Abstract
In animal breeding programs, deoxyribonucleic acid (DNA) markers can be used to identify sires that are less susceptible to disease. These DNA markers are typically discovered in populations that display differences in susceptibility. To find those differences, it was hypothesized that sires influence their offspring responses to infection with H. parasuis. To identify differences in susceptibility, colostrum-deprived pigs derived from 6 sires were inoculated with a virulent strain of H. parasuis serovar 5. Pigs were infected at 21-d of age and euthanized 1, 2, or 3 days post-infection. Rectal temperatures, bacterial detection, clinical signs, and lesions were measured by comparing disease susceptibility in the offspring from each sire. The effect of the sire on the severity of disease in the offspring was statistically analyzed using to a 2-way ANOVA with sire and test day as fixed effects. Significant differences among sires were found for lesions, rectal temperatures from days 0–1 and 0–2 (P < 0.05) and marginal effects for clinical signs (P = 0.08). On average, the offspring of sire H94 was the most susceptible to challenge. Responses to infection were categorized to determine the clinical responses and analyzed by Chi square. Overall, 10% of all pigs infected were fully resistant to H. parasuis infection. Boar H94 didn’t produce any fully resistant offspring. Differences in susceptibility to H. parasuis were observed, and the results support the hypothesis that sires influence their offspring’s response to infection. Tissues from this population could be used to identify DNA markers for genetic selection of sires that produce offspring more resistant to H. parasuis infection.
Résumé
Dans les programmes de reproduction animale, les marqueurs ADN peuvent être utilisés pour identifier les géniteurs qui sont moins susceptibles à la maladie. Ces marqueurs ADN sont typiquement découverts dans des populations qui présentent des susceptibilités différentes. Pour trouver ces différences, l’hypothèse a été émise que les géniteurs influencent la réponse de leurs rejetons à une infection par Haemophilus parasuis. Afin d’identifier ces différences dans la susceptibilité, des porcs privés de colostrum et provenant de 6 géniteurs ont été inoculés avec une souche virulente de H. parasuis sérovar 5. Les porcs ont été infectés à 21 jours d’âge et euthanasiés 1, 2 et 3 jours post-infection. La température rectale, la détection bactérienne, les signes cliniques et les lésions ont été mesurés en comparant la susceptibilité à la maladie chez les rejetons de chaque géniteur. L’effet du géniteur sur la sévérité de la maladie chez les rejetons a été analysé à l’aide d’un test ANOVA à 2 voies avec le géniteur et le jour du test comme effets fixés. Des différences significatives entre les géniteurs ont été trouvées pour les lésions, la température rectale pour les jours 0 à 1 et 0 à 2 (P < 0,05) et des effets marginaux pour les signes cliniques (P = 0,08). En moyenne, les rejetons du géniteur H94 étaient les plus susceptibles à une infection. Les réponses à l’infection étaient caractérisées afin de déterminer les réponses cliniques et analyser le tout par un test de Chi carré. De manière globale, 10 % de tous les porcs infectés étaient complètement résistants à l’infection par H. parasuis. Le verrat H94 n’a pas eu de rejeton complètement résistant. Des différences dans la susceptibilité à H. parasuis ont été observées, et les résultats confirment l’hypothèse que les géniteurs influencent la réponse à une infection de leur rejeton. Les tissus de cette population pourraient être utilisés afin d’identifier les marqueurs ADN pour une sélection génétique des géniteurs qui produisent des rejetons plus résistants à l’infection par H. parasuis.
(Traduit par Docteur Serge Messier)
Introduction
Development of infectious disease in animals is attributed to the environment, the pathogenicity of the microbe, and a range of host factors. The environment plays an important role in the outcome of pathogen and host interactions, and differences in microbe pathogenicity exist that are important in causing clinical signs and lesions. Host-microbe interactions are complex and influenced by multiple factors such as the age of the host, the presence of passive immunity (antibodies), innate and acquired immunity, and concurrent infections (1–6). Many of these factors can be controlled in experimental infections, but interestingly, it is not uncommon to observe that some animals develop disease while others do not express disease under identical challenge environments (7). It has become clear that factors inherent to the host influence disease susceptibility even in the presence of virulent microbes. The role of host genetic factors and their importance in the development of disease have been reported for various animal species including swine (8–19).
Haemophilus parasuis, a Gram-negative bacterium, is one of the earliest and most prevalent colonizers of piglets in the farrowing house and a common isolate from nasal secretions in pigs (3). Disease caused by H. parasuis continues to be one of the significant bacterial infections in nursery and finishing pigs in multisite systems and it is becoming increasingly important as those systems are expanded (3,20). In the Americas, Glässer’s disease or polyserositis caused by H. parasuis is a disease common in nearly all farms and particularly in high health herds. Current management and vaccination strategies often do not work, and alternative control methods are necessary (20). The presentation of H. parasuis infection ranges from a subclinical infection to severe classic Glässer’s disease, which manifests as polyserositis, septicemia, arthritis, and meningitis (3,20). Although some H. parasuis strains are known to cause pneumonia (20), the presence of H. parasuis in the lung is generally thought to be secondary to other primary pathogens (21,22). Infections with porcine reproductive and respiratory syndrome virus (PRRSV) in young piglets are frequently associated with secondary infections including H. parasuis (22). Haemophilus parasuis has been reported in association with 32% of cases of postweaning multisystemic wasting syndrome (PMWS) (21). These examples illustrate how this bacterium coexists with the most relevant syndromes that the swine industry faces today.
It was hypothesized that the genetic makeup of the sire influences the outcome of experimental infections when progenies are infected with H. parasuis. To determine if the genetics of the pig has influence on the susceptibility to infection, the development of disease was assessed in animals derived from related boars with similar genetic background. Genetic influences are easier to assess in boars because multiple litters can be analyzed, traits can be measured in a short time frame, and differences between sires are separated from maternal effects on litters.
Materials and methods
Animal infections and experimental design
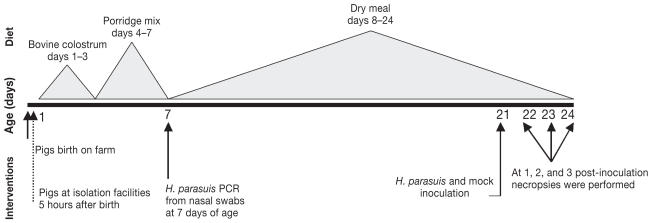
Two hundred colostrum-deprived (CD) pigs born on a commercial farm in Spain were used for experimental challenge studies. Groups of 20 pigs originating from 2 different sires were used during each of the 10 consecutive experiments. A total of 6 sires were used across the 10 experiments. On average, there were 3.8 litters represented per sire in every experiment. Immediately after birth, piglets were collected from the birth canal, separated from the mother without allowing them to ingest colostrum, and their skin disinfected with a 2% iodine solution. Animals were transported to isolation facilities at the Centro de Investigación en Sanidad Animal (CISA) in Madrid, Spain. At this research facility, CD piglets were bottle-fed with bovine colostrum (milk obtained during the 1st day after calving, from a nearby dairy farm) for 3 days; with a porridge mixture (Kwikstart; SCA Iberica, Zaragoza, Spain) from days 3–7; and with piglet dry meal formula (Startrite 100; SCA Iberica, Zaragoza, Spain) for the rest of the study. The management and diet of the CD pigs has been described previously (23–24). Nasal swabs were obtained when the pigs were 7-d of age to determine if they were negative to H. parasuis prior to inoculation. At 21-d of age, pigs were allocated to either an infected or a control group and inoculated with H. parasuis [range, 1 × 106 to × 108 colony-forming units (CFU)] or mock-inoculated with phosphate buffered saline (PBS), respectively. Infected and control pigs were housed in 2 separate rooms and managed under similar conditions (21). Pigs from each sire were represented in infected and control groups, and to avoid cross contamination, control rooms were always attended to first. Because 80.5% of CD pigs survived to the challenge date, there were 4–5 infected pigs and 1 control pig euthanized at each of the 1, 2, and 3 days post-inoculation time points (Figure 1). All animal procedures followed regulations of the European Directive 86/609/CEE (Nov 24, 1986).
Figure 1.
Experimental time-line indicating interventions, age, and diet.
Pedigree
Six boars from the same commercial line (Landrace-Duroc synthetic) were used to produce the offspring by mating them with Landrace × Large White sows. On average, 33 pigs from each boar (H77, H70, H78, H84, H92, and H94) were included in the study. Sires H78, H70, and H77 were half siblings, and H94 and H92 were also half siblings.
Bacterial inoculum and challenge
The H. parasuis serovar 5, strain 29755 was used as the inoculum. The strain was kindly provided by Dr. Pijoan (CAPS, University of Minnesota, 1988 Fitch Avenue, St. Paul, Minnesota, USA). Bacteria were cultured on blood agar plates with a nurse strain of Staphyloccocus aureus for 24 h and re-isolated on chocolate agar and and 37°C (20,25). Pigs were inoculated grown for 18 h at 5% CO2 intratracheally (IT) as described by Solano (26) with a 3 mL suspension of bacteria in PBS containing a target of 5 × 107 CFU/mL. Control pigs were inoculated IT with PBS alone. For inoculation, pigs were sedated with 10–15 mg/kg of a combination of tiletamine and zolacepam (Zoletil; Virbac, Madrid, Spain) and 50–100 μg/kg medetomidona (Domtor; Pfizer, Barcelona, Spain). After inoculation, sedation was reversed using atipamozole (Antisedan; Pfizer) following the manufacturer’s recommendations. Animal procedures followed the regulations of the European Directive 86/609/CEE.
Clinical signs
A veterinarian monitored the pigs each morning from birth to the day of inoculation (21 days), for the presence of clinical signs. Clinical signs associated with H. parasuis infection and rectal temperatures were recorded 1 day prior to inoculation and for 3 days after inoculation. This was done for scoring purposes, and to monitor disease severity to allow moribund animals to be euthanized humanely. Four clinical traits relevant to H. parasuis infection were scored including demeanor (0 = normal; 1 = slight lack of alertness; 2 = refusal to move; 3 = prostrate or moribund); apparent respiratory signs (0 = normal respiratory rate; 1 = slightly increased rate; 2 = increased rate; 3 = rapid rate); lameness (0 = normal; 1 = lameness in 1 limb; 2 = lameness in 2 limbs; 3 = severe lameness); and neurological signs (0 = normal; 1= muscular rigidity or tremors; 2 = convulsion; 3 = paralysis). A total score for clinical signs was calculated for each pig based on the sum of the scores for all 4 traits. Rectal temperatures were not included in the clinical signs score, and were evaluated separately.
Bacterial recovery
Nasal swabs were obtained when the pigs were 7 d of age to determine by polymerase chain reaction (PCR) if pigs were negative to H. parasuis prior to inoculation. Pigs were euthanized at days 1, 2, or 3 post-inoculation using an intravenous injection of pentobarbital. A tonsil swab was obtained from each pig at necropsy and tested by PCR. The total number of positive tonsil swabs was compared to the total number of pigs that had at least 1 positive bacterial recovery from one or more internal tissues to determine how consistently tonsils are colonized in experimentally infected pigs. A necropsy was performed, swabs were collected for bacterial recovery, and macroscopic lesions were recorded. Bacterial isolation and PCR were used for H. parasuis detection from 6 tissue surfaces: pleura, pericardium, peritoneum, carpal joint, knee joint, and meninges. Swabs for bacterial isolation and DNA extraction were collected from each of the tissues and processed within 2 h after collection. For bacterial isolation, the swabs were cultured on blood agar plates with a nurse strain of Staphylococcus aureus as a source of nicotinamide adenine dinucleotide (NAD) for growth and incubated overnight at 37°C. To confirm isolation of bacteria belonging to the genus Haemophilus, a few colonies were re-isolated in NAD-supplemented chocolate agar grown in an atmosphere containing 5% CO2 at 37°C for 24 h (20,25).
PCR detection of Haemophilus parasuis
A DNA extraction protocol was used to detect H. parasuis DNA (22). A modification was made to have 35 cycles rather than the published 30. The H. parasuis amplification primers used for PCR were: HPS-forward (5′GTG ATG AGG AAG AAG GGT GGT GT 3′) and HPS-reverse (5′GGC TTC GTC ACC CTC TGT 3′). A 20 μL total reaction mixture was used containing 9 μL of distilled water, 10 ng of template DNA or 1 μL of extracted DNA samples, 0.2 μM of each primer, 50 μM potassium chloride (KCl), 10 mM tris-hydrogen chloride (tris-HCl), 1.5 mM magnesium chloride (MgCl2), 240 μM of each denatured nucleoside triphosphate (dNTP) and 0.5 U of Taq DNA polymerase (Roche). The PCR amplification was performed after denaturation at 94°C for 5 min, with 35 cycles of denaturation for 30 s, 94°C, annealing for 30 s at 59°C, extension for 30 s at 72°C, and a final extension step for 7 min at 72°C. The amplified product was 821 base pairs (bp) and was resolved by electrophoresis of 5 μL of PCR product in a 2% agarose gel. The sensitivity of the PCR was found to be 102 CFU/mL, consistent with the results previously reported (22). For DNA extraction, a positive control was used by diluting H. parasuis in 300 μL of sterile PBS, and a negative control with sterile PBS only. For PCR, a known and well-characterized sample of extracted DNA was used as a positive control and a sample without DNA was used as a negative control.
Haemophilus parasuis detection from internal organs
A scoring system was created to measure H. parasuis recovery from internal tissues. Isolation and PCR were considered positive or negative for each tissue. An H. parasuis detection score was calculated for each animal by dividing the number of positive tissues by the total number of tissue samples collected (6 tissues). If a pig had 5 out of 6 tissues positive by isolation or PCR, the score assigned was 0.83.
Correlations between bacterial isolation and PCR detection of Haemophilus parasuis
Correlations between bacterial isolation and PCR were estimated to determine which of the 2 methods had more successful H. parasuis identifications (JMP software; SAS Institute, Cary, North Carolina, USA). Also, the number of positive bacterial isolations and PCR positive reactions were compared for each of the tissues tested to determine from which tissues H. parasuis could be detected most often. Finally, to determine how quickly bacteria spreads through internal tissues, the number of positive isolations from all tissue of pigs killed at day 1 post-inoculation was investigated.
Lesions detected at necropsy
Sites typically affected by H. parasuis infection were monitored for lesions: pleura (pleuritis), pleural cavity (hydrothorax), pericardium (pericarditis), pericardial space (hydropericardium), peritoneum (peritonitis), peritoneal cavity (ascites), meninges (meningitis), joint (arthritis), and lung (pneumonia). A score of 0–3 was assigned for pleura, pericardium, peritoneum, or lung, based on the following ranking: 0 for normal, 1 for slight, few strands of fibrin deposition, 2 for moderate or several strands of fibrin deposition, and 3 for copious fibrin deposits and purulent exudate. A score of 0–2 was assigned for pleural, pericardium, and peritoneal spaces based on the following ranking: 0 for normal, 1 for moderate fluid accumulation, and 2 for severe fluid accumulation and change of color.
Statistical analysis
First, to evaluate the degree of susceptibility in the offspring of all boars, data was subjected to a 2-way ANOVA with Sire and Test Day as fixed effects. Sire by test day interaction was not significant; therefore, it was omitted from the model. Student’s t-test was used for multiple comparisons among least mean squares (LMS) of the sires. Statistical analyses were carried out using JMP software (SAS Institute). The traits examined were individual pig scores for bacterial isolation (data for 123 animals tested), PCR (123 animals), clinical signs (118 animals), lesion (123 animals), temperature from day 0–1 (101 animals), 0–2 (69 animals), and 0–3 (37 animals). Statistical differences were considered significant when P < 0.05. Secondly, to characterize the response to H. parasuis infection, each pig was assigned a specific disease category (fully resistant, less resistant, less susceptible, fully susceptible) taking into consideration, bacterial isolation, PCR detection, clinical signs, and lesions post-infection following the definitions listed in Table I. Disease responses between boars under this categorization were compared by Chi square.
Table I.
Definitions for each disease category given to all pigs infected with H. parasuis
| Disease Category | Definition |
|---|---|
| Fully resistant (FR) | No samples positive by PCR or bacterial isolation (out of 6 tissues tested), clinical signs or lesions with a score of 1 in no more than 2 tissues (or a total lesion score of no more than 3). |
| Less resistant (LR) | Two or less samples positive by PCR and no bacterial isolation. Clinical signs and lesions with a total score of 3 or less. |
| Less susceptible (LS) | Two or less samples positive by PCR positive or isolation. Clinical signs and lesions with a total score of 3 or less. |
| Fully susceptible (FS) | Five or 6 samples positive by PCR and isolation positive. Signs and lesions typical of H. parasuis infection with a score of 3 or more. |
Results
Animal infections
Of the 200 pigs that began the experiment, 80.5% (162) survived to the challenge date. From those, 133 were infected with H. parasuis, 4 of which were removed from the study due to technical issues, and 29 that were used as controls. All the nasal swabs tested by PCR from pigs at 7 d of age were negative, confirming that pigs were free of H. parasuis prior to infection. Control pigs did not show any clinical signs, postmortem lesions, and H. parasuis was not isolated or detected by PCR from the internal organs of these animals.
Results of the statistical analysis indicated that there were differences in offspring’s susceptibility by sire for clinical signs (just above significance level, P = 0.08), lesions (P < 0.05), and temperatures from days 0–1 (P < 0.005), 0–2 (P < 0.05). No significant differences were found for bacterial recovery by isolation (P > 0.05) or PCR (P > 0.05) or for temperatures from days 0–3 (P > 0.05).
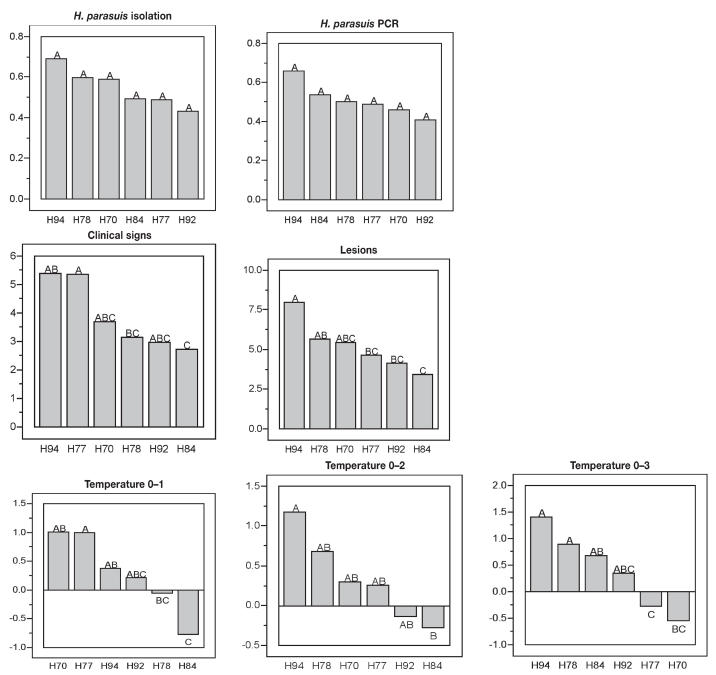
Offspring from sire H94 had the highest H. parasuis isolation and PCR scores but those differences were not significantly different to the offspring from the other sires (Figure 2). Offspring from sire H94 had, on average, the highest clinical signs scores, although only significantly higher than sire H84 (P < 0.05). The offspring of sire H94 also had the highest H. parasuis lesion scores and those differences were statistically significant when compared with the offspring of sires H77, H92, and H84 (P < 0.05) (Figure 2). Pigs sired by boar H94 had significantly higher rectal temperatures from days 0–1 and 0–2 compared with pigs from sire H84, and on day 0–3 compared with pigs from sires H77 and H70 (Figure 2). Consequently, piglets derived from boar H94 showed an overall increased susceptibility to H. parasuis infection.
Figure 2.
Ranking for the susceptibility to H. parasuis for the offspring of 6 boars of similar genetic background. A 2-way ANOVA model was used with Sire and Test Day as fixed effects. Student’s t-test was used for multiple comparisons among least mean squares (LMS). The LMS of the score is shown for each trait measured (isolation, PCR) in the Y-axis. Sires, shown in the X-axis, with similar letter did not differ significantly at P < 0.05, in the X-axis.
In contrast, offspring of boars H92 and H84 had on average the lowest isolation, PCR, and clinical signs and lesions scores (Figure 2). The offspring of sire H92 had significantly different lesion scores than the offspring of sire H94. Offspring of sire H84 had significantly lower clinical signs and lesion scores and lower rectal temperatures at days 0–1 and 0–2 than the offspring of sire H94. These results indicate that offspring of sire H92 and H84 were the most resistant to H. parasuis infection.
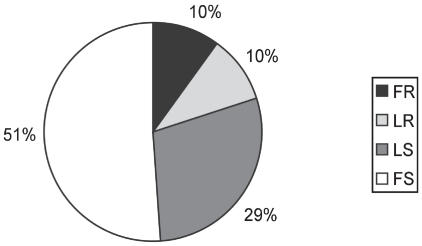
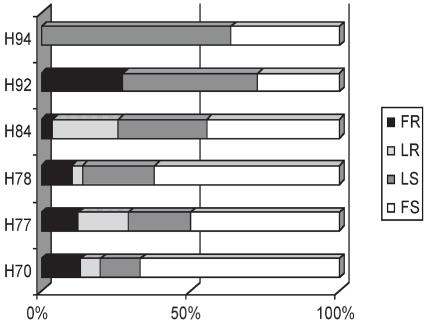
When classified into categories for relative resistance or susceptibility, 10% of the pigs were completely resistant to disease under the conditions of this study and did not show any clinical sign or lesions associated with H. parasuis infection up to 3 days post-inoculation, while the other 90% developed disease (Figure 3). Haemophilus parasuis was not detected in any of the internal organs evaluated in those pigs. The disease-resistant pigs were challenged at the same time as the diseased pigs, but H. parasuis was not detected in any of the internal organs evaluated during the course of the experiment. Fully resistant pigs were found in 6 out of 10 experimental challenges. Offspring of all sires except boar H94 (P < 0.05) had a proportion of offspring categorized as fully resistant (Figure 4); sire H94 produced more susceptible offspring. This is in contrast to sire H92, which had the highest proportion of fully resistant offspring. All sires, except H92 and H94, had some offspring categorized as less resistant, and all boars had progeny classified as either “fully susceptible” or “less susceptible” (Figure 4).
Figure 3.
Distribution of disease categories in response to a H. parasuis infection for all 126 pigs that were challenged in the study independently of which boar was used to sire them. Categories include fully susceptible (FS), less susceptible (LS), less resistant (LR), and fully resistant (FR). Disease categories were made based on results from H. parasuis isolation, PCR detection, clinical signs, and lesions post-infection.
Figure 4.
Distribution of disease category in response to a H. parasuis infection by sire. Pigs were categorized as: fully resistant (FR), less resistant (LR), less susceptible (LS), and fully susceptible (FS) following inoculation with H. parasuis. Differences in disease category distributions were found among all boars by Chi square (P < 0.05).
Correlation between bacteria isolation and PCR detection of Haemophilus parasuis
There was a correlation of 0.78 between H. parasuis isolation and PCR when scores from positive samples of all internal tissues were compared. The correlations between H. parasuis isolation and lesions was 0.50, and between PCR and lesions 0.54. At the time of necropsy only 50% of the pigs (63/126) that had had H. parasuis detected in an internal tissue were also positive by PCR in the tonsil.
Bacterial recovery and PCR detection of Haemophilus parasuis
Detection was attempted in 126 of the 129 pigs that were infected because 3 pigs were found dead at day 1 post-infection, and began to decompose; therefore, those tissues were not used. From the 126 pigs left, H. parasuis was detected by isolation more often from the meninges (59%), the peritoneum and pleura (57%), followed by the carpal joint (50%), the knee joint (51%), and the pericardium (49%). Haemophilus parasuis was also detected by PCR most commonly from the meninges (56%), the peritoneum (54%), the pleura and the knee joint (52%), the carpal joint (50%), and the pericardium (48%). When sampling a 3-tissue combination such as the meninges, peritoneum, and pleura, H. parasuis was detected in 80% of the cases by isolation and/or PCR. When sampling the meninges, peritoneum, and carpal joint, H. parasuis was detected in 78% of individuals. By comparison, H. parasuis was detected more often at the carpal joint (61%) than the knee joint (55%). Thirty-eight pigs were euthanized 1 d post-inoculation, and at that time positive isolations were made for all tissues investigated including: 50% from the meninges, 45% from the peritoneum, 42% from the pericardium, 39% from the pleura, 14% from the carpal joint, and 29% at the knee joint. Bacteria were present in all tissues tested from 7/38 pigs euthanized 1 d post-inoculation, and all tissues were negative for isolation in 10/38 pigs. These results indicate that by 24 h post- infection, the H. parasuis were capable of reaching all tissues investigated in certain pigs.
There was a significant test day [1, 2 or 3 days post-inoculation (DPI)] effect on isolation (P < 0.001), PCR (P < 0.01), clinical signs (P < 0.05), and an effect just above significance on lesion (P = 0.087). The number of lesions went up linearly from day 1 to day 3 post-inoculation while the level of infection, as measured by isolation and PCR, went down after peaking at day 2. Clinical signs continued to be high at day 3 (Table II).
Table II.
Test day effects on disease traits
| Test Day (DPI)
|
|||
|---|---|---|---|
| TRAIT | 1 | 2 | 3 |
| Isolation | 0.431b | 0.719a | 0.493b |
| Lesion | 4.039b | 5.455a,b | 6.152a |
| PCR | 0.377b | 0.639a | 0.513a,b |
| Clinical signs | 2.515b | 4.596a | 4.553a |
Means with uncommon superscript differed significantly (P < 0.05).
Discussion
Susceptibility to disease in an animal is determined by genetics and environment. Often the environment can affect the pig’s performance as much or more so than the animal’s genetic makeup (27,28). In this study, the influence of the sires on their offsprings’ response to infection was evaluated using repetitive challenges in standardized conditions to minimize environmental factors and in the absence of passive immunity from sows. Combining data from bacterial recovery, clinical signs, lesion scores, and rectal temperature, differences in susceptibility to H. parasuis infection were observed. Offspring of sires H92 and H84 were the most resistant, and offspring of H94 were the most susceptible to Glässer’s disease. A limitation of the study was the number of offspring, and therefore addressing genetic susceptibility to Glässer’s disease should be corroborated using a greater number of animals.
Evidence of host genetics influencing infectious and noninfectious diseases has been documented for several diseases in pigs. For instance, resistance to F18 and K88 Escherichia coli is conferred by a lack of bacterial receptors (11,29– 31). Sires have been reported to influence innate immunity traits that are crucial for defense against infection to Salmonella choleraesuis in an experiment using limited number of boars and experimental challenge infections in a pig resource population (17). The genetic mutation responsible for the porcine stress syndrome is well known in the swine industry and commonly used for genetic selection (32). Differences in susceptibility to PRRSV in different pig breeds (33,34) have also been reported.
Ten percent of the pigs in this study were not affected by challenge with H. parasuis, as they were negative for bacterial isolation and PCR detection in internal organs, and did not have clinical signs or lesions. These findings were observed across most of the groups challenged. It could be argued that pigs evaluated this early (up to 3 d post-infection) had the potential to manifest the disease later had they been left to live, and should not be categorized as truly resistant. Although that is a possibility, it is unlikely, particularly considering that the bacterial challenge contained a large number of infectious organisms, and that colostrum-deprived pigs are very susceptible animals. Pigs deprived of sow colostrum do not acquire the maternal immunity necessary for protection against several pathogens, notably H. parasuis, and often succumb to infections caused by the normal pig flora. Evidence of this was seen in the loss of almost 20% of the pigs in this study that did not survive to the challenge date.
Immune traits were not measured in this study due to the short interval between challenge and euthanasia. Typically active immune response to H. parasuis and other microbes is not expected to develop within 3 d post-exposure. Blanco et al (23) were unable to reproduce H. parasuis disease in pigs of the same genetic background, age, and farm of origin when the piglets had ingested colostrum for 2 days and remained with a sow that had a history of natural exposure to H. parasuis. Those findings indicated that antibodies provided to the pig by passive immunity are instrumental in protecting against Glässer’s disease as reported by Solano-Aguilar et al (5). Blanco et al (23) also determined that pigs fed with bovine colostrum under the same conditions of the present study did not show blood antibody titers to H. parasuis detectable by using a commercial ELISA kit (Biovet Canada, Saint-Hyacinthe, Quebec). Therefore, the bovine colostrum used during the experiments likely did not provide any protective antibodies to H. parasuis.
All pigs that were susceptible to H. parasuis infection developed serositis. Only 18% developed arthritis, and 3% developed meningitis in spite of H. parasuis being detected by PCR and isolation was 63–64% from joints and 57% from meninges. It is interesting that only a fraction of the pigs developed lesions of meningitis or lameness in spite of bacteria often being detected in meninges and joints. It is possible that given more time, those pigs could have developed more obvious lesions, or perhaps the host is able to control bacterial growth and prevent the lesions from developing in those tissues. The dynamics of how the bacteria interact with joint tissues and meninges remains to be determined.
To be able to cause meningitis, bacteria have to cross the blood-brain barrier (BBB). Vanier et al (35) evaluated adherence and invasion of H. parasuis in porcine brain microvascular endothelial cells, the endothelium lining of brain capillaries that forms the BBB. The authors reported that Nagasaki serovar 5, the same strain used in this study, has a high level of adherence and was able to adhere to and invade the endothelial cells. Although intracellular viable H. parasuis was found in the endothelial cells up to 6 h after antibiotic treatment, the microvascular endothelial cells infected did not show any abnormality (35). Further studies are necessary to understand the steps that lead to development of lesions.
A correlation of 0.78 between bacterial isolation and the PCR suggests that the PCR is an appropriate tool for detection of H. parasuis. Although H. parasuis detection was more successful by using bacterial isolation than PCR, the H. parasuis PCR is still useful because the fastidious nature of H. parasuis often limits isolation from clinical samples, particularly in the cases where pigs are found dead and bacterial isolation is not possible due to contamination with other bacteria. The best chances for bacterial detection by isolation or PCR were observed when a combination of meninges, peritoneum, and pleura were used (80% of cases). This information can be of use to the diagnostician, as these tissues should be used as primary sampling sites. The results from this study indicate that the level of infection, as measured by isolation and PCR, lowers after peaking at day 2; therefore, care should be taken about when to attempt bacterial detection.
Detection of H. parasuis from tonsils was possible in 50% of cases when bacteria were also detected in at least 1 internal tissue (results not shown). Consistent isolation from tonsil swabs in experimentally infected pigs has been reported (25, 36) but results from this study were inconsistent with those findings. However, in an experimental challenge using intranasal infection of H. parasuis in cesarean-derived colostrum-deprived pigs, Vahle et al (37) were unable to isolate H. parasuis consistently from the tonsil. The interpretation of colonization of H. parasuis in the upper respiratory tract, particularly the tonsil, remains controversial as suggested by Oliveira (20).
Identifying gene markers associated with differences in susceptibility to disease is one way of enabling selective breeding of pigs that are more resistant to Glässer’s disease. A population that varies in susceptibility to H. parasuis was found in this study, which reports differences in susceptibility to H. parasuis in pigs and host genetic factors that influence the progression of H. parasuis infection. Tissue derived from these animals can be used for identification of genes and DNA markers associated with those differences in susceptibility. The DNA markers can be used to increase the accuracy of selection for complex breeding objectives, which may include resistance to Glässer’s and other pig diseases, that will be expressed as robust growth and decreased mortality in commercial environments.
Acknowledgments
This project was funded by the European Community contract number QLK2-2000-00726. The authors are grateful to Drs. Simone Oliveira and Carlos Pijoan, and the members of the PathoCHIP team for the support provided for this project. Our thanks are also extended to the Vall Companys Grupo Castellano for their cooperation.
References
- 1.Nielsen R. Pathogenicity and immunity studies of Haemophilus parasuis serotypes. Acta Vet Scand. 1993;34:193–198. doi: 10.1186/BF03548209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rapp-Gabrielson VJ, Gabrielson DA, Schamber GJ. Comparative virulence of Haemophilus parasuis serovars 1 to 7 in guinea pigs. Am J Vet Res. 1992;53:987–993. [PubMed] [Google Scholar]
- 3.Rapp-Gabrielson VJ. Haemophilus parasuis. In: Straw BE, editor. Diseases of Swine. 7. Ames, Iowa: Iowa State Univ Pr; 1999. pp. 475–482. [Google Scholar]
- 4.Rosendal S. Comparative virulence of porcine Haemophilus bacteria. Can J Comp Med. 1985;49:68–74. [PMC free article] [PubMed] [Google Scholar]
- 5.Solano-Aguilar GI, Pijoan C, Rapp-Gabrielson V, Collins J, Carvalho LF, Winkelman N. Protective role of maternal antibodies against Haemophilus parasuis infection. Am J Vet Res. 1999;60:81–87. [PubMed] [Google Scholar]
- 6.Solano GI, Baustista E, Molitor TW, Segales J, Pijoan C. Effect of porcine reproductive and respiratory syndrome virus infection on the clearance of Haemophilus parasuis by porcine alveolar macrophages. Can J Vet Res. 1998;62:251–256. [PMC free article] [PubMed] [Google Scholar]
- 7.Galina L, Pijoan C, Sitjar M, Christianson WT, Rossow K, Collins JE. Interaction between Streptococcus suis serotype 2 and porcine reproductive and respiratory syndrome virus in specific pathogen-free pigs. Vet Rec. 1994;134:60–64. doi: 10.1136/vr.134.3.60. [DOI] [PubMed] [Google Scholar]
- 8.Vasco SG. Specific resistance to the African swine fever virus (ASFV) RPCV. 1991;86:76–82. [Google Scholar]
- 9.Oura CA, Powell PP, Anderson E, Parkhouse RM. The pathogenesis of African swine fever in the resistant bush pig. J Gen Virol. 1998;79:1439–1443. doi: 10.1099/0022-1317-79-6-1439. [DOI] [PubMed] [Google Scholar]
- 10.Meijernk E, Fries R, Vögeli P, et al. Two alpha (1,2) fucosyltranferase genes on porcine chromosome 6q11 are closely linked to the blood group inhibitor (s) and Escherichia coli F18 receptor (ECF-8R) loci. Mamm Genome. 1997;10:736–741. doi: 10.1007/s003359900556. [DOI] [PubMed] [Google Scholar]
- 11.Vögeli P, Meijerink E, Fries R, et al. A molecular test for the detection of E. coli F18 receptors: A breakthrough in the struggle against edema disease and post weaning diarrhea in swine. Schweiz Arch Tierheilkd. 1977;139:479–484. [PubMed] [Google Scholar]
- 12.Sellwood R. Escherichia coli diarrhoea in pigs with or without the K88 receptor. Vet Record. 1979;105:228–230. doi: 10.1136/vr.105.10.228. [DOI] [PubMed] [Google Scholar]
- 13.Meeker DL, Rothschild MF, Christian LL, Warner CM, Hill HT. Genetic control of immune response to pseudorabies and atrophic rhinitis vaccines: II. Comparison of additive direct and maternal genetic effects. J Anim Sci. 1987;64:414–419. doi: 10.2527/jas1987.642414x. [DOI] [PubMed] [Google Scholar]
- 14.Lunney JK, Butler JE. Immunogenetics. In: Rothschild MF, Ruvinsky A, editors. Genetics of the Pig. Wallingford, UK: CAB Intnl; 1998. pp. 163–197. [Google Scholar]
- 15.Mallard BA, Wilkie BN, Kennedy BW, Quinton M. Use of estimated breeding values in a selection index to breed yorkshire pigs for high and low immune and innate resistance factors. Anim Biotechnol. 1992;3:257–280. [Google Scholar]
- 16.Magnusson U, Wilkie BN, Mallard BA, Rosendal S, Kennedy B. Mycoplasma hyorhinis infection of pigs selectively bred for high and low immune response. Vet Immunol Immunopathol. 1998;61:83–96. doi: 10.1016/s0165-2427(97)00132-3. [DOI] [PubMed] [Google Scholar]
- 17.Van Diemen PM, Kreukniet MB, Galina L, Bumstead N, Wallis TS. Characterization of a resource population of pigs screened for resistance to salmonellosis. Vet Immunol and Immunopath. 2002;88:183–189. doi: 10.1016/s0165-2427(02)00165-4. [DOI] [PubMed] [Google Scholar]
- 18.Berthelot F, Beaumont C, Mompart F, Girard-Santosuosso O, Pardon P, Duchet-Suchaux M. Estimated heritability of the resistance to cecal carrier state of Salmonella enteritidis in chickens. Poult Sci. 1998;77:797–801. doi: 10.1093/ps/77.6.797. [DOI] [PubMed] [Google Scholar]
- 19.Cole RK, Hutt FB. Genetic difference in resistance to Newcastle disease. Avian Dis. 1961;5:205–214. [Google Scholar]
- 20.Oliveira S, Pijoan C. Haemophilus parasuis: New trends on diagnosis, epidemiology and control. Vet Microbial. 2004;99:1–12. doi: 10.1016/j.vetmic.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 21.Kim J, Chung HK, Junt T, Cho WS, Choi C, Chae C. Postweaning multisystemic wasting syndrome of pigs in Korea: prevalence, microscopic lesions and coexisting microorganisms. J Vet Med Sci. 2002;64:57–62. doi: 10.1292/jvms.64.57. [DOI] [PubMed] [Google Scholar]
- 22.Oliveira S, Galina L, Pijoan C. Development of a PCR test to diagnose Haemophilus parasuis infections. J Vet Diagn Invest. 2001;13:495–501. doi: 10.1177/104063870101300607. [DOI] [PubMed] [Google Scholar]
- 23.Blanco I, Galina-Pantoja L, Oliveira S, Pijoan C, Sánchez C, Canals A. Comparison of Haemophilus parasuis infection in colostrum-deprived and sow reared piglets. Vet Microbiol. 2004;103:21–27. doi: 10.1016/j.vetmic.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 24.Oliveira S, Galina L, Blanco I, Canals A, Pijoan C. Naturally-farrowed, artificially-reared pigs as an alternative model for experimental infection by Haemophilus parasuis. Can Vet J Res. 2003;67:146–150. [PMC free article] [PubMed] [Google Scholar]
- 25.Segalés J, Domingo M, Solano GI, Pijoan C. Immunohistochemical detection of Haemophilus parasuis serovar 5 in formalin-fixed, paraffin-embedded tissues of experimentally infected swine. J Vet Diagn Invest. 1997;9:237–243. doi: 10.1177/104063879700900303. [DOI] [PubMed] [Google Scholar]
- 26.Solano G, Pijoan C. A simple technique for tracheal culture to detect respiratory pathogens in live pigs. Swine Health Prod. 1997;5:30–31. [Google Scholar]
- 27.Frank JW, Richert BT, Schinckel AP, Belstra BA, Ellis M, Grant AL. Purdue University Swine Report. 1997. Effects of environment, genotype, sex, and antibiotic treatment on pig growth, carcass characteristics, and pork quality. [Google Scholar]
- 28.Williams NH, Stahly TS, Zimmerman DR. Effect of level of chronic immune system activation on the growth and dietary lysine needs of pigs fed from 6 to 112 kg. J Anim Sci. 1997;75:2481–2496. doi: 10.2527/1997.7592481x. [DOI] [PubMed] [Google Scholar]
- 29.Frydendahl K, Kåre Jensen T, Strodel-Andersen TK, Fredholm M, Evans G. Association between the porcine Escherichia coli F18 receptor genotype and phenotype and susceptibility to colonisation and post-weaning diarrhoea caused by E. coli O138:F18. Vet Microbiol. 2003;93:39–51. doi: 10.1016/s0378-1135(02)00348-6. [DOI] [PubMed] [Google Scholar]
- 30.Meijerink E, Neuenschwander E, Fries R, et al. A DNA polymorphism influencing alpha (1,2) fucosyltransferase activity of the pig FUT1 enzyme determines susceptibility of small intestinal epithelium to Escherichia coli F18 adhesion. Immunogenetics. 2000;52:129–136. doi: 10.1007/s002510000263. [DOI] [PubMed] [Google Scholar]
- 31.Sellwood R. Escherichia coli diarrhoea in pigs with or without the K88 receptor. Vet Rec. 1979;105:228–230. doi: 10.1136/vr.105.10.228. [DOI] [PubMed] [Google Scholar]
- 32.O’Brien PJ, Bal RO. Porcine Stress Syndrome. In: Straw BE, editor. Diseases of Swine. Ames, Iowa: Iowa State Univer Pr; 1999. pp. 475–482. [Google Scholar]
- 33.Christopher-Hennings J, Holler LD, Benfield DA, Nelson EA. Detection and duration of porcine reproductive and respiratory syndrome virus in semen, serum, peripheral blood mononuclear cells, and tissues from Yorkshire, Hampshire, and Landrace boars. J Vet Diagn Invest. 2001;13:133–142. doi: 10.1177/104063870101300207. [DOI] [PubMed] [Google Scholar]
- 34.Halbur PG, Rothschild MF, Thacker BJ, Meng XJ, Paul PS, Bruna JD. Differences in susceptibility of Duroc, Hampshire, and Meishan pigs to infection with a high virulence strain (VR2385) of porcine reproductive and respiratory syndrome virus (PRRSV) J Anim Breed Genet. 1998;115:181–189. [Google Scholar]
- 35.Vanier G, Szczotka A, Friedl P, Lacouture S, Jaques M, Gottschalk M. Haemophilus parasuis invades porcine brain microvascular endothelial cells. Microbiology. 2006;152:135–142. doi: 10.1099/mic.0.28312-0. [DOI] [PubMed] [Google Scholar]
- 36.Amano H, Shibata M, Kajio N, Morozumi T. Pathologic observations of pigs intranasally inoculated with serovar 1, 4 and 5 of Haemophilus parasuis using immunoperoxidase method. J Vet Med Sci. 1994;56:639–644. doi: 10.1292/jvms.56.639. [DOI] [PubMed] [Google Scholar]
- 37.Vahle JL, Haynes JS, Andrews JJ. Experimental reproduction of Haemophilus parasuis infection in swine: Clinical, bacteriological, and morphologic findings. J Vet Diagn Invest. 1995;7:476–480. doi: 10.1177/104063879500700409. [DOI] [PubMed] [Google Scholar]