Abstract
Cyanovirin (CV-N) is a small lectin with potent HIV neutralization activity, which could be exploited for a mucosal defense against HIV infection. The wild-type (wt) protein binds with high affinity to mannose-rich oligosaccharides on the surface of gp120 through two quasi-symmetric sites, located in domains A and B. We recently reported on a mutant of CV-N that contained a single functional mannose-binding site, domain B, showing that multivalent binding to oligomannosides is necessary for antiviral activity. The structure of the complex with dimannose determined at 1.8 Å resolution revealed a different conformation of the binding site than previously observed in the NMR structure of wt CV-N. Here, we present the 1.35 Å resolution structure of the complex, which traps three different binding conformations of the site and provides experimental support for a locking and gating mechanism in the nanoscale time regime observed by molecular dynamics simulations.
Keywords: cyanovirin, lectins, sugar binding, antiviral protein, X-ray structure analysis
Cyanovirin is an 11-kDa cyanobacterial lectin currently under investigation as a viral microbicide to prevent human immunodeficiency virus (HIV) transmission via the mucosal membrane (Boyd et al. 1997; De Clercq 2005; Reeves and Piefer 2005; Balzarini 2007). It is part of a larger group of non-homologous algal lectins that includes Microcystis viridis lectin (Williams Jr. et al. 2005), scytovirin (Bokesch et al. 2003), and griffithsin (Ziolkowska et al. 2006), which have potent antiviral activity against HIV. Although there are differences in the binding specificities of the algal lectins, the mechanism of action involves high affinity binding to the N-linked oligomannosides that cover a vast portion of the viral envelope glycoprotein gp120 (Botos and Wlodawer 2005; Ziolkowska et al. 2006; Scanlan et al. 2007).
Cyanovirin, the first member of this group to be discovered, has been extensively characterized with respect to its antiviral activity (O'Keefe et al. 2003; Barrientos and Gronenborn 2005; Helle et al. 2006), biophysical properties (Bewley and Otero-Quintero 2001; Barrientos and Gronenborn 2002; Barrientos et al. 2002, 2004; Shenoy et al. 2002), and structure. The structure of cyanovirin in the free and carbohydrate-bound forms have been determined by NMR (Bewley et al. 1998; Bewley 2001) and X-ray crystallography (Yang et al. 1999; Botos et al. 2002). The solution structures show a monomeric protein with a novel beta-sheet fold that comprises two quasi-symmetric domains, A (residues 1–38/90–101) and B (residues 39–89), connected on each side by a short hinge region. Each domain comprises a carbohydrate-binding pocket, which binds linear termini in branched high-mannose oligosaccharides. In some conditions, cyanovirin can form a domain-swapped dimer (Bennett et al. 1994; Liu and Eisenberg 2002) in which two CV-N molecules exchange part of each domain to form a dimeric structure containing four carbohydrate-binding domains (Yang et al. 1999; Bewley and Clore 2000; Barrientos and Gronenborn 2002; Botos et al. 2002; Clore and Bewley 2002; Barrientos et al. 2004). All the structures of wt CV-N obtained by X-ray crystallography contain the dimer (Yang et al. 1999; Barrientos et al. 2002; Botos et al. 2002).
We recently reported the first crystal structure of a cyanovirin mutant (P51G-m4-CVN) that contains the monomer, in the free and dimannose-bound form (Fromme et al. 2007). P51G-m4-CVN was designed to clarify whether the presence of one mannose-binding site is sufficient for antiviral activity, or whether multivalent interactions, either via the two sites on each cyanovirin molecule or via the four possible sites on the domain swapped form, are necessary. This issue had been somewhat controversial, due to the difficulties in clearly discriminating between the possible models, and further complicated by the slight selectivity of each domain for a different linear oligomannoside (Bewley and Otero-Quintero 2001; Chang and Bewley 2002; Kelley et al. 2002; Shenoy et al. 2002; Barrientos et al. 2004, 2006). P51G-m4-CVN recapitulates two sets of mutations introduced by other groups: the m4-CVN set, which abolishes domain A (Chang and Bewley 2002), and P51G, which stabilizes the monomer (Barrientos et al. 2002). As predicted, P51G-m4-CVN contains a single functional carbohydrate-binding site (domain B) and folds exclusively as a monomer under physiological conditions (Fromme et al. 2007).
The complete loss of antiviral activity in P51G-m4-CVN demonstrated the importance of multivalent interactions with the viral gp120, confirming one of the mechanisms that had been hypothesized for antiviral lectins (Botos et al. 2002; Barrientos and Gronenborn 2005; Botos and Wlodawer 2005; Ziolkowska and Wlodawer 2006). Our conclusions were corroborated by independent studies of a complementary mutant, CV-N(mutDB), in which the binding site on domain B had been abolished on the background of P51G: This mutant is also inactive against HIV (Barrientos et al. 2006).
The 1.8 Å structure of P51G-m4-CVN showed some important differences when compared to the solution structure of the dimannose complex, 1 IIY (Bewley 2001; Bewley and Otero-Quintero 2001), in particular in the conformation and hydrogen bonding pattern of Arg 76. In the NMR structure, the side chain forms two direct hydrogen bonds to the ligand, while in our structure Arg 76 occupies a different rotamer and forms a single water-mediated hydrogen bond to the ligand. Each of these binding modes had been observed in molecular dynamics studies of cyanovirin, which showed gating and locking of the binding by two residues, Glu 41 and Arg 76 in domain B (Margulis 2005), and it is quite likely that both modes may exist.
Here, we present the structure of P51G-m4-CVN at 1.35 Å resolution (Fig. 1), which clarifies important details of the sugar-binding sites. We were able to observe three different conformers of Arg 76 and multiple flexible amino acids, thereby providing the first experimental evidence for a conformational gating of the sugar-binding site of cyanovirin.
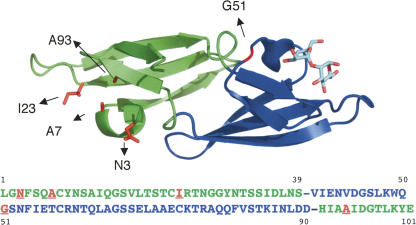
Figure 1.
Crystal structure of P51G-m4-CVN at 1.35 Å resolution: cartoon representation of one monomer in complex with dimannose. Domain A (green) contains four mutations (K3N, T7A, E23I, N93A, highlighted as red sticks) in the binding site, resulting in loss of binding (Chang and Bewley 2002). Domain B (blue) is unaltered and binds to dimannose. A P51G mutation in the hinge region stabilizes the monomer (Barrientos et al. 2002). The sequence of P51G-m4-CVN is reported below; domains A and B are represented in green and blue, respectively, while mutations from wt are in red. Figure generated with PyMOL (DeLano Scientific; http://www.pymol.org).
Results and Discussion
Overall structure description
The best native data set obtained from improved P51G-m4-CVN crystals was solved to a resolution of 1.35 Å (PDB code 2RDK); the statistics of the data evaluation are summarized in Table 1. The space group and unit cell constants are identical to the previously reported structure at 1.8 Å resolution (Fromme et al. 2007), with dense packing in the unit cell and a low solvent content of 43%. Two monomers associate in the asymmetric unit in a parallel arrangement that is distinct from the tilt observed in the domain-swapped dimer in solution and X-ray structures (Barrientos et al. 2002), which results in an orthogonal geometry between the two halves (domains AB′, A′B).
Table 1.
Statistics of data collection and structure refinement
The structure is characterized by low main chain B factors (mean B value 17.5, range 10–36), with the exception of a loop in domain A (R24 to G28) that has average main chain B factors of 32.5, and to a lesser extent a loop in domain B (Arg 76 to Gln 79), with average main chain B factors of 20. Differences in the crystal packing contacts of the two monomers affect the B factors observed in the two monomers: In particular, the B factors of loop R24–G28 of domain A are lowered to 11–16 in monomer 2. Overall, the flexibility observed is consistent with other structures of cyanovirin (Bewley et al. 1998; Bewley 2001; Botos et al. 2002) and points to a higher mobility of the loops compared with the body of the structure. The main differences with the NMR structure of wt are observed in the loops and in the hinge region.
Effect of the hinge region mutation
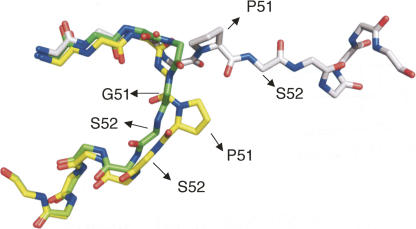
The P51G mutation stabilizes the monomeric form of the protein in solution, as observed in wt (Barrientos and Gronenborn 2002) and in P51G-m4-CVN (Fromme et al. 2007), and allows its crystallization in P51G-m4-CVN. Figure 2 shows an overlay of the structures of wt cyanovirin, in the monomeric (NMR) and dimeric (X-ray) forms, and of P51G-m4-CVN. In the wild-type monomer, the proline forces the following residue, S52, to assume unfavorable backbone dihedral angles (φ 174.4 and ψ 12.4, calculated with MolProbity) (Davis et al. 2004). Substituting glycine for proline results in a local rearrangement of the hinge region: All amino acids are now in favorable areas of the Ramachandran plot. As previously suggested, an alternate solution to relieving the strain of the hinge region is observed in the domain swapped dimer: The hinge is extended in a conformation that places the amino acids involved in allowed areas of the Ramachandran plot (Barrientos and Gronenborn 2002).
Figure 2.
Overlay of the backbone hinge region (residues 46–57) of cyanovirin, monomer and dimer, and of P51G-m4-CVN. The monomeric NMR structure of wt CV-N (PDB code 2EZM) is represented in yellow, while the domain-swapped crystal structure (PDB code 1M5M) is represented in gray, and the monomeric crystal structure of P51G-m4-CVN in green (PDB code 2RDK; this work). Only the side chain of P51 (wt) is shown. In 2EZM, S52 assumes disallowed backbone dihedral angles.
Carbohydrate-binding site
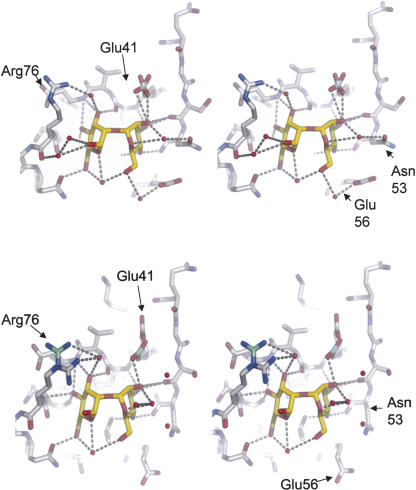
The dimannose-binding site of P51G-m4-CVN revealed a unique mode of binding, involving a water-mediated hydrogen bond between Arg 76 and the ligand. While this binding mode was very clear in monomer 1 (chain A), an indication of the flexibility of the site had come from the lowered resolution obtained for monomer 2 (chain B). In the current structure, the position of Arg 76 in each monomer is well defined. However, we could see some extra electron density by carefully inspecting the electron density map at a low contour level. We calculated the total omit map of the binding sites using the procedure of Bhat (1988) as integrated in the program Sfcheck. This procedure was extended systematically to all side chains in each binding site (monomer 1 and 2). We found five side chains with conformational flexibility: Thr 19, Thr 31, and Glu 41 in monomer 1, and Arg 76 and Glu 41 in monomer 2. Thr 19 and Thr 31 are located at the start and end of the flexible loop in domain A, while Glu 41 and Arg 76 are located in the well-defined sugar-binding pocket in domain B. Three different conformers of Arg 76 were trapped. In monomer 1, Arg 76 has a unique conformation with 100% occupancy. It interacts with the dimannose by hydrogen bonding via a tightly bound water molecule. This conformation is identical to the position identified in the 1.8 Å structure but different from the direct double hydrogen bond to the ligand (locking position) observed in the NMR structure (Bewley 2001). In monomer 2, Arg 76 is present in two different conformations that place the side chain in an either proximal or distal position in relation to the ligand (Fig. 3). The distal side has occupancy of 80% and is already visible at a contour level of 1.5 σ, while the proximal position has occupancy of 20% and is only visible at a low contour level of 0.85 σ. The water molecule that bridges Arg 76 to the mannose in monomer 1 is not observed in monomer 2, and no direct hydrogen bonds are observed. The side chain of Glu 41 can occupy two conformers, with approximately equal occupancies. As the hydrogen bond between Glu 41 and the anomeric atom of the dimannose ligand is highly directional, it has been proposed as a key determinant of the specificity of cyanovirin for Manα(1–2)Manα dimers (Margulis 2005). The hydrogen bonding interactions between the ligand and the binding site in each monomer are shown in Figure 3.
Figure 3.
Comparison of the dimannose-binding site in the two monomers, top and bottom panel, each in stereoview. Amino acids lining the binding site are depicted in sticks and colored by atom (carbon, gray; oxygen, red; nitrogen, blue), while the dimannose ligand is depicted in sticks (carbon, yellow; oxygen, red), and crystallographic water molecules participating in ligand binding are shown as red spheres; direct or water-mediated hydrogen bonds are indicated as yellow dashed lines. Figure generated with PyMOL (DeLano Scientific; http://www.pymol.org).
These findings are consistent with the gating mechanism observed by molecular dynamics simulations of the cyanovirin–dimannose complex (Margulis 2005) based on the NMR structure (Bewley 2001). In the simulations, Arg 76 undergoes a large conformational change that brings the side chain from an unlocked position, far from the ligand, to a locked position in which two direct hydrogen bonds to the ligand are observed. This change happens on a nanoscale time range, and the locked position is stable for several nanoseconds. Furthermore, modeling studies also suggested a critical role of Glu 41 in the tight binding of the sugar and possibly in the selectivity for Manα(1–2)Manα.
Conclusions
The high-resolution crystal structure of P51G-m4-CVN presented here reveals atomic details of the Arg 76 gating mechanism and shows details of the hydrogen bonding network and its changes in response to alternate conformations of Arg 76, which are summarized in Figure 3.
The crystallographic studies presented here, interpreted together with the molecular dynamics studies (Margulis 2005) and the NMR data (Bewley 2001) previously available, unravel a flexible hydrogen-bonding network that may gate the dimannose ligand in the binding site, locked by Arg 76 and Glu 41. This mechanism provides insight into the high affinity and specificity of cyanovirin for its target ligands. The atomic detail of the site in different states will form the basis for the design of new mutants with optimized binding sites.
Materials and Methods
Expression, purification, and crystallization
P51G-m4CVN, containing a six-His tag at the C terminus, was cloned into a pET-26b (+) vector (Novagen) and expressed in Escherichia coli BL-21 (DE3) cells as described before (Fromme et al. 2007). Briefly, the cell culture was induced with IPTG at OD600 of 0.7 and harvested after 3 h. The protein was purified by affinity chromatography using Ni Sepharose Fast Flow resin (Novagen). Crystals were grown by the hanging-drop vapor diffusion method with crystal drops containing a 2 + 2 μL mix of 10 mg/mL protein, 2 mM Man2, and a reservoir buffer containing 100 mM HEPES, pH 6, 100 mM Mg2SO4 using 28%–32% (w/v) PEG 8000 as precipitant. The crystals were frozen in liquid nitrogen directly from the mother liquor.
X-ray data collection and refinement
High-resolution X-ray diffraction data sets of the crystals were measured at beamline 8.2.2 at Advanced Light Source, Berkeley. Data were collected with an ADSC 315 CCD detector; the wavelength was set to 1 Å. The oscillation range was 1° per frame; the total rotation range was 120°. The data were evaluated with the software package HKL 2000 (Collaborative Computational Project, Number 4 1994). For the primary data evaluation of X-ray data we used the software package HKL 2000 version 1.93 (Otwinowski and Minor 1997; Minor et al. 2006). The indexed and scaled data were further evaluated with the software kit CCP4i (Collaborative Computational Project, Number 4 1994). The 1.8 Å structure of P51G-m4-CVN (Fromme et al. 2007) (PDB entry 2PYS) was used as the starting model for molecular replacement. The program Phaser 1.3.1 (McCoy et al. 2005) was used for molecular replacement. The calculated model from Phaser was used as input file to ARP/warp version 6.1.1 (Perrakis et al. 1999, 2001) and the structure was rebuilt based on the 1.35 Å data. All residues from the protein of the final model were determined by wARP/ARP. The mannose was refined with the ligand subprogram of wARP/ARP (Zwart et al. 2004). Last refinement was made manually with Xtalview (McRee 1999) and several rounds of Refmac 5.2.0019 (Murshudov et al. 1997). After inspecting the 2Fo-Fc map and correcting minor misalignments in side chains, the Fo-Fc map was used to find residues with alternative conformers; the program Coot (Emsley and Cowtan 2004) was used to assign the two conformers. The model was validated by calculating the omit map (Bhat 1988) with the program Sfcheck (Vaguine et al. 1999). In difficult cases the occupancy of the interesting residue(s) was set to zero to obtain the most possible unbiased phase.
Acknowledgment
We would like to thank Dr. Corie Ralston (Beamline 8.2.2 at the Advanced Light Source, Berkeley) for help with data collection.
Footnotes
Reprint requests to: Giovanna Ghirlanda, Department of Chemistry and Biochemistry, Arizona State University, 1711 South Rural Road, Tempe, Arizona 85250-1604, USA; e-mail: gghirlanda@asu.edu; fax: (480) 965-2747; or Petra Fromme, Department of Chemistry and Biochemistry, Arizona State University, 1711 South Rural Road, Tempe, Arizona 85287-1604, USA; e-mail: Pfromme@asu.edu; fax: (480) 965-2747.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.083472808.
References
- Balzarini, J. Carbohydrate-binding agents: A potential future cornerstone for the chemotherapy of enveloped viruses? Antivir. Chem. Chemother. 2007;18:1–11. doi: 10.1177/095632020701800101. [DOI] [PubMed] [Google Scholar]
- Barrientos, L.G., Gronenborn, A.M. The domain-swapped dimer of cyanovirin-N contains two sets of oligosaccharide-binding sites in solution. Biochem. Biophys. Res. Commun. 2002;298:598–602. doi: 10.1016/s0006-291x(02)02489-0. [DOI] [PubMed] [Google Scholar]
- Barrientos, L.G., Gronenborn, A.M. The highly specific carbohydrate-binding protein cyanovirin-N: Structure, anti-HIV/Ebola activity and possibilities for therapy. Mini Rev. Med. Chem. 2005;5:21–31. doi: 10.2174/1389557053402783. [DOI] [PubMed] [Google Scholar]
- Barrientos, L.G., Louis, J.M., Botos, I., Mori, T., Han, Z., O'Keefe, B.R., Boyd, M.R., Wlodawer, A., Gronenborn, A.M. The domain-swapped dimer of cyanovirin-N is in a metastable folded state: Reconciliation of X-ray and NMR structures. Structure. 2002;10:673–686. doi: 10.1016/s0969-2126(02)00758-x. [DOI] [PubMed] [Google Scholar]
- Barrientos, L.G., Lasala, F., Delgado, R., Sanchez, A., Gronenborn, A.M. Flipping the switch from monomeric to dimeric CV-N has little effect on antiviral activity. Structure. 2004;12:1799–1807. doi: 10.1016/j.str.2004.07.019. [DOI] [PubMed] [Google Scholar]
- Barrientos, L.G., Matei, E., Lasala, F., Delgado, R., Gronenborn, A.M. Dissecting carbohydrate-cyanovirin-N binding by structure-guided mutagenesis: Functional implications for viral entry inhibition. Protein Eng. Des. Sel. 2006;19:525–535. doi: 10.1093/protein/gzl040. [DOI] [PubMed] [Google Scholar]
- Bennett, M.J., Choe, S., Eisenberg, D. Domain swapping: Entangling alliances between proteins. Proc. Natl. Acad. Sci. 1994;91:3127–3131. doi: 10.1073/pnas.91.8.3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewley, C.A. Solution structure of a cyanovirin-N:Manα1–2Manα complex structural basis for high-affinity carbohydrate-mediated binding to gp120. Structure. 2001;9:931–940. doi: 10.1016/s0969-2126(01)00653-0. [DOI] [PubMed] [Google Scholar]
- Bewley, C.A., Clore, G.M. Determination of the relative orientation of the two halves of the domain-swapped dimer of cyanovirin-N in solution using dipolar couplings and rigid body minimization. J. Am. Chem. Soc. 2000;122:6009–6016. [Google Scholar]
- Bewley, C.A., Otero-Quintero, S. The potent anti-HIV protein cyanovirin-N contains two novel carbohydrate-binding sites that selectively bind to Man8 D1D3 and Man9 with nanomolar affinity: Implications for binding to the HIV envelope protein gp120. J. Am. Chem. Soc. 2001;123:3892–3902. doi: 10.1021/ja004040e. [DOI] [PubMed] [Google Scholar]
- Bewley, C.A., Gustafson, K.R., Boyd, M.R., Covell, D.G., Bax, A., Clore, G.M., Gronenborn, A.M. Solution structure of cyanovirin-N, a potent HIV-inactivating protein. Nat. Struct. Biol. 1998;5:571–578. doi: 10.1038/828. [DOI] [PubMed] [Google Scholar]
- Bhat, T.N. Calculation of an omit map. J. Appl. Crystallogr. 1988;21:279–281. [Google Scholar]
- Bokesch, H.R., O'Keefe, B.R., McKee, T.C., Pannell, L.K., Patterson, G.M., Gardella, R.S., Sowder R.C., II, Turpin, J., Watson, K., Buckheit R.W., Jr, et al. A potent novel anti-HIV protein from the cultured cyanobacterium Scytonema varium . Biochemistry. 2003;42:2578–2584. doi: 10.1021/bi0205698. [DOI] [PubMed] [Google Scholar]
- Botos, I., Wlodawer, A. Proteins that bind high-mannose sugars of the HIV envelope. Prog. Biophys. Mol. Biol. 2005;88:233–282. doi: 10.1016/j.pbiomolbio.2004.05.001. [DOI] [PubMed] [Google Scholar]
- Botos, I., O'Keefe, B.R., Shenoy, S.R., Cartner, L.K., Ratner, D.M., Seeberger, P.H., Boyd, M.R., Wlodawer, A. Structures of the complexes of a potent anti-HIV protein cyanovirin-N and high mannose oligosaccharides. J. Biol. Chem. 2002;277:34336–34342. doi: 10.1074/jbc.M205909200. [DOI] [PubMed] [Google Scholar]
- Boyd, M.R., Gustafson, K.R., McMahon, J.B., Shoemaker, R.H., O'Keefe, B.R., Mori, T., Gulakowski, R.J., Wu, L., Rivera, M.I., Laurencot, C.M., et al. Discovery of cyanovirin-N, a novel human immunodeficiency virus-inactivating protein that binds viral surface envelope glycoprotein gp120: Potential applications to microbicide development. Antimicrob. Agents Chemother. 1997;41:1521–1530. doi: 10.1128/aac.41.7.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, L.C., Bewley, C.A. Potent inhibition of HIV-1 fusion by cyanovirin-N requires only a single high affinity carbohydrate-binding site: Characterization of low affinity carbohydrate-binding site knockout mutants. J. Mol. Biol. 2002;318:1–8. doi: 10.1016/S0022-2836(02)00045-1. [DOI] [PubMed] [Google Scholar]
- Clore, G.M., Bewley, C.A. Using conjoined rigid body/torsion angle simulated annealing to determine the relative orientation of covalently linked protein domains from dipolar couplings. J. Magn. Reson. 2002;154:329–335. doi: 10.1006/jmre.2001.2489. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4. The CCP4 Suite—programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Davis, I.W., Murray, L.W., Richardson, J.S., Richardson, D.C. MOLPROBITY: Structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 2004;32:W615–W619. doi: 10.1093/nar/gkh398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Clercq, E. Emerging anti-HIV drugs. Expert Opin. Emerg. Drugs. 2005;10:241–273. doi: 10.1517/14728214.10.2.241. [DOI] [PubMed] [Google Scholar]
- Emsley, P., Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Fromme, R., Katiliene, Z., Giomarelli, B., Bogani, F., Mahon, J.M., Mori, T., Fromme, P., Ghirlanda, G. A monovalent mutant of cyanovirin-N provides insight into the role of multiple interactions with gp120 for antiviral activity. Biochemistry. 2007;46:9199–9207. doi: 10.1021/bi700666m. [DOI] [PubMed] [Google Scholar]
- Helle, F., Wychowski, C., Vu-Dac, N., Gustafson, K.R., Voisset, C., Dubuisson, J. Cyanovirin-N inhibits hepatitis C virus entry by binding to envelope protein glycans. J. Biol. Chem. 2006;281:25177–25183. doi: 10.1074/jbc.M602431200. [DOI] [PubMed] [Google Scholar]
- Kelley, B.S., Chang, L.C., Bewley, C.A. Engineering an obligate domain-swapped dimer of cyanovirin-N with enhanced anti-HIV activity. J. Am. Chem. Soc. 2002;124:3210–3211. doi: 10.1021/ja025537m. [DOI] [PubMed] [Google Scholar]
- Liu, Y., Eisenberg, D. 3D domain swapping: As domains continue to swap. Protein Sci. 2002;11:1285–1299. doi: 10.1110/ps.0201402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulis, C.J. Computational study of the dynamics of mannose disaccharides free in solution and bound to the potent anti-HIV virucidal protein cyanovirin. J. Phys. Chem. B. 2005;109:3639–3647. doi: 10.1021/jp0406971. [DOI] [PubMed] [Google Scholar]
- McCoy, A.J., Grosse-Kunstleve, R.W., Storoni, L.C., Read, R.J. Likelihood-enhanced fast translation functions. Acta Crystallogr. D Biol. Crystallogr. 2005;61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- McRee, D.E. XtalView Xfit—a versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol. 1999;125:156–165. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- Minor, W., Cymborowski, M., Otwinowski, Z., Chruszcz, M. HKL-3000: The integration of data reduction and structure solution—from diffraction images to an initial model in minutes. Acta Crystallogr. D Biol. Crystallogr. 2006;62:859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- Murshudov, G.N., Vagin, A.A., Dodson, E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- O'Keefe, B.R., Smee, D.F., Turpin, J.A., Saucedo, C.J., Gustafson, K.R., Mori, T., Blakeslee, D., Buckheit, R., Boyd, M.R. Potent anti-influenza activity of cyanovirin-N and interactions with viral hemagglutinin. Antimicrob. Agents Chemother. 2003;47:2518–2525. doi: 10.1128/AAC.47.8.2518-2525.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski, Z., Minor, W. Processing of X-ray diffraction data collected in oscillation mode. In: Carter C.W., Sweet R.M., editors. Macromolecular Crystallography, part A. Vol. 276. Academic Press; New York: 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- Perrakis, A., Morris, R., Lamzin, V.S. Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- Perrakis, A., Harkiolaki, M., Wilson, K.S., Lamzin, V.S. ARP/wARP and molecular replacement. Acta Crystallogr. D Biol. Crystallogr. 2001;57:1445–1450. doi: 10.1107/s0907444901014007. [DOI] [PubMed] [Google Scholar]
- Reeves, J.D., Piefer, A.J. Emerging drug targets for antiretroviral therapy. Drugs. 2005;65:1747–1766. doi: 10.2165/00003495-200565130-00002. [DOI] [PubMed] [Google Scholar]
- Scanlan, C.N., Offer, J., Zitzmann, N., Dwek, R.A. Exploiting the defensive sugars of HIV-1 for drug and vaccine design. Nature. 2007;446:1038–1045. doi: 10.1038/nature05818. [DOI] [PubMed] [Google Scholar]
- Shenoy, S.R., Barrientos, L.G., Ratner, D.M., O'Keefe, B.R., Seeberger, P.H., Gronenborn, A.M., Boyd, M.R. Multisite and multivalent binding between cyanovirin-N and branched oligomannosides: Calorimetric and NMR characterization. Chem. Biol. 2002;9:1109–1118. doi: 10.1016/s1074-5521(02)00237-5. [DOI] [PubMed] [Google Scholar]
- Vaguine, A.A., Richelle, J., Wodak, S.J. SFCHECK: A unified set of procedures for evaluating the quality of macromolecular structure-factor data and their agreement with the atomic model. Acta Crystallogr. D Biol. Crystallogr. 1999;55:191–205. doi: 10.1107/S0907444998006684. [DOI] [PubMed] [Google Scholar]
- Williams D.C., Jr, Lee, J.Y., Cai, M., Bewley, C.A., Clore, G.M. Crystal structures of the HIV-1 inhibitory cyanobacterial protein MVL free and bound to Man3GlcNAc2: Structural basis for specificity and high-affinity binding to the core pentasaccharide from N-linked oligomannoside. J. Biol. Chem. 2005;280:29269–29276. doi: 10.1074/jbc.M504642200. [DOI] [PubMed] [Google Scholar]
- Yang, F., Bewley, C.A., Louis, J.M., Gustafson, K.R., Boyd, M.R., Gronenborn, A.M., Clore, G.M., Wlodawer, A. Crystal structure of cyanovirin-N, a potent HIV-inactivating protein, shows unexpected domain swapping. J. Mol. Biol. 1999;288:403–412. doi: 10.1006/jmbi.1999.2693. [DOI] [PubMed] [Google Scholar]
- Ziółkowska, N.E., Wlodawer, A. Structural studies of algal lectins with anti-HIV activity. Acta Biochim. Pol. 2006;53:617–626. [PubMed] [Google Scholar]
- Ziółkowska, N.E., O'Keefe, B.R., Mori, T., Zhu, C., Giomarelli, B., Vojdani, F., Palmer, K.E., McMahon, J.B., Wlodawer, A. Domain-swapped structure of the potent antiviral protein griffithsin and its mode of carbohydrate binding. Structure. 2006;14:1127–1135. doi: 10.1016/j.str.2006.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwart, P.H., Langer, G.G., Lamzin, V.S. Modelling bound ligands in protein crystal structures. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2230–2239. doi: 10.1107/S0907444904012995. [DOI] [PubMed] [Google Scholar]