Abstract
A rice genic male-sterility gene ms-h is recessive and has a pleiotropic effect on the chalky endosperm. After fine mapping, nucleotide sequencing analysis of the ms-h gene revealed a single nucleotide substitution at the 3′-splice junction of the 14th intron of the UDP-glucose pyrophosphorylase 1 (UGPase1; EC2.7.7.9) gene, which causes the expression of two mature transcripts with abnormal sizes caused by the aberrant splicing. An in vitro functional assay showed that both proteins encoded by the two abnormal transcripts have no UGPase activity. The suppression of UGPase by the introduction of a UGPase1-RNAi construct in wild-type plants nearly eliminated seed set because of the male defect, with developmental retardation similar to the ms-h mutant phenotype, whereas overexpression of UGPase1 in ms-h mutant plants restored male fertility and the transformants produced T1 seeds that segregated into normal and chalky endosperms. In addition, both phenotypes were co-segregated with the UGPase1 transgene in segregating T1 plants, which demonstrates that UGPase1 has functional roles in both male sterility and the development of a chalky endosperm. Our results suggest that UGPase1 plays a key role in pollen development as well as seed carbohydrate metabolism.
Keywords: Oryza sativa, genic male sterility, chalky endosperm, UGPase1, RNAi, complementation test
Introduction
Male sterile (ms) mutants have been reported in many species of higher plants as the result of both spontaneous and induced mutations (Kaul, 1998). The mutations are associated with a range of different phenotypes, including structural aberrations such as short filaments (Mulligan et al., 1994), lack of dehiscence (Dawson et al., 1999) or pollen with a smooth surface (Ariizumi et al., 2003), and also with functional defects associated with gametogenesis, specifically meiosis (Dawson et al., 1993; Glover et al., 1998; He et al., 1996; Moritoh et al., 2005; Peirson et al., 1996; Ross et al., 1997; Sanders et al., 1999). All of these mutations result in non-functional pollen. A number of genes associated with male sterility have been identified in diverse plant species such as Arabidopsis (Aarts et al., 1993; Ariizumi et al., 2003, 2004; Thorlby et al., 1997; Wilson et al., 2001), wheat (Block et al., 1997; Klindworth et al., 2002), Chinese cabbage (Miao et al., 2003), soybean (Jin et al., 1998), tomato (Gorman et al., 1996), sunflower (Chen et al., 2006; Perez-Vich et al., 2005) and chives (Engelke and Tatlioglu, 2000).
Male sterility is conditioned by either cytoplasmic-specific (CMS) or genetic (chromosomal) male sterility (GMS) genes. In rice, male sterility is classified into four major groups: male sterility caused by CMS, photoperiod-sensitive GMS (PGMS), thermo-sensitive GMS (TGMS) and other genic male sterilities (Kurata et al., 2005). The CMS lines require a combination of male-sterile and fertility restorer lines to maintain a hybrid system, whereas an alteration of environmental conditions, such as day length and/or temperature, can restore fertility in PGMS and TGMS lines (Liu et al., 2001; Wang et al., 2003). However, the prediction and control of environmental factors, especially of temperature, is not always possible in the field. Abnormal weather can bring the temperature down below the critical level that is required to regain fertility in TGMS lines, which is simply called fertility conversion. This results in a potential problem for the seed production of two-line hybrid rice, such as the mixture of real hybrids with selfed seeds. To ensure high-quality hybrid seed production from P/TGMS lines, molecular markers can be used to help remove false hybrids from the mixture. In the past, several morphological markers, such as pale leaves (Dong et al., 1995) or purple leaves (Mou et al., 1995), have been employed for marking P/TGMS lines. However, removing false hybrid seedlings must be performed manually, which is labor-intensive and cannot ensure that false hybrids have been completely eliminated.
In a previous publication, Koh and Heu (1995) reported on the discovery of a new, chemically induced GMS gene, ms-h, and showed that it was recessive and associated with the chalky endosperm character. They suggested that the gene might be useful in a hybrid seed production system, and discussed its effectiveness compared with other systems. The ms-h gene was mapped to the distal region of chromosome 9 and was demonstrated to have a pleiotropic effect on the chalky endosperm (Koh et al., 1999). Most reported male sterility genes are closely linked to, or have pleiotropic effects on, deleterious characteristics, making them poor candidates for use in economically viable hybrid seed production. On the contrary, because the pleiotropic effect of the recessive ms-h gene is expressed only in the seeds of the homozygous male-sterile (mother) plants, this character is useful for predicting which individuals will produce heterozygous F1 hybrid progeny, based on an examination of the seeds prior to planting.
The rice genome contains two homologous UDP-glucose pyrophosphorylase (UGPase) genes, UGPase1 on chromosome 9 (Abe et al., 2002) and UGPase2 on chromosome 2 (GenBank accession number AF249880). The UGPase2 gene is 80% similar at the cDNA nucleotide sequence level, and is 88% identical at the amino acid sequence level, to UGPase1. Both UGPase1 and UGPase2 are ubiquitously expressed throughout rice development, and UGPase1 is expressed at much higher levels than UGPase2. Remarkably, UGPase1 transcripts are present at higher levels in florets before flowering, suggesting that it plays a special role in rice flower development (Chen et al., 2007).
In this paper, we report on the map-based isolation of the ms-h gene, and on the identification of a single nucleotide substitution in the UGPase1 gene that leads to the production of nonfunctional proteins with abnormal sizes, and results in male sterility and the chalky endosperm character.
Results
High-resolution mapping of the ms-h gene
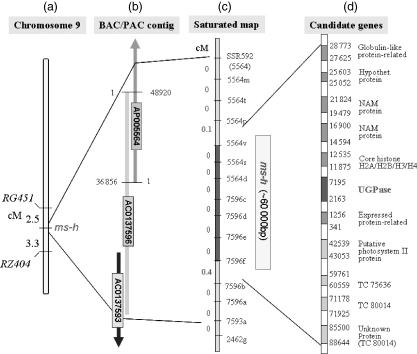
The ms-h gene was previously mapped to the long arm of chromosome 9 in the interval delimited by RFLP markers, RG451 and RZ404, at a distance of 2.5 and 3.3 cM, respectively (Figure 1a; Koh et al., 1999). For fine mapping of the ms-h gene, an F2 population was derived from a cross between the Hwacheong ms-h mutant (temperate japonica) and Milyang 23 (an indica-like Tongil-type variety), and 1051 F2 plants were evaluated for phenotypic segregation of male fertility and sterility by examining spikelet fertility and chalky endosperm in F3 seeds. To identify additional markers closely linked to the ms-h gene, we designed 15 STS (sequence-tagged site) and 12 CAPS (cleaved amplified polymorphic sequence) markers based on available rice genome sequences within the interval containing the ms-h gene (Table 1). To identify genomic targets for CAPS marker design, we first compared publicly available rice genome sequences in the target region between the japonica variety, Nipponbare and the indica variety, 9311, using the Gramene database (http://www.gramene.org) and NCBI Blast (http://www.ncbi.nlm.nih.gov). Subsequently, only those sequences with differences in the recognition sites of restriction enzymes were used as templates for designing CAPS primers. The STS and CAPS markers were used to survey F2 plants, and the ms-h gene was found to be flanked by STS markers, 5564p and 7596b, at a distance of 0.1 and 0.4 cM, respectively. The interval spanned a region defined by two overlapping PAC/BAC clones, AP005564 and AC137596, on chromosome 9 (Figure 1b). Nine recombinant individuals were identified between markers 7596f and 7596b within an interval of 14 451 bp. Seven STS markers co-segregated with the ms-h locus in all the mutant plants. As a result of this map-based cloning experiment, the region containing the ms-h gene was narrowed down to a 60-kb region flanked by STS markers, 5564v and 7596f (Figure 1c).
Figure 1.
- Linkage map of ‘ms-h’ with flanking markers on chromosome 9 (Koh et al., 1999).
- The PAC/BAC contigs encompassing the ms-h gene region.
- The saturated linkage map of the region containing the ‘ms-h’ locus based on a genetic analysis of the F2 population (1051 plants). The ms-h gene was flanked by the STS (sequence-tagged site) markers, 5564p and 7596b, at a distance of 0.1 and 0.4 cM, respectively.
- The grey boxes indicate a total of 11 candidate genes contained in the approximately 60-kb DNA region between two STS markers, 5564p and 7596b. Sequence comparison of candidate genes between the original parent and the mutant revealed a single nucleotide substitution in the ‘UGPase1’ gene. NAM is the abbreviation for No Apical Meristem.
Table 1.
The PCR-based molecular markers designed for fine mapping
| CAPS | Forward primer (5′→3′) | Reverse primer (5′→3′) | Fragment size amplified in japonica (bp) | Restriction enzyme | Originated clone |
|---|---|---|---|---|---|
| 869a | CTTCCCCGAGGTAGGTGCTA | CAGGCACATCAACAATTCCA | 1296 | RsaI | AP006548 |
| 869c | TCCAGCAGAGTCTCCATCAA | CACAGTCATCACATGCATCATT | 1377 | AluI, MspI | AP006548 |
| RG451 | TCCATAAGATCGTTCATCTGG | GTGTAAACCCTGGATGTGATG | 550 | MnlI | AP005862 |
| 18420b | TTTTGGTCGTGACCGTGTAA | AGGCTCATATCAACGCGAAA | 1311 | AluI, MnlI, Tru9I, | AP006149 |
| 2505a | AAAAATCTTGGCACCAGAGG | GAATTTTGATGTGGGAGCTG | 1582 | XbaI, DraI, Sau3AI | AP006548 |
| STS | Forward primer (5′→3′) | Reverse primer (5′→3′) | Fragment size amplified in japonica (bp) | Originated clone |
|---|---|---|---|---|
| SSR592 | ACATCATGGGCTTTCCAAAC | GCTATCCGATCGATACCTTCC | 269 | AP005742 |
| 5564m | CACTTTGGTTAGGCCGACTC | GCGTAAGACCTCCCTCCAAT | 165 | AP005564 |
| 5564t | CAGGTGACCAGGTGGAATTT | TGCCTACTTTGGGTTTGTTTG | 422 | AP005564 |
| 5564p | CGGATCAGCTAAGAGCGATT | ACCACGCGAGGTATGAGC | 156 | AP005564 |
| 5564v | TCTCCATGACCAACCTATTGC | CAAGGGAGAGTTTCCTCACG | 160 | AP005564 |
| 5564s | CTCTTGCCGTGCTATGTGAA | TCAAACTCCAAAACCCAAGC | 396 | AP005564 |
| 5564d | CCTGCCATCTCTTCAAGCAT | TCAAGTTCACACAGCCAAGT | 292 | AP005564 |
| 7596c | CAAAGCGGACAGAAAACGAT | TCTGGTTTTTAGCTATGCCGTA | 136 | AC137596 |
| 7596d | CGGCTTCTTTCCTCTTTCG | GGAGTATGAGGAGGGGAAGG | 178 | AC137596 |
| 7596e | TCGCTACTTTTACCGCATCC | CAAAACCAGTGGGCTACACC | 221 | AC137596 |
| 7596f | GAACTTTAGAAAAGGTAAGGCTTCT | CAGTTTGATTGCACCATTGC | 188 | AC137596 |
| 7596b | GATGACGCCCAACAATCTCT | GGACTATAGGCCGTTCCTTG | 143 | AC137596 |
| 7596a | TCTGAGTGGTTGGTTTGTCG | GGGTGTACTGTGGGATTTCG | 178 | AC137596 |
| 7593a | AAGAACATGTACCCTACGAACACA | TTTTTCTTCCTCACCAGAACAA | 279 | AC137593 |
| 2462d | TTCTTTTTCATGCCCCACTC | GATCCGGACAGGTTCGTTTA | 1526 | AC137593 |
UGPase1 is the candidate for the ms-h gene
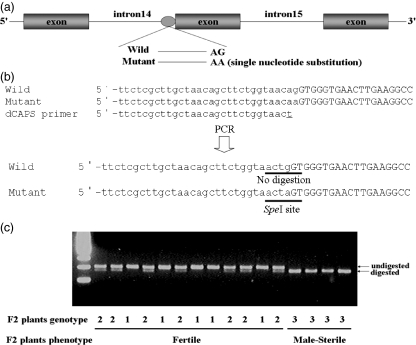
Eleven candidate genes were identified in the 60-kb target interval based on in silico genome annotation (http://rgp.dna.affrc.go.jp; http://www.tigr.org/tdb/e2k1/osa1; Figure 1d). To identify the best candidate for the ms-h gene among these genes, we sequenced all 11 gene candidates in the ms-h mutant and in the wild-type (wt), Hwacheong, and compared them with the corresponding sequences in the publicly available genome sequence for cv. Nipponbare. This comparison identified a point mutation in the UGPase1 gene that distinguished the ms-h mutant from both Hwacheong and Nipponbare. The critical polymorphism was a single nucleotide substitution of Guanine to Adenine, corresponding to the final nucleotide at the 3′-splice junction of the 14th intron of the UGPase1 gene (Figure 2a).
Figure 2.
- The mutation within the UGPase1 gene in the ms-h mutant.
- dCAPS marker development for detection of the 1-bp substitution at the 3′-splice junction of the 14th intron. The dCAPS marker using a mismatch primer selectively generates a restriction site (SpeI) in the mutant, but not in the wild-type parent. Lowercase letters of sequences indicate the 14th intron and uppercase letters indicate the 15th exon.
- dCAPS marker genotype of F2 plants from Hwacheong ms-h mutant × Hwacheong, classified by phenotype. After digestion with restriction enzyme SpeI, a single, 196-bp PCR product was observed in fertile homozygotes (genotype 1), whereas in the male-sterile homozygotes (genotype 3), a shorter, 169-bp PCR product was observed, resulting from the generation of a new SpeI recognition site resulting from the single nucleotide substitution. In fertile heterozygotes (genotype 2), both fragments are observed. Genotype 1, Ms-h/Ms-h; genotype 2, Ms-h/ms-h; genotype 3, ms-h/ms-h.
To further explore the association between this single nucleotide polymorphism (SNP) in the UGPase1 gene and the male-sterile phenotype of the ms-h mutant, we designed a dCAPS marker to detect the functional base substitution, and used it to trace the inheritance of the ms-h mutation in an F2 population derived from a cross between the Hwacheong ms-h mutant and wt Hwacheong. dCAPS analysis offers a robust and accurate tool for detecting SNPs without sequencing, and it is particularly valuable for analyzing F2 segregation because dCAPS markers are co-dominant and can readily distinguish heterozygotes from homozygotes (Michaels and Amasino, 1998; Neff et al., 1998). We constructed a dCAPS marker consisting of a mismatch primer UGP1-CAPS-F that generated a SpeI site specifically in the ms-h mutant (Figure 2b). Using the primer set UGP1-CAPS-F and UGP1-CAPS-R a 196-bp DNA fragment could be amplified from all F2 plants. When the 196-bp fragments were digested with SpeI (recognition sequence, A/CTAGT), F2 plants showing spikelet sterility displayed a short fragment as a result of digestion with SpeI, whereas F2 plants showing spikelet fertility had a longer, undigested fragment (Figure 2c). Some F2 plants showing spikelet fertility contained both fragments, indicating that these plants were heterozygous for the alleles of both wt and mutant UGPase1.
To examine whether the G-to-A mutation at the 3′-splice junction of the 14th intron of the UGPase1 was present as a natural variant in other cultivars, we performed dCAPS analysis on seven additional cultivars, including four japonicas (Ilpum, Dongjin, Nagdong and TR22183), two Tongil types (Dasan and Milyang 23) and one indica type (IR36). All seven cultivars demonstrated only one undigested fragment (data not shown). This supported our hypothesis that the 1-bp mutation identified in the UGPase1 gene is responsible for the male sterility of the ms-h mutant.
Analysis of ms-h transcripts and enzyme activity assays based on deduced amino acid sequences
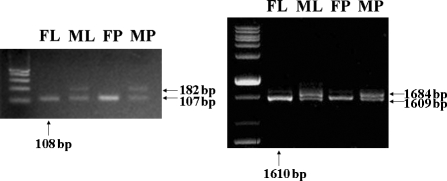
As this point mutation occurred at the splice site between the 14th intron and 15th exon, we performed RT-PCR analysis of the ms-h mutant and wt Hwacheong using two sets of UGPase1-specific primers to investigate whether the pre-mRNA splice site was altered. One set of UGPase1-specific primers, UGP1-PRT primers, was designed from the sequences of the 14th and 15th exons, to fully span the 14th intron region, and the second set, UGP1-FRT primers, was used to amplify a full-length UGPase1 cDNA. As shown in Figure 3, wt Hwacheong displayed only a 108-bp fragment in RT-PCR with UGP1-PRT primers, whereas the ms-h mutant contained two fragments, a 182-bp fragment as well as a shorter fragment that appeared identical to the 108-bp fragment observed in the wt, Hwacheong. When the shorter fragment amplified from the ms-h mutant was cloned and sequenced, the RT-PCR product showed a 1-bp deletion in the spliced message, compared with the corresponding sequence of the wt RT-PCR product, although this 1-bp difference was not detectable on the PAGE gel. Moreover, the 182-bp fragment amplified from the ms-h mutant was revealed to contain the entire, unspliced 14th intron (74 bp).
Figure 3.
RT-PCR analysis with UGPase1-specific primers. As a result of each RT-PCR using the UGP1-PRT primer set (left) and the UGP1-FRT primer set (right), wild-type (wt) Hwacheong displayed only the expected size fragment in both reactions; whereas the ms-h mutant contained two fragments, one similar to the wt and a second, longer fragment. The same banding pattern was observed in both leaf and panicle. The arrows pointing upwards indicate the fragment size of the wt Hwacheong, and the arrows pointing left represent the fragment size of the ms-h mutant. Abbreviations: FL, fertile leaf; ML: male-sterile leaf; FP, fertile panicle; MP, male-sterile panicle.
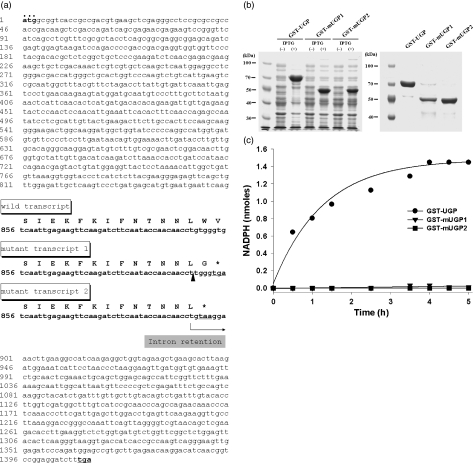
The deduced amino acid sequences of the ms-h mutant transcripts displaying two abnormal sizes suggests that both the 1-bp deletion and the 74-bp insertion cause frame shifts that generate two independent stop codons in the process of translation, resulting in truncated 299- and 298-aa proteins, instead of the 469-aa protein encoded by the wt UGPase1 transcript (Figure 4a). To further confirm whether two C-terminal deleted UGPase1 proteins of the ms-h mutant are nonfunctional, we performed enzyme activity assays in vitro. The UGPase1 cDNAs encoding full-length protein of the wt Hwacheong and two truncated proteins of the ms-h mutant were amplified by PCR with each specific primer, and were transformed into Escherichia coli after vector construction. Glutathione S-transferase (GST)-tagged three-recombinant proteins were purified and separated by SDS-PAGE (Figure 4b). UGPase activity assays were performed with recombinant proteins by monitoring the NADPH formation at 340 nm. As expected, enzyme activity of GST-UGP containing the full-length UGPase1 protein appeared, whereas both GST-mUGP1 and GST-mUGP2 did not show any enzyme activity, indicating that two C-terminal deleted proteins were nonfunctional (Figure 4c). Thus, this single base substitution in the splice site found in the ms-h mutant appears to cause unstable splicing, leading to the presence of two mature transcripts, both with abnormal sizes and, because of the corresponding stop codons, the mRNA transcripts are translated into a truncated and a nonfunctional UGPase1 protein, respectively.
Figure 4.
- Nucleotide sequence of the UGPase1 open reading frame, and alignment of the nucleotide and deduced amino acid sequence of the 14th exon 3′-region derived from wild-type (wt) Hwacheong and from the ms-h mutant. Nucleotide numbering starts from the start of translation; the protein sequence is derived from the nucleotide sequence. Wild transcript denotes the UGPase1 transcript of wt Hwacheong, whereas mutant transcripts 1 and 2 represent the 1-bp deleted transcript and the 74-bp inserted transcript found in the ms-h mutant, respectively. The deduced amino acid sequences of the two ms-h mutant transcripts with abnormal sizes demonstrate that both the 1-bp deletion and the 74-bp insertion cause a frame shift and generate stop codons, resulting in truncated 299- and 298-aa proteins, instead of the full-length 469-aa protein. The start codon is set in bold with a row of dots above it. The underlined sequences and asterisk indicate termination codons. The 1-bp deletion site is marked by an arrowhead.
- SDS-PAGE of recombinant proteins induced with isopropyl-beta-D-thiogalactopyranoside (left) and purified-recombinant UGPase1 proteins (right). The molecular weight of glutathione S-transferase (GST) (23-kDa)-tagged recombinant UGPase1 proteins agrees with the estimated values of 74 and 56 kDa for the full-length UGPase1 protein (469 aa) and the two C-terminal deleted proteins (299 and 298 aa).
- Activity assays of three recombinant UGPase1 proteins. The formation of NADPH was calculated from the absorption changes at 340 nm monitored for 5 h using an NADPH molar extinction coefficient of 6.22 × 103 m−1 cm−1. •, GST-UGP, GST-tagged recombinant protein containing full-length UGPase1; ▴, GST-mUGP1, GST-tagged recombinant protein containing C-terminal deleted 299-aa UGPase1; ▪, GST-mUGP2, GST-tagged recombinant protein containing C-terminal deleted 298-aa UGPase1.
RNAi-mediated silencing of the UGPase1 gene causes male sterility
To confirm that the UGPase1 gene is causally related to male fertility, we generated UGPase1-RNAi transgenic plants by exploiting double-stranded RNA (dsRNA)-mediated interference to silence the target gene (Baulcombe, 2002; Moritoh et al., 2005; Prasad and Vijayraghavan, 2003). The RNAi construct included 473-bp of the gene-specific sequence, corresponding to the full-length UGPase1 cDNA, which was linked with the intronic sequence in the antisense and sense configurations, and then placed under the control of the constitutive 35S promoter (Figure 5a). The UGPase1-RNAi construct was introduced into calli derived from wt Hwacheong immature embryos by Agrobacterium-mediated transformation, with an empty vector used as a control. Thirty-three independent transformants were regenerated, and the presence of the RNAi construct was confirmed by PCR using Bar-F and Bar-R primers (data not shown). At the spikelet ripening stage, five of the transformed lines displayed low fertility, and two lines were male sterile (Figure 5b–e). Moreover, these transformed lines showed pleiotropic developmental abnormalities similar to the Hwacheong ms-h mutant phenotype, including reduced culm length and retarded growth (Table 2).
Figure 5.
- Schematic diagrams of the pUGP1RNAi construct for double-stranded RNA interference and the pUGP1COM construct used for the complementation test. In pUGP1RNAi, the 473-bp gene-specific fragment of the UGPase1 gene was linked with the intron in both antisense and sense orientations, such that the transcripts were expected to create a dsRNA stem with a single-stranded loop. Phenotype of UGPase1-RNAi plants (b–g).
- Hwacheong plants after ripening: containing the empty vector (left) and transformed by pUGP1RNAi (right).
- Photograph (c) is the double enlargement of a part of the photo (b).
- Panicles of a vector-transformed plant and a UGPase1-RNAi plant at anthesis (left) and after ripening (right).
- Flower and anther morphology of a vector-transformed plant (left) and UGPase1-RNAi plant (right) at the heading stage. To view the anthers, the lemma was ripped off.
- I2-KI staining of pollen grains from a vector-transformed plant at the heading stage, showing the presence of normal, round and starch-filled grains.
- I2-KI staining of pollen grains from a UGPase1-RNAi plant at the heading stage, showing the presence of abnormal, small and non-stained grains caused by the lack of starch. Phenotypic complementation by introduction of the UGPase1 gene (h–m).
- Phenotype of Hwacheong ms-h mutants after ripening: plants containing the empty vector (left) and complemented by the introduction of pUGP1COM (right).
- Photograph (i) is the triple enlargement of a part of the photo (h).
- Panicles of a vector-transformed plant and the complemented plant at anthesis (left) and after ripening (right).
- Flower and anther morphology of an empty vector-transformed plant (left) and the complemented plant (right) at heading stage.
- I2-KI staining of pollen grains from a empty vector-transformed plant at heading, showing the presence of abnormal and non-stained grains.
- I2-KI staining of pollen grains from the complemented plant at heading. This photograph shows the presence of normal grains, indicating the restoration of fertility. The scale bar corresponds to 100 μm.
Table 2.
Morphological characteristics of transgenic plantsa
| Line | Heading date | Culm length (cm) | Panicle length (cm) | Spikelet fertility |
|---|---|---|---|---|
| wt Hwacheong | August 23 | 87.4 | 18.3 | Fertile |
| r23b | August 22 | 63.8 | 18.1 | Sterile |
| Difference | ns | ** | ns | |
| Hwacheong gms | August 21 | 62.5 | 18.0 | Sterile |
| c10c | August 22 | 85.5 | 18.2 | Fertile |
| Difference | ns | ** | ns |
The original T0 plant was grown in the field by crown division.
r23: UGPase1 silenced transformant.
c10: UGPase1 complemented transformant.
Significant at the 0.01 probability level.
ns, not significant.
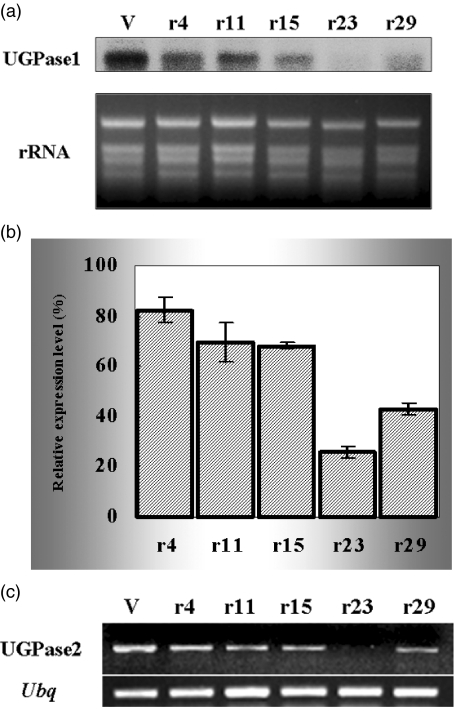
When pollen viability was compared in high- and low-fertility lines, by staining for starch with I2-KI solution, the five low-fertility transformants showed light pollen staining as compared with empty vector-transformed plants that displayed normal starch accumulation, and pollens from the two male-sterile transformants (r23 and r29) did not stain for starch (Figure 5f,g). If the low or no-staining phenotypes of UGPase1-RNAi transformants were caused by the introduced dsRNA, we would expect to see reduced UGPase1 transcription levels in these transgenic plants. When Northern blot analysis was used to examine UGPase1 expression levels in RNA samples harvested from spikelets at the booting stage in transgenic lines, it could be seen that transcription was most severely suppressed in the two male-sterile transgenic lines (r23 and r29), and was partially suppressed in the low-fertility transformants, compared with empty vector-transformed plants (Figure 6a). Subsequently, we examined the expression of UGPase1 transcripts in more detail in seven transgenic lines using real-time quantitative RT-PCR analysis. First-strand cDNAs that were reverse-transcribed with oligo (dT) were used as the template for the quantitative PCR analysis. Results were computed to show relative expression levels in UGPase1-RNAi transformants compared with a vector-transformed plant using ubiquitin as a standard. As can be seen in Figure 6b, UGPase1 transcriptional levels were slightly suppressed in the low-fertility lines (r4, r11 and r15), whereas the levels were severely reduced in the ms transformants, to 26% in r23 and 43% in r29. To determine whether the expression of a dsRNA interference construct towards UGPase1 affects the expression of UGPase2, we analyzed UGPase2 expression by semi-quantitative RT-PCR of the total RNA extracted from spikelets. As shown in Figure 6c, UGPase2 transcription in most RNAi transformants was just slightly suppressed, but UGPase2 transcripts of the r23 line were completely suppressed, similar to the suppression pattern of UGPase1 transcripts.
Figure 6.
- Northern blot analysis of UGPase1 gene expression. The upper panel shows the RNA gel blot probed with the NotI fragments of UGP1 i pGEMT containing the 473-bp gene-specific region of the UGPase1 gene. The lower panel shows ethidium bromide stained rRNA as a loading control.
- Quantitative RT-PCR analysis of UGPase1 gene expression. The expression value was normalized with a ubiquitin control, and the results represent the average values of duplicate experiments shown as relative expression levels compared with empty vector-transformed plants.
- Semi-quantitative RT-PCR analysis of the expression of the UGPase2 gene. Amplification of the ubiquitin gene was used as a control. V, empty vector-transformed plant; r4, r11, r15, r23 and r29: UGPase1-RNAi plants.
Taken together, the results indicate that the RNA interference of UGPase1 causes male sterility in proportion to the transcriptional suppression of UGPase1, and that the endogenous UGPase mRNAs, including UGPase1 and UGPase2, are degraded globally in UGPase1-RNAi transformants, leading to developmental growth retardation. The existence of homologous UGPase genes in rice raises the question as to whether each has a unique and independent function, or whether they share related or redundant functions. The incomplete co-suppression of UGPase1 and UGPase2 in RNAi-silenced transformants in this study implies that there may be a complementary interaction between the two homologous genes, despite the fact that virtually nothing is known about an interaction between the two homologous UGPase genes in rice.
Transgenic complementation of ms-h mutation
To further confirm that the point mutation in the UGPase1 gene causes male sterility in rice, we complemented the ms-h phenotype by introducing an overexpression construct containing the wt UGPase1 sequence into homozygous ms-h mutants (Figure 5a). An empty vector was again introduced as a control. Transformants containing the complementation vector were selected on hygromycin, and 29 transgenic lines were regenerated. PCR screening using HPT-F and HPT-R primers identified 11 transgenic lines that contained the expression construct, and these were grown in a greenhouse and investigated for spikelet fertility and other morphological characteristics at maturity.
Figure 5h–m shows that the introduction of the wt UGPase1 gene complements the mutant phenotype. This finding is confirmed by the formation of filled grains, pollens that stain clearly with I2-KI solution and the normal formation of anthers and fertile panicles. The number of filled and empty spikelets was counted on two representative panicles per plant. Overexpression of the UGPase1 gene in transgenic ms-h plants resulted in spikelet fertility that ranged from 33.4% to 10.2%. Although the degree of fertility restoration differs among plants, the occurrence of filled seeds in the ms-h background is a significant indicator of complementation. Moreover, all of these transformants recovered a wt Hwacheong phenotype with normal morphology (Table 2). RT-PCR analysis with the UGP1-PRT-F and UGP1-PRT-R primer set showed amplification of a single, strong-intensity fragment that was similar in size to that seen in the fertile wt Hwacheong control (Figure 7a). When this fragment was subcloned and sequenced, it was found to include a mixture of two fragments: one that was identical to the wt UGPase1 gene transcript and one that corresponded to the abnormal fragment derived from the ms-h mutant. In the meantime, complementation of UGPase1 has no effect on the expression of UGPase2 (data not shown). Accordingly, this complementation test further confirmed that functional disruption of the UGPase1 gene is responsible for male sterility in rice.
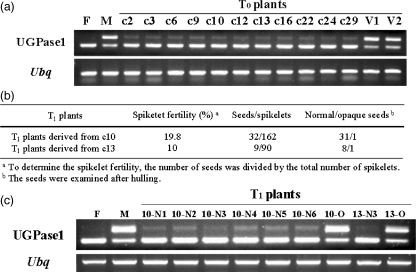
Figure 7.
- RT-PCR analysis of T0 plants.
- Spikelet fertility and the ratio of normal to opaque seed of T1 plants derived from T0 plants (c10 and c13).
- RT-PCR analysis of T1 plants derived from T0 plants (c10 and c13). The upper panel shows a gel electrophoresis pattern resulting from RT-PCR using the UGP1-PRT primer set. The lower panel shows gel electrophoresis following RT-PCR of the ubiquitin gene as a control. F, wild-type plant (fertile Hwacheong); M, Hwacheong gms mutant; c2, c3, c6, c9, c10, c12, c13, c16, c22, c24 and c29, complemented plants; V1 and V2, empty vector transformed plants; 10-N1, 10-N2, 10-N3, 10-N4, 10-N5 and 10-N6, T1 plants derived from normal seeds of the c10 transformant; 10-O, T1 plant derived from opaque seeds of the c10 transformant; 13-N3, T1 plant derived from a normal seed of the c13 transformant; 13-O, T1 plant derived from an opaque seed of the c13 transformant; Ubq, ubiquitin.
The ms-h gene has a pleiotropic effect on chalky endosperm
Koh et al. (1999) previously reported on the co-segregation of the ms-h gene and the development of a chalky endosperm. To confirm that the chalky endosperm results from a pleiotropic effect of the ms-h gene, we evaluated both male-sterile and male-fertile transgenic progeny to determine whether opaque seeds were always associated with ms-h. In the case of UGPase1-RNAi transformants, two male-sterile transgenic T0 lines (r23 and r29) were crossed with wt Hwacheong as the male parent to obtain F1 progeny. Seven F1 seeds were planted and observed for spikelet fertility and the occurrence of opaque seeds (chalky endosperm) after ripening of the spikelets. From a cross between r23 × Hwacheong, only one F1 progeny was obtained, which was male sterile. This result was predicted based on the Northern blot analysis of the r23 line that demonstrated the efficiency with which the RNAi construct suppressed UGPase1. At the same time, two out of six F1 progenies derived from r29 × Hwacheong were male sterile, one had a low spikelet fertility (21.5%), and three were male fertile, with spikelet fertilities ranging from 78.1% to 80.8%. Remarkably, three fertile or partially fertile F1 progenies produced a small number of opaque F2 seeds, at an average ratio of one-fiftieth (Table 3). This suggests that expression of functional UGPase1 may be intermittently suppressed in these second-generation RNAi transgenic lines, or that the RNAi construct is imperfectly transmitted from one generation to the next.
Table 3.
Spikelet fertility and the ratio of normal to opaque seeds of UGPase-RNAi transformant F1 progenies
| No. | Cross combination | Spikelet fertility (%)a | Seeds/ spikelets | Normal/opaque seedsb |
|---|---|---|---|---|
| r23 | r23 × Hwacheong | 0 | 0/259 | 0 |
| r29 | r29 × Hwacheong | 0 | 0/191 | 0 |
| r29 | r29 × Hwacheong | 0 | 0/130 | 0 |
| r29 | r29 × Hwacheong | 21.5 | 83/386 | 81/2 |
| r29 | r29 × Hwacheong | 80.8 | 156/193 | 153/3 |
| r29 | r29 × Hwacheong | 78.1 | 171/219 | 168/3 |
| r29 | r29 × Hwacheong | 78.5 | 128/163 | 128/0 |
To determine the spikelet fertility, the number of seeds was divided by the total number of spikelets.
The seeds were examined after hulling.
The T1 seeds harvested from 11 T0 transgenic lines produced in the complementation test were also examined for chalkiness after hulling. Chalky grains were segregated in two (c10 and c13) of the 11 plants (Figure 7b). The opaque T1 seeds harvested from c10 and c13 transformants were planted with other normal T1 seeds to verify the pleiotropism with male sterility. After maturing, we confirmed the co-segregation of the ms-h gene and seed opaqueness, based on phenotypic examination and molecular analysis of UGPase1-RNA expression patterns (Figure 7c). These results clarify that the ms-h gene has a pleiotropic effect on the chalky endosperm, which is consistent with the previous study (Koh et al., 1999).
Discussion
A single nucleotide substitution within a splice site generates abnormal-size transcripts as a result of unstable splicing
In this study, we showed that the male-sterile phenotype observed in the ms-h mutant resulted from a point mutation within the UGPase1 gene. Splicing depends on the presence of signal sequences in the pre-mRNA. In almost all genes the first two nucleotides at the 5′-end of an intron are GT, and the last two at the 3′-end are AG (Green, 1991; Moore and Sharp, 1993). According to this GT-AG rule for RNA splicing (Cai et al., 1998; Isshiki et al., 1998), it is reasonable to expect that a single base change from G to A, where the first base of the 15th exon is a G, would cause a one-base downstream shift of the AG site at the 3′-end of the 14th intron in the UGPase1 gene. The result of this SNP is the formation of a 1-bp deleted transcript resulting from alternative splicing (Figure 4a). However, the formation of a 74-bp inserted transcript indicates that pre-mRNA splicing of the UGPase1 gene in the ms-h mutant is an unstable process. Although introns are ubiquitous and share a high degree of structural/sequence similarity across species, the signals that specifically define splice sites are not completely understood. Two models for splice site selection have been suggested in the initial recognition of exon/intron borders: exon definition and intron definition. Although most of the early studies of splice site mutants in vertebrates favored the exon definition of splicing (Berget, 1995), initial reports from plants tend to favor the intron definition (Goodall and Filipowicz, 1989; Lou et al., 1993; McCullough et al., 1993). The escape from splicing according to the intron definition may be a step towards the interpretation of the 74-bp intron insertion in the UGPase1 transcripts reported in the ms-h mutant in this study.
The point mutation of the UGPase1 gene causes the loss of UGPase activity
UGPase presents in all prokaryotic and eukaryotic organisms. It catalyzes the reversible conversion of Glc-1-P and UTP into UDP-Glc (UDPG) and pyrophosphate (PPi), a key precursor for polysaccharide synthesis (Feingold and Avigad, 1980;Kleczkowski, 1994). The putative three-dimensional structure of the barley UGPase monomer is bowl-like, with an active site positioned in a central groove (Kleczkowski et al., 2004). This shape is common for AGPase and UGPase, and perhaps for all pyrophosphorylase-like proteins (Peneff et al., 2001). The active site of UGPase contains several amino acid residues that have been shown to be important for substrate binding and catalysis of the enzyme. For example, five lysyl residues (Lys263, Lys329, Lys367, Lys409 and Lys410) participate in substrate binding and catalysis in potato (Katsube et al., 1991; Kazuta et al., 1991). Other results also showed that several lysine residues are necessary for UGPase activity, although positions are somewhat variable among species (Eimert et al., 1996; Pua et al., 2000). Interestingly, rice also has similar positioning of five lysine residues (Lys257, Lys263, Lys323, Lys329, Lys361 and Lys367) as in potato, except for additional Lys440 for substrate binding in rice. In addition, position 168 (NQS) and 307 (NLS) are putative glycosylation sites, and position 420 (SER) is a phosphorylation site (Sowokinos et al., 2004). In addition, previous results suggested that oligomerization of UGPase plays a regulatory role in any process requiring UDPG as a substrate, and that the C-terminus is responsible for oligomeric conformation (Geisler et al., 2004). Incidentally, the mutant UGPase1 encodes two C-terminal deleted 299- and 298-aa proteins, instead of one functional full-length (469-aa) protein. Thereby, we assume that the truncated two proteins do not have enzymatic activity. In fact, our enzymatic assay showed that these two mutant proteins have no UGPase activity (Figure 4c), supporting the hypothesis that the C-terminus is required for the enzymatic activity of UGPase.
UGPase plays a key role in pollen and endosperm development
Most male-sterile mutants are controlled by monogenic recessive genes, and have defects in sporogenic tissues, tapetal cells, pollen mother cells, microspores and/or pollen at the pre-meiotic, meiotic and post-meiotic stages of anther and pollen development (Singh, 2003; Twell, 2002). Many male-sterile lines are characterized by a perturbed carbohydrate metabolism (Dorion et al., 1996). Carbohydrates are considered to play a critical role in anther and pollen development. They are not only energy sources that sustain growth but they also take part in cell-wall biosynthesis during pollen development (Clément and Audran, 1995; Goetz et al., 2001). UDPG, a key substrate/product of the enzyme for carbohydrate metabolism in both the source and sink tissues, is used directly or indirectly in the biosynthesis of cell-wall polysaccharides, reflecting the key role of UDPG as a precursor for cell-wall biogenesis (Gibeaut, 2000). An unloading pathway via the functional coupling of cell-wall invertase with a monosaccharide transporter is prominent in symplastically isolated pollen cells (Ji et al., 2005; Oliver et al., 2005; Roitsch et al., 2003), and the subsequent conversion of Glc-1-P metabolized from apoplastically cleaved sucrose into UDPG by UGPase is a vital process for pollen cell-wall biosynthesis. These results indicated that UGPase participates in an essential process for pollen development. More recently, it has been reported that rice UGPase is essential for pollen callose deposition, and its co-suppression results in a TGMS (Chen et al., 2007). Our previous results showed that pollen development in the ms-h mutant was arrested at the binucleate or trinucleate microspore stage because of uneven meiosis, and implied interference with cell-wall formation during pollen meiosis (Koh and Heu, 1995). Thus, our findings support that UGPase is a key component that controls pollen development.
Our previous study also showed that starch granules in the endosperm of ms-h mutants are more roundish, polyhedral and smaller than those of wt Hwacheong. The starch structure of the ms-h mutant has a higher frequency of long glucose chain amylose and a shorter branching of amylopectin than wt Hwacheong (Sohn et al., 1997). Starch is synthesized by apoplastic or symplastic pathways (Kleczkowski et al., 2004), and the ADP-Glc (ADPG) that is required for its synthesis is provided via two mechanisms (Denyer et al., 1996). First, it is synthesized via the cytosolic ADP glucose pyrophosphorylase (AGPase), in which case a transporter is required to transfer ADPG into the plastid. Second, it is synthesized via the plastidial AGPase, in which case a supply of plastidial Glc-1-P is required (Denyer et al., 1996). However, starch biosynthesis in the endosperm cells of cereals such as maize and barley mostly starts with the cytosolic synthesis of ADPG followed by the subsequent import of this compound into the storage plastid, which is dependent on an extra plastidial AGPase (Denyer et al., 1996). Besides, the production of UDPG by UGPase is coupled to the activity of cytosolic AGPase in the cytosol of cereal seed endosperm, which implies that UGPase directly regulates ADPG levels by affecting its synthesis by AGPase (Kleczkowski, 1994). Chlamydomonas mutants with lesions in the pathway of ADPG synthesis, which presumably have reduced levels of ADPG, lack the long-chain fraction of amylopectin that is present in normal starch (Van den Koornhuyse et al., 1996).
Therefore, our study on the endosperm of ms-h mutants with short branching of amylopectin provides clues indicating that the opaque phenotype arises from the alteration of the starch structure by the insufficient supply of long amylopectin chains, as a result of the ADPG synthesis reduction caused by the disorder of UGPase. The interaction between UGPase activity and starch in the opaque phenotype must be closely related, although the mechanism remains unclear. Therefore, the way in which UGPase participates in carbohydrate metabolism during endosperm development is worthy of further study.
Experimental procedures
Plant materials and genotype evaluation
A male-sterile mutant, Hwacheong ms-h, was induced via chemical mutagenesis using N-methyl-N-nitrosourea from a Korean japonica cultivar, Hwacheongbyeo (Koh and Heu, 1995). The F2 population used for fine mapping was derived from a cross between the Hwacheong ms-h mutant (japonica) and Milyang 23 (tongil-type rice, derived from an indica × japonica cross, and which was similar to indica). F2 plants (1051) were classified as either male sterile or fertile (wt) based on an examination of spikelet fertility, and to distinguish heterozygotes from homozygous wt plants, F3 seeds harvested from fertile F2 plants were evaluated for the presence of a chalky endosperm after hulling.
Genetic mapping
Total genomic DNA was extracted from the leaves of both parents and from each F2 individual using the method of McCouch et al. (1988). Based on results from a prior mapping experiment, closely linked RFLP (restriction fragment length polymorphism) markers were used to identify DNA sequences within the ms-h region using an in silico approach (http://www.gramene.org; http://rgp.dna.affrc.go.jp; http://www.tigr.org/tdb/e2k1/osa1; http://www.genome.arizona.edu). To fine-map the ms-h gene, 15 STS and 12 CAPS markers were developed based on available rice genome sequence data. The STS and CAPS primers used in this work, along with the corresponding restriction enzymes for the CAPS markers, are listed in Table 1. PCR products were digested completely with specific restriction enzymes, and were then size-separated on 1–2% agarose gels containing 0.15 μg ml−1 ethidium bromide and 0.5× Tris-Borate-EDTA running buffer.
Linkage analyses were performed with the segregation data in the F2 populations using map maker version 3.0 (Lander et al., 1987). Genetic distances between markers were calculated in Kosambi centi Morgans (cM).
Sequence alignments and dCAPS analysis
Overlapping DNA fragments across the ms-h region were amplified by PCR. PCR fragments were then purified and analyzed by direct sequencing with a Big Dye Terminator Cycle sequencing kit using an ABI 377 sequencer (Applied Biosystems, http://www.appliedbiosystems.com). The results of sequencing were aligned with the original parent. For dCAPS analysis, PCR amplification with the primer set UGP1-CAPS-F (5′-TTCTCGCTTGCTAACAGCTTCTGGTAACT-3′) and UGP1-CAPS-R (5′-ATCAACTTCCTGTGAATACCAACTGCTTT-3′) was performed using 10 ng of extracted DNA in a total volume of 25 μl containing 1X reaction buffer, 0.5 mm deoxyribonucleotide triphosphate, 0.4 μm of each primer, and 1 U of Taq DNA polymerase (Bioneer, http://www.bioneer.com). A total reaction of 35 cycles was programmed for 30 sec at 94°C, 30 sec at 65°C and 1 min at 72°C in a Thermal Cycler (Bio-Rad, http://www.bio-rad.com). Each PCR product (5 μl) was digested with SpeI in a total volume of 20 μl at 37°C overnight. After digestion, 5 μl of each digest was electrophoresed in a 3% agarose gel.
RT-PCR and amino acid annotation
Total RNA was isolated using the SV Total RNA Isolation kit (Promega, http://www.promega.com) following the manufacturer's instructions. A 1-μg aliquot of total RNA was reverse-transcribed using an oligo (dT) primer and an M-MLV Reverse Transcriptase kit (Promega). Of the synthesized first-strand cDNAs, 2% was used for PCR analysis with two sets of UGPase1-specific primers: for target region amplification, UGP1-PRT-F (5′-CCCTGATGAGCATGTGAATG-3′) and UGP1-PRT-R (5′-TCAGCTTCTACCAGCCTCTTG-3′) primers were used; for full-length cDNA amplification, UGP1-FRT-F (5′-CATATCTCCCGTCCTTTC-3′) and UGP1-FRT-R (5′-ATGAAATACAACGCCCTTGG-3′) primers were used. The amplification reaction was carried out using the following conditions: 5 min at 94°C, 35 cycles of 1 min at 94°C, 1 min at 60°C and 2 min at 72°C, with a final extension step of 10 min at 72°C. Amplification products were recovered and sequenced. The amino acid sequences of RT-PCR products were deduced and compared with the original parents using NCBI BLAST (http://www.ncbi.nlm.nih.gov/blast/Blast.cgi).
Preparation of recombinant UGPase proteins and enzyme activity assay
The UGPase1 cDNAs encoding the full-length protein of the wt Hwacheong and the two truncated proteins of the ms-h mutant were amplified by PCR and inserted into pGEX4T-1 (Amersham, http://www.amersham.com) to produce the GST-UGP containing full-length UGPase1 (469 aa), the GST-mUGP1 containing the C-terminal deleted 299-aa protein and the GST-mUGP2 containing the C-terminal deleted 298-aa protein. Three constructs were transformed into Escherichia coli BL21/DE3 (pLysS) cells. The transformed cells were treated with isopropyl-beta-D-thiogalactopyranoside to induce recombinant protein expression. The recombinant proteins were purified according to the supplier's instructions. The protein concentrations were determined by the Bradford assay (Bio-Rad).
UGPase activity assays were performed with the recombinant proteins of GST-UGP, GST-mUGP1 and GST-mUGP2 using a one-step spectrophotometric method (Sowokinos et al., 1993). Reaction mixtures (pH 7.5) contained (in 1 ml): 5 μmol of MgCl2, 0.6 μmol of NADP, 1 μmol of UDP-glucose, 1 U of phosphoglucomutase, 1 U of glucose-6-phosphate dehydrogenase, 20 μmol of Cys, 80 μmol of glycylglycine, 0.02 μmol of Glc-1,6-diP and 1 μg (0.1 μg μl−1) of a recombinant protein. Reactions were initiated with 2.5 μmol of PPi. The formation of NADPH (340 nm) was monitored continuously at 30°C until the reaction rate was no longer linear. All assays were run with minus PPi blanks to correct for any contaminating NADPH production.
Vector constructs and rice transformation
To generate the UGPase1-RNAi construct for UGPase1 gene suppression, a 473-bp fragment of UGPase1 cDNA was amplified using first primers UGP1-RNAi-F (5′-AAAAAGCAGGCTACCACCTGATCCATAACCAG-3′) and UGP1-RNAi-R (5′-AGAAAGCTGGGTGTTTGATGGGTTTGTTCTGG-3′), and was subcloned into pGEM-T (Promega). This construct was denoted as UGP1 i pGEM-T and its sequence was verified. The UGP1 i pGEM-T was amplified using second primers attB1 (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCT-3′) and attB2 (5′-GGGGACCACTTTGTACAAGAAAGCTGGGT-3′), and the resulting attB-PCR products were cloned into the GatewayTM pDONR 201 cloning vector, which carries two recombination sites (attL1 and attL2), by BP clonase reaction (Invitrogen, http://www.invitrogen.com). Subsequently, these entry clones with UGPase1 were inserted in opposite directions into two regions, each flanked by recombination sites (attR1 and attR2) in the destination vector, pB7GWIWG2(II) (VIB-Ghent University, Belgium), using an LR clonase reaction (Invitrogen; http://www.invitrogen.com). The resulting RNAi construct was denoted as pUGP1RNAi. For the complementation test using the UGPase1 gene, a PCR-amplified UGPase1 full-length cDNA was digested with SacI and inserted into the pCamLA overexpression vector, a pCambia 1300-modified vector containing a 35S promoter and Tnos terminator. The resulting overexpression construct was denoted pUGP1COM. Agrobacterium strain LBA 4404 harboring pUGP1RNAi and pUGP1COM was used to transform rice calli induced from the mature embryos of the normal Hwacheong and Hwacheong ms-h mutants, respectively, according to the method described by Hiei et al. (1994). UGPase1-RNAi plants were regenerated from transformed calli by selecting for phosphinotricin resistance, and the transformants for the complementation test were selected for hygromycin resistance. The regenerated plants were confirmed by PCR analysis with each antibiotic resistance-specific primer: for the Bar gene, Bar-F (5′-CATCGCAAGACCGGCAACAGGATTCAA-3′) and Bar-R (5′-GCTCCACTGACGTTCCATAAATTCCCC-3′) primers were used; for the HPT gene, HPT-F (5′-GTAAATAGCTGCGCCGATGG-3′) and HPT-R (5′-TACTTCTACACAGCCATCGG-3′) primers were used.
Pollen and spikelet fertility
Pollen fertility was determined at anthesis using a 1% iodine-potassium iodide (I2-KI) solution, as described by Shinjyo (1969). The numbers of dark blue (stainable) and reddish brown (unstainable) pollen grains in each individual were counted under an optical microscope. Plants with <5% stainable pollen and zero seed setting of bagged panicles were classified as sterile, and all others were regarded as fertile. At the same time, fertility/sterility was confirmed by self-pollination tests.
Real-time quantitative RT-PCR and Northern blot analysis
For real-time quantitative RT-PCR analysis of RNAi plants, QuantiTectTM SYBR Green PCR kit (Qiagen, http://www.qiagen.com) and the Rotor-Gene 2000 (Corbett Research, http://www.corbettlifescience.com) were used according to the manufacturer's instructions. RNA isolation from spikelets at the booting stage was carried out using the TRI-ZOLTM reagent from Invitrogen. A 1-μg aliquot of total RNA treated with DNaseI (Invitrogen) was reverse-transcribed using an oligo (dT) primer and AMV Reverse Transcriptase (Promega). Of the synthesized first-strand cDNAs, 10% were used for PCR analysis with different sets of gene-specific primers: for UGPase1, real-RNAi-UGP1-F (5′-CCCTGATGAGCATGTGAATG-3′) and real-RNAi-UGP1-R (5′-CTGCAGTTTCGAGTTGCAGA-3′) primers were used; for ubiquitin, RUB2-F1 (5′-AATCAGCCAGTTTGGTGGAGCTG-3′) and RUB2-R1 (5′-ATGCAAATGAGCAAATTGAGCACA-3′) primers were used as a control (Wang et al., 2000). For semiquantitative RT-PCR of UGPase2 cDNA, primer pair UGP2-F (5′-TCATCAGATCAGCGTGAAGC-3′) and UGP2-R (5′-GCCCACTCACAAGGAGAAAA-3′), based on the 5′- and 3′-untranslated regions of rice UGPase2, were used (Chen et al., 2007). For Northern blot analysis, a 10-μg aliquot of total RNAs was separated by electrophoresis in a 1.5% (w/v) formaldehyde agarose gel and then transferred to a Hybond-N+ nylon membrane (Amersham). The membrane was hybridized with 32P-radiolabeled partial UGPase1 cDNA probes, a NotI fragment of UGP1 i pGEMT and was then washed using standard procedures (Sambrook and Russell, 2001).
Acknowledgments
This research was supported by a grant (code#CG3111) from the Crop Functional Genomics Center of the 21st Century Frontier Research Program funded by the Ministry of Science and Technology, Republic of Korea. The corresponding author extends acknowledgements to IAEA/FAO for Fellowship support (IAEA/RCA, RAS/5/037) during the early phase of this study at Cornell University in 2002.
References
- Aarts MGM, Dirkse WG, Stiekema WJ, Pereira A. Transposon tagging of a male sterility gene in Arabidopsis. Nature. 1993;363:715–717. doi: 10.1038/363715a0. [DOI] [PubMed] [Google Scholar]
- Abe A, Niiyama H, Sasahara T. Cloning of cDNA for UDP-glucose pyrophosphorylase and the expression of mRNA in rice endosperm. Theor. Appl. Genet. 2002;105:216–221. doi: 10.1007/s00122-002-0927-z. [DOI] [PubMed] [Google Scholar]
- Ariizumi T, Hatakeyama K, Hinata K, Sato S, Kato T, Tabata S, Toriyama K. A novel male-sterile mutant of Arabidopsis thaliana, faceless pollen-1, produces pollen with a smooth surface and an acetolysis-sensitive exine. Plant Mol. Biol. 2003;53:107–116. doi: 10.1023/B:PLAN.0000009269.97773.70. [DOI] [PubMed] [Google Scholar]
- Ariizumi T, Hatakeyama K, Hinata K, Inatsugi R, Nishida I, Sato S, Kato T, Tabata S, Toriyama K. Disruption of the novel plant protein NEF1 affects lipid accumulation in the plastids of the tapetum and exine formation of pollen, resulting in male sterility in Arabidopsis thaliana. Plant J. 2004;39:170–181. doi: 10.1111/j.1365-313X.2004.02118.x. [DOI] [PubMed] [Google Scholar]
- Baulcombe D. RNA silencing. Curr. Biol. 2002;12:R82–R84. doi: 10.1016/s0960-9822(02)00665-6. [DOI] [PubMed] [Google Scholar]
- Berget SM. Exon recognition in vertebrate splicing. J. Biol. Chem. 1995;270:2411–2414. doi: 10.1074/jbc.270.6.2411. [DOI] [PubMed] [Google Scholar]
- Block MD, Debrouwer D, Moens T. The development of a nuclear male sterility system in wheat. Expression of the barnase gene under the control of tapetum specific promoters. Theor. Appl. Genet. 1997;95:125–131. [Google Scholar]
- Cai X, Wang Z, Xing Y, Zhang J, Hong M. Aberrant splicing of intron I leads to the heterogeneous 5′ UTR and decreased expression of waxy gene in rice cultivars of intermediate amylase content. Plant J. 1998;14:459–465. doi: 10.1046/j.1365-313x.1998.00126.x. [DOI] [PubMed] [Google Scholar]
- Chen J, Hu J, Vick BA, Jan CC. Molecular mapping of a nuclear male-sterility gene in sunflower (Helianthus annuus L.) using TRAP and SSR markers. Theor. Appl. Genet. 2006;113:122–127. doi: 10.1007/s00122-006-0278-2. [DOI] [PubMed] [Google Scholar]
- Chen R, Zhao X, Shao Z, Wei Z, Wang Y, Zhu L, Zhao J, Sun M, He R, He G. Rice UDP-glucose pyrophosphorylase1 is essential for pollen callose deposition and its cosuppression results in a new type of thermosensitive genic male sterility. Plant Cell. 2007;19:847–861. doi: 10.1105/tpc.106.044123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clément C, Audran JC. Anther wall layers control pollen sugar nutrition in Lilium. Protoplasma. 1995;187:172–181. [Google Scholar]
- Dawson J, Wilson ZA, Aarts MGM, Braithwaite AF, Briarty LG, Mulligan BJ. Microspore and pollen development in six male-sterile mutants of Arabidopsis thaliana. Can. J. Bot. 1993;71:629–638. [Google Scholar]
- Dawson J, Sozen E, Vizir I, Van Waeyenberge S, Wilson ZA, Mulligan BJ. Characterization and genetic mapping of a mutation (ms35) which prevents anther dehiscence in Arabidopsis thaliana by affecting secondary wall thickening in the endothecium. New Phytol. 1999;144:213–222. [Google Scholar]
- Denyer K, Dunlap F, Thorbjørnsen T, Keepling P, Smith AM. The major form of ADP-glucose pyrophosphorylase in maize endosperm is extra-plastidial. Plant Physiol. 1996;112:779–785. doi: 10.1104/pp.112.2.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong F, Zhu X, Xiong Z, Cheng S, Sun Z, Min S. Breeding of photo-thermoperiod sensitive genic male-sterile indica rice with a pale-green-leaf marker. Chin. J. Rice Sci. 1995;9:65–70. [Google Scholar]
- Dorion S, Lalonde S, Saini HS. Induction of male sterility in wheat by meiotic-stage water deficit is preceded by a decline in invertase activity and changes in carbohydrate metabolism in anthers. Plant Physiol. 1996;111:137–145. doi: 10.1104/pp.111.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eimert K, Villand P, Kilian A, Kleczkowski LA. Cloning and characterization of several cDNAs for UDP-glucose pyrophosphorylase from barley (Hordeum vulgare) tissues. Gene. 1996;170:227–232. doi: 10.1016/0378-1119(95)00873-x. [DOI] [PubMed] [Google Scholar]
- Engelke T, Tatlioglu T. Genetic analyses supported by molecular methods provide evidence of a new genic (St1) and a new cytoplasmic (St2) male sterility in Allium schoenoprasum L. Theor. Appl. Genet. 2000;101:478–486. [Google Scholar]
- Feingold DS, Avigad G. Sugar nucleotide transformation in plants. In: Stumpf PK, Conn EE, editors. The Biochemistry of Plants. Vol. 3. New York, USA: Academic Press; 1980. pp. 101–170. [Google Scholar]
- Geisler M, Wilczynska M, Karpinski S, Kleczkowski LA. Toward a blueprint for UDP-glucose pyrophosphorylase structure/function properties: homology-modeling analyses. Plant Mol. Biol. 2004;56:783–794. doi: 10.1007/s11103-004-4953-x. [DOI] [PubMed] [Google Scholar]
- Gibeaut DM. Nucleotide sugars and glucosyltransferases for synthesis of cell wall matrix polysaccharides. Plant Physiol. Biochem. 2000;38:69–80. [Google Scholar]
- Glover J, Grelon M, Craig S, Chaudhury A, Dennis E. Cloning and characterization of MS5 from Arabidopsis: a gene critical in male meiosis. Plant J. 1998;15:345–356. doi: 10.1046/j.1365-313x.1998.00216.x. [DOI] [PubMed] [Google Scholar]
- Goetz M, Godt DE, Guivarch A, Kahmann U, Chriqui D, Roistch T. Induction of male sterility in plants by metabolic engineering of the carbohydrate supply. Proc. Natl Acad. Sci. USA. 2001;98:6522–6527. doi: 10.1073/pnas.091097998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodall GJ, Filipowicz W. The AU-rich sequences present in the introns of plant nuclear pre-mRNAs are required for splicing. Cell. 1989;58:473–483. doi: 10.1016/0092-8674(89)90428-5. [DOI] [PubMed] [Google Scholar]
- Gorman SW, Banasiak D, Fairley C, McCormick S. A 610 kb YAC clone harbors 7cM of tomato (Lycopersicon esculentum) DNA that includes the male sterile 14 gene and a hotspot for recombination. Mol. Gen. Genet. 1996;251:52–59. doi: 10.1007/BF02174344. [DOI] [PubMed] [Google Scholar]
- Green MR. Biochemical mechanisms of constitutive and regulated pre-mRNA splicing. Annu. Rev. Cell Biol. 1991;7:559–599. doi: 10.1146/annurev.cb.07.110191.003015. [DOI] [PubMed] [Google Scholar]
- He CP, Tirlapur V, Cresti M, Peja M, Crone DE, Mascarenhas JP. An Arabidopsis mutant showing aberrations in male meiosis. Sex. Plant Reprod. 1996;9:54–57. [Google Scholar]
- Hiei Y, Ohta S, Komori T, Kumashiro T. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence of analysis of the boundaries of the T-DNA. Plant J. 1994;6:271–282. doi: 10.1046/j.1365-313x.1994.6020271.x. [DOI] [PubMed] [Google Scholar]
- Isshiki M, Morino K, Nakajima M, Okagaki RO, Wessler SR, Izawa T, Shimamoto K. A naturally occurring functional allele of the rice waxy locus has a GT to TT mutation at the 5′ splice site of the first intron. Plant J. 1998;15:133–138. doi: 10.1046/j.1365-313x.1998.00189.x. [DOI] [PubMed] [Google Scholar]
- Ji XM, Van den Ende W, Van Laere A, Cheng SH, Bennett J. Structure, evolution, and expression of the two invertase gene families of rice. J. Mol. Evol. 2005;60:615–634. doi: 10.1007/s00239-004-0242-1. [DOI] [PubMed] [Google Scholar]
- Jin W, Palmer RG, Horner HT, Shoemaker RC. Molecular mapping of a male-sterile gene in soybean. Crop Sci. 1998;38:1681–1685. [Google Scholar]
- Katsube T, Kazuta T, Tanizawa K, Fukui T. Expression in Escherichia coli of UDP-glucose pyrophosphorylase cDNA from potato tuber and functional assessment of the five lysyl residues located at the substrate-binding site. Biochemistry. 1991;30:8546–8551. doi: 10.1021/bi00099a008. [DOI] [PubMed] [Google Scholar]
- Kaul MLH. Male Sterility in Higher Plants. 1. Vol. 10. Berlin, Germany: Springer-Verlag; 1998. [Google Scholar]
- Kazuta Y, Omura Y, Tagaya M, Nakano K, Fukui T. Identification of lysyl residues located at the substrate-binding site in UDP-glucose pyrophosphorylase from potato tuber: affinity labeling with uridine di- and triphosphopyridoxals. Biochemistry. 1991;30:8541–8545. doi: 10.1021/bi00099a007. [DOI] [PubMed] [Google Scholar]
- Kleczkowski LA. Glucose activation and metabolism through UDP-glucose pyrophosphorylase in plants. Phytochemistry. 1994;37:1507–1515. [Google Scholar]
- Kleczkowski LA, Geisler M, Ciereszko I, Johansson H. UDP-glucose pyrophosphorylase – an old protein with new tricks. Plant Physiol. 2004;134:912–918. doi: 10.1104/pp.103.036053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klindworth DL, Williams ND, Maan SS. Chromosomal location of genetic male sterility genes in four mutants of hexaploid wheat. Corp Sci. 2002;42:1447–1450. [Google Scholar]
- Koh HJ, Heu MH. Agronomic characteristics of a mutant for genic male sterility-chalky endosperm and its utilization on F1 hybrid breeding system in rice. Korean J. Crop Sci. 1995;40:684–696. [Google Scholar]
- Koh HJ, Son YH, Heu MH, Lee HS, McCouch SR. Molecular mapping of a new genic male-sterility gene causing chalky endosperm in rice (Oryza sativa L.) Euphytica. 1999;106:57–62. [Google Scholar]
- Kurata N, Miyoshi K, Nonomura KI, Ymazaki Y, Ito Y. Rice mutants and genes related to organ development, morphogenesis and physiological traits. Plant Cell Physiol. 2005;46:48–62. doi: 10.1093/pcp/pci506. [DOI] [PubMed] [Google Scholar]
- Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newburg L. MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics. 1987;1:174–181. doi: 10.1016/0888-7543(87)90010-3. [DOI] [PubMed] [Google Scholar]
- Liu N, Shan Y, Wang FP, Xu CG, Peng KM, Li XH, Zhang Q. Identification of an 85-kb fragment containing pms1, a locus for photoperiod-sensitive genic male sterility in rice. Mol. Genet. Genomics. 2001;266:271–275. doi: 10.1007/s004380100553. [DOI] [PubMed] [Google Scholar]
- Lou H, McCullough AJ, Schuler MA. 3′ Splice site selection in dicot plant nuclei is position dependent. Mol. Cell. Biol. 1993;13:4485–4493. doi: 10.1128/mcb.13.8.4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCouch SR, Kochert G, Yu ZH, Wang ZY, Khush GS, Coffman WR, Tanksley SD. Molecular mapping of rice chromosomes. Theor. Appl. Genet. 1988;76:815–829. doi: 10.1007/BF00273666. [DOI] [PubMed] [Google Scholar]
- McCullough AJ, Lou H, Schuler MA. Factors affecting authentic 5′ splice site selection in plant nuclei. Mol. Cell. Biol. 1993;13:1323–1331. doi: 10.1128/mcb.13.3.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y, Dreyer F, Cai A, Jung C. Molecular markers for genic male sterility in Chinese cabbage. Euphytica. 2003;132:227–234. [Google Scholar]
- Michaels SD, Amasino RM. A robust method for detecting single-nucleotide changes as polymorphic markers by PCR. Plant J. 1998;14:381–385. doi: 10.1046/j.1365-313x.1998.00123.x. [DOI] [PubMed] [Google Scholar]
- Moore MJ, Sharp PA. Evidence of two active sites in the spliceosome provided by stereochemistry of pre-mRNA. Nature. 1993;365:364–368. doi: 10.1038/365364a0. [DOI] [PubMed] [Google Scholar]
- Moritoh S, Miki D, Akiyama M, Kawahara M, Izawa T, Maki H, Shimamoto K. RNAi-mediated silencing of OsGEN-L (OsGEN-like), a new member of the RAD2/XPG nuclease family, causes male sterility by defect of microspore development in rice. Plant Cell Physiol. 2005;46:699–715. doi: 10.1093/pcp/pci090. [DOI] [PubMed] [Google Scholar]
- Mou T, Li C, Yang G, Lu X. Genetic studies on seeding leaf color in purple rice. Chin. J. Rice Sci. 1995;9:45–48. [Google Scholar]
- Mulligan BJ, Wilson ZA, Dawson J, Kalantidis K, Vizir I, Briarty LG, Thorlby G, Morroll S, Shlumukov L. The use of male sterile mutants of Arabidopsis to identify genes essential for male gametophyte development. Flowering Newsl. 1994;17:12–20. [Google Scholar]
- Neff MM, Neff JD, Chorky J, Pepper AE. dCAPS, a simple technique for the genetic analysis of single nucleotide polymorphisms: experimental applications in Arabidopsis thaliana genetics. Plant J. 1998;14:387–392. doi: 10.1046/j.1365-313x.1998.00124.x. [DOI] [PubMed] [Google Scholar]
- Oliver SN, Van Dongen JT, Alfred SC, et al. Cold-induced repression of the rice anther-specific cell wall invertase gene OSINV4 is correlated with sucrose accumulation and pollen sterility. Plant Cell Environ. 2005;28:1534–1551. [Google Scholar]
- Peirson BN, Owen HA, Feldmann KA, Makaroff CA. Characterization of three male-sterile mutants of Arabidopsis thaliana chromosome 5: III. Sequence features of the regions of 1, 191, 918bp covered by seventeen physically assigned P1 clones. DNA Res. 1996;4:401–414. doi: 10.1093/dnares/4.6.401. [DOI] [PubMed] [Google Scholar]
- Peneff C, Ferrari P, Charrier V, Taburet Y, Monnier C, Zamboni V, Winter J, Harnois M, Fassy F, Bourne Y. Crystal structures of two human pyrophosphorylase isoforms in complexes with UDPGlc (Gal) NAc: role of the alternatively spliced insert in the enzyme oligomeric assembly and active site architecture. EMBO J. 2001;20:6191–6202. doi: 10.1093/emboj/20.22.6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Vich B, Berry ST, Velasco L, Fernandez-Martinez JM, Gandhi S, Freeman C, Heescacker A, Knapp SJ, Leon AJ. Molecular mapping of nuclear male-sterility genes in sunflower. Crop Sci. 2005;54:1851–1857. [Google Scholar]
- Prasad K, Vijayraghavan U. Double-stranded RNA interference of a rice PI/GLO paralog, OsMADS2, uncovers its second-whorl-specific function in floral organ pattering. Genetics. 2003;165:2301–2305. doi: 10.1093/genetics/165.4.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pua EC, Lim SS, Liu P, Liu JZ. Expression of a UDP glucose pyrophosphorylase cDNA during fruit ripening of banana (Musa acuminata) Aust. J. Plant Physiol. 2000;27:1151–1159. [Google Scholar]
- Roitsch T, Balibrea ME, Hofmann M, Proels R, Sinba AK. Extracellular invertase: key metabolic enzyme and PR protein. J. Exp. Bot. 2003;54:513–524. doi: 10.1093/jxb/erg050. [DOI] [PubMed] [Google Scholar]
- Ross KJ, Fransz P, Armstrong SJ, Vizir I, Mulligan BJ, Franklin FCH, Jones GH. Cytological characterization of four meiotic mutants of Arabidopsis isolated from T-DNA transformed lines. Chromosome Res. 1997;5:551–559. doi: 10.1023/a:1018497804129. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. 3. New York, USA: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Sanders PM, Bui AQ, Weterings K, McIntire KN, Hsu YC, Lee PY, Truong MT, Beals TP, Goldberg RB. Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sex. Plant Reprod. 1999;11:297–322. [Google Scholar]
- Shinjyo C. Cytoplasmic-genetic male sterility in cultivated rice, Oryza sativa L.: II. The inheritance of male sterility. Jpn J. Genet. 1969;44:149–156. [Google Scholar]
- Singh RJ. Plant Cytogenesis. 2. Boca Raton, FL, USA: CRC Press; 2003. pp. 73–92. [Google Scholar]
- Sohn YH, Koh HJ, Lee HS, Hen MH. Physicochemical characteristics of endosperm in a white-core mutant line ‘Hwacheong ms-h’ causing male sterility in rice. Korean J. Breed. 1997;29:424–430. [Google Scholar]
- Sowokinos JR, Spychalla JP, Desborough L. Pyrophosphorylases in Solanum tuberosum: IV. Purification, tissue localization, and physicochemical properties of UDP-glucose pyrophosphorylase. Plant Physiol. 1993;101:1073–1080. doi: 10.1104/pp.101.3.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowokinos JR, Vigdorovich V, Abrahamsen M. Molecular cloning and sequence variation of UDP-glucose pyrophosphorylase cDNAs from potatoes sensitive and resistant to cold sweetening. J. Plant Physiol. 2004;161:947–955. doi: 10.1016/j.jplph.2004.04.006. [DOI] [PubMed] [Google Scholar]
- Thorlby GJ, Shlumukov L, Vizir I, Yang CY, Mulligan BJ, Wilson ZA. Fine-scale molecular genetic (RFLP) and physical mapping of a 8.9cM region on the top arm of Arabidopsis chromosome 5 encompassing the male sterility gene ms1. Plant J. 1997;12:471–479. doi: 10.1046/j.1365-313x.1997.12020471.x. [DOI] [PubMed] [Google Scholar]
- Twell D. The developmental biology of pollen. In: O'Neill SD, Roberts JA, editors. Plant Reproduction. Sheffield, UK: Sheffield Academic Press; 2002. pp. 86–153. [Google Scholar]
- Van den Koornhuyse N, Libessart N, Delrue B, Zabawinski C, Decq A, Iglesias A, Carton A, Preiss J, Ball S. Control of starch composition and structure through substrate supply in the monocellular alga Chlamydomonas reinhardtii. J. Biol. Chem. 1996;271:16281–16287. doi: 10.1074/jbc.271.27.16281. [DOI] [PubMed] [Google Scholar]
- Wang J, Jiang J, Oard JH. Structure, expression and promoter activity of two polyubiquitin genes from rice (Oryza sativa L.) Plant Sci. 2000;156:201–211. doi: 10.1016/s0168-9452(00)00255-7. [DOI] [PubMed] [Google Scholar]
- Wang YG, Xing QH, Deng QY, Liang FS, Yuan LP, Weng ML, Wang B. Fine mapping of the rice thermo-sensitive genic male-sterile gene tms5. Theor. Appl. Genet. 2003;107:917–921. doi: 10.1007/s00122-003-1327-8. [DOI] [PubMed] [Google Scholar]
- Wilson ZA, Morroll SM, Dawson J, Swarup R, Tighe PJ. The Arabidopsis MALE STERILITY1 (MS1) gene is a transcriptional regulator of male gametogenesis, with homology to the PHD-finger family of transcription factors. Plant J. 2001;28:27–39. doi: 10.1046/j.1365-313x.2001.01125.x. [DOI] [PubMed] [Google Scholar]