1 for steady-state accumulation and therefore function.
The bacterial type III effector proteins AvrB and AvrRpm1 each trigger strong disease resistance through RPM1. Yet the activation of TAO1 by AvrB suggests that additional layers of recognition in response to infection can effectively add to the plant's defense response. In a mechanistic sense, if a given type III effector has multiple cellular targets, then direct or indirect recognition by more than one NB-LRR may add to both the flexibility and the amplitude of the host's overall response. These scenarios provide hosts with greater evolutionary capacity, by allowing NB-LRR proteins to continually fine-tune their responses to a single pathogen effector while maintaining an intermediate level of disease resistance. This possibility may explain recent examples of genetic requirements for combined action of (i) a TIR-NB-LRR and a CC-NB-LRR protein (26) and (ii) a TIR-NB-LRR and an atypical CC-containing NB-LRR (27) against a virus and an isolate of the oomycete parasite Hyaloperonospora parasitica, respectively. Our findings further the concept that a CC-NB-LRR and a TIR-NB-LRR, which typically trigger different response pathways in plants (28), can additively contribute to disease resistance.
Methods
A. tumefaciens-Mediated, DEX-Inducible Transient Expression Assays.
Transient transformation assays were performed as described (4). Arabidopsis chlorosis response in this assay was observed at 72 h after induction. Protein was extracted 8 h after induction from four transformed leaf discs.
Generation of Stable Transgenic Lines, Mutagenesis, and Screening.
Stable transgenic plant lines were generated by the floral dip method (29). Segregation of hygromycin resistance was used to isolate lines containing single-copy insertions of both the DEX:avrB-HA (4) and RPM1-myc (17) transgenes in Mt-0. For ethane methyl sulfonate (EMS) mutagenesis, ≈30,000 seeds were incubated in a 0.25% EMS solution for 8 h, washed in distilled water, and dried on filter paper. Fast neutron mutagenesis was performed by the International Atomic Energy Agency research laboratory in Vienna, Austria. Approximately 700,000 M2 seeds in 100 lots (80 EMS, 20 fast neutron) were sprayed with 20 μM DEX as described (30). Putative mutants were isolated and allowed to self-pollinate. M3 progeny were retested by Agrobacterium transient delivery of DEX:avrB-HA as described above.
Positional Cloning of TAO1 and Out-Crossing of the DEX:avrB-HA Transgene.
Mapping of TAO1 was performed as described (31). Candidate genes within the TAO1 interval were PCR-amplified from the tao1 mutants, and PCR products were sequenced directly. To obtain tao1 lines lacking the DEX:avrB-HA transgene, M3 tao1 lines were crossed to nontransgenic TAO1 Mt-0 plants. F2 plants containing the DEX:avrB-HA transgene were selected and sprayed with 20 μM DEX. Nonchlorotic (tao1) lines were selected and allowed to self-pollinate. F3 individuals that lacked the DEX:avrB-HA transgene were selected and allowed to self-pollinate. F4 seed was confirmed for tao1 genotype by Agrobacterium transient transformation of DEX:avrB-HA. Subsequent confirmation of tao1 mutant alleles was performed by using dominant cleaved amplified polymorphic sequences specific to the tao1 allele in question.
Bacterial Infections.
For Fig. 3A, 4-week-old plants were hand-infiltrated with bacteria at a concentration of 105 cfu/ml. For each sample, four leaf disks were pooled per time point (16 total). Leaf disks were ground in 10 mM MgCl2, subject to serial dilutions, and plated on King's Broth (KB) plates with rifampicin selection. For Fig. 4A and Fig. S4B, dip inoculations were performed as described (32). Briefly, 2- to 3-week-old seedlings were inoculated by dipping in solutions of bacteria at a concentration of 2.5 × 107 cfu/ml. Three seedlings were placed in 1 ml of 10 mM MgCl2 with 0.2% Silwet L-77. The weights of the tubes with seedlings were measured, and the tubes were then shaken for 1 h at 28°C. Serial dilutions in 10 mM MgCl2 were made and plated on KB plates with rifampicin selection.
Protein Analysis.
For detection of AvrB-HA, PR-1, and RPM1-myc, total protein extracts were prepared by grinding ≈200 mg of leaf tissue in buffer [50 mM Tris·HCl (pH 8.0), 1% SDS, 1 mM EDTA, 1 mM 2-mercaptoethanol, and plant protease inhibitor mixture (Sigma)]. Protein concentrations were determined by using the Bradford-Lowry method and quantification buffer from BioRad. Ten microliters of 6× Laemmli buffer (final concentration 1×) was added to all measured samples after quantification. For immunodetection of AvrB-HA and PR-1, 10-μg protein samples were electrophoresed on 14% SDS polyacrylamide gels. For immunodetection of RPM1-myc, 40-μg protein samples were electrophoresed on 8% SDS polyacrylamide gels. Western blots were performed by using standard methods. Anti-PR-1 serum (gift of Robert A. Dietrich, Syngenta, Research Triangle Park, NC) was used at a dilution of 1:10,000. The detection of the HA epitope tag was with supernatants from cultures of hybridoma 3F10 monoclonal anti-HA antibody (Roche) at a dilution of 1:1,000. Detection of the myc epitope tag was with supernatants from hybridoma 9E10 monoclonal anti-myc antibody at a dilution of 1:10 (17).
RT-PCR.
RNA was isolated from various plants lines by using TRIZOL Reagent (GIBCO/BRL) according to the manufacturer's instructions. RT-PCR (RETROscript; Ambion) analysis in Fig. S1 and Fig. S3 was performed according to the manufacturer's instructions. Plant 18S Competimer primers (Ambion) were used to coamplify the 18S internal loading control. For Fig. S1, primers flanking the first intron of TAO1 5′-tatgaaatgcagagaagagttgg-3′ and 3′-ataaccgttctgctggtagag-5′ were used with the PCR conditions 94°C 10 s, 55°C 60 s, 72°C 60 s, 31 cycles. A ratio of 9:1 Competimer/18S was used. For Fig. S3, the RPM1 transcript primers 5′-caccatggcttcggctactgttgattttg-3′ and 3′-cactttgcatcgccatcatcaatagg-5′ RPM1 transcript primers were used with the PCR conditions 94°C 10 s, 55°C 60 s, 72°C 60 s, 31 cycles. A ratio of 9.5:1 Competimer/18S was used. Products were separated on a 1% agarose gel.
Supplementary Material
Acknowledgments.
We thank Drs. David Mackey, Ai-Jiuan Wu, and Marc Nishimura for critical reading of the manuscript, and Dr. David A. Hubert and Dr. Petra Epple for technical suggestions. This work was supported by Department of Energy Grant DE-FG05-95ER20187 (to J.L.D.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. EU031442 and EU031443).
This article contains supporting information online at www.pnas.org/cgi/content/full/0802157105/DCSupplemental.
References
- 1.Jones JDG, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 2.Bisgrove SR, Simonich MT, Smith NM, Sattler NM, Innes RW. A disease resistance gene in Arabidopsis with specificity for two different pathogen avirulence genes. Plant Cell. 1994;6:927–933. doi: 10.1105/tpc.6.7.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grant MR, et al. Structure of the Arabidopsis RPM1 gene enabling dual specificity disease resistance. Science. 1995;269:843–846. doi: 10.1126/science.7638602. [DOI] [PubMed] [Google Scholar]
- 4.Nimchuk Z, et al. Eukaryotic fatty acylation drives plasma membrane targeting and enhances function of several type III effector proteins from Pseudomonas syringae. Cell. 2000;101:353–363. doi: 10.1016/s0092-8674(00)80846-6. [DOI] [PubMed] [Google Scholar]
- 5.Mackey D, Holt BF, III, Wiig A, Dangl JL. RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated disease resistance in Arabidopsis. Cell. 2002;108:743–754. doi: 10.1016/s0092-8674(02)00661-x. [DOI] [PubMed] [Google Scholar]
- 6.Desveaux D, et al. Type III effector activation via nucleotide binding, phosphorylation, and host target interaction. PLoS Pathog. 2007;3:e48. doi: 10.1371/journal.ppat.0030048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ashfield T, Keen NT, Buzzell RI, Innes RW. Soybean resistance genes specific for different Pseudomonas syringae avirulence genes are allelic, or closely linked, at the RPG1 locus. Genetics. 1995;141:1597–1604. doi: 10.1093/genetics/141.4.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ashfield T, Ong LE, Nobuta K, Schneider CM, Innes RW. Convergent evolution of disease resistance gene specificity in two flowering plant families. Plant Cell. 2004;16:309–318. doi: 10.1105/tpc.016725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Belkhadir Y, Nimchuk Z, Hubert DA, Mackey D, Dangl JL. Arabidopsis RIN4 negatively regulates disease resistance mediated by RPS2 and RPM1 downstream or independent of the NDR1 signal modulator and is not required for the virulence functions of bacterial type III effectors AvrRpt2 or AvrRpm1. Plant Cell. 2004;16:2822–2835. doi: 10.1105/tpc.104.024117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shang Y, et al. RAR1, a central player in plant immunity, is targeted by Pseudomonas syringae effector AvrB. Proc Natl Acad Sci USA. 2006;103:19200–19205. doi: 10.1073/pnas.0607279103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bieri S, et al. RAR1 positively controls steady-state levels of barley MLA resistance proteins and enables sufficient MLA6 accumulation for effective resistance. Plant Cell. 2004;16:3480–3495. doi: 10.1105/tpc.104.026682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tornero P, et al. RAR1 and NDR1 contribute quantitatively to disease resistance in Arabidopsis and their relative contributions are dependent on the R gene assayed. Plant Cell. 2002;14:1005–1015. doi: 10.1105/tpc.001032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holt BF, III, Belkhadir Y, Dangl JL. Antagonistic control of disease resistance protein stability in the plant immune system. Science. 2005;309:929–932. doi: 10.1126/science.1109977. [DOI] [PubMed] [Google Scholar]
- 14.Grant MR, et al. Independent deletions of a pathogen-resistance gene in Brassica and Arabidopsis. Proc Natl Acad Sci USA. 1998;95:15843–15848. doi: 10.1073/pnas.95.26.15843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aoyama T, Chua NH. A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 1997;11:605–612. doi: 10.1046/j.1365-313x.1997.11030605.x. [DOI] [PubMed] [Google Scholar]
- 16.Meyers BC, Kozik A, Griego A, Kuang H, Michelmore RW. Genomewide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell. 2003;15:809–834. doi: 10.1105/tpc.009308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boyes DC, Nam J, Dangl JL. The Arabidopsis thaliana RPM1 disease resistance gene product is a peripheral plasma membrane protein that is degraded coincident with the hypersensitive response. Proc Natl Acad Sci USA. 1998;95:15849–15854. doi: 10.1073/pnas.95.26.15849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Glazebrook J, et al. Phytoalexin-deficient mutants of Arabidopsis reveal that PAD4 encodes a regulatory factor and that four PAD genes contribute to downy mildew resistance. Genetics. 1997;146:381–392. doi: 10.1093/genetics/146.1.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feys BJ, et al. Arabidopsis senescence-associated gene101 stabilizes and signals within an enhanced disease susceptibility1 complex in plant innate immunity. Plant Cell. 2005;17:2601–2613. doi: 10.1105/tpc.105.033910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glazebrook J, Rogers EE, Ausubel FM. Isolation of Arabidopsis mutants with enhanced disease susceptibility by direct screening. Genetics. 1996;143:973–982. doi: 10.1093/genetics/143.2.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ong LE, Innes RW. AvrB mutants lose both virulence and avirulence activities on soybean and Arabidopsis. Mol Microbiol. 2006;60:951–962. doi: 10.1111/j.1365-2958.2006.05162.x. [DOI] [PubMed] [Google Scholar]
- 22.Ritter C, Dangl JL. The avrRpm1 gene of Pseudomonas syringae pv. maculicola is required for virulence on Arabidopsis. Mol Plant-Microbe Interact. 1995;8:444–453. doi: 10.1094/mpmi-8-0444. [DOI] [PubMed] [Google Scholar]
- 23.Hubert DA, et al. Cytosolic HSP90 associates with and modulates the Arabidopsis RPM1 disease resistance protein. EMBO J. 2003;22:5679–5689. doi: 10.1093/emboj/cdg547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Warren RF, Henk A, Mowery P, Holub E, Innes RW. A mutation within the leucine-rich repeat domain of the Arabidopsis disease resistance gene RPS5 partially suppresses multiple bacterial and downy mildew resistance genes. Plant Cell. 1998;10:1439–1452. doi: 10.1105/tpc.10.9.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen H, et al. Firefly luciferase complementation imaging assay for protein–protein interactions in plants. Plant Physiol. 2008;146:368–376. doi: 10.1104/pp.107.111740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peart JR, Mestre P, Lu R, Malcuit I, Baulcombe DC. NRG1, a CC-NB-LRR protein, together with N, a TIR-NB-LRR protein, mediates resistance against tobacco mosaic virus. Curr Biol. 2005;15:968–973. doi: 10.1016/j.cub.2005.04.053. [DOI] [PubMed] [Google Scholar]
- 27.Sinapidou E, et al. Two TIR:NB:LRR genes are required to specify resistance to Peronospora parasitica isolate Cala2 in Arabidopsis. Plant J. 2004;38:898–909. doi: 10.1111/j.1365-313X.2004.02099.x. [DOI] [PubMed] [Google Scholar]
- 28.Aarts N, et al. Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene-mediated signaling pathways in Arabidopsis. Proc Natl Acad Sci USA. 1998;95:10306–10311. doi: 10.1073/pnas.95.17.10306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 30.McNellis TW, et al. Glucocorticoid-inducible expression of a bacterial avirulence gene in transgenic Arabidopsis thaliana induces hypersensitive cell death. Plant J. 1998;14:247–258. doi: 10.1046/j.1365-313x.1998.00106.x. [DOI] [PubMed] [Google Scholar]
- 31.Lukowitz W, Gillmour CS, Scheible W-R. Positional cloning in Arabidopsis: Why it feels good to have a genome initiative working for you. Plant Physiol. 2000;123:795–806. doi: 10.1104/pp.123.3.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tornero P, Dangl JL. A high throughput method for quantifying growth of phytopathogenic bacteria in Arabidopsis thaliana. Plant J. 2001;28:475–481. doi: 10.1046/j.1365-313x.2001.01136.x. [DOI] [PubMed] [Google Scholar]
- 33.Li X. Manhattan, KS: Kansas State University; 2006. Interplay between bacterial virulence and plant innate immunity in Pseudomonas–Arabidopsis interactions. PhD thesis. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.