Abstract
C3-like exoenzymes are ADP-ribosyltransferases that specifically modify some Rho GTPase proteins, leading to their sequestration in the cytoplasm, and thus inhibiting their regulatory activity on the actin cytoskeleton. This modification process goes through three sequential steps involving NAD-hydrolysis, Rho recognition, and binding, leading to Rho ADP-ribosylation. Independently, three distinct residues within the ARTT loop of the C3 exoenzymes are critical for each of these steps. Supporting the critical role of the ARTT loop, we have shown previously that it adopts a distinct conformation upon NAD binding. Here, we present seven wild-type and ARTT loop-mutant structures of C3 exoenzyme of Clostridium botulinum free and bound to its true substrate, NAD, and to its NAD-hydrolysis product, nicotinamide. Altogether, these structures expand our understanding of the conformational diversity of the C3 exoenzyme, mainly within the ARTT loop.
Keywords: ADP-ribosyltransferase, C3 exoenzyme, ARTT loop, plasticity, crystal structures
C3-like exoenzymes are bacterial ADP-ribosyltransferase toxins used as tools to understand the diverse regulatory functions of their cellular targets, the mammalian Rho GTPases (Aktories et al. 2004; Ménétrey and Ménez 2006). Rho proteins are known for their pivotal role in regulating the actin cytoskeleton, but also in influencing cell polarity, microtubule dynamics, membrane transport pathways, and transcription-factor activity (Etienne-Manneville and Hall 2002). The manner by which C3-like exoenzymes catalyzed ADP-ribosylation and inactivation of Rho GTPases is now well understood. These exoenzymes exhibit both a glycohydrolytic activity that splits NAD into nicotinamide and ADP-ribose and an ADP-ribosyltransferase activity that transfers the ADP-ribose moiety to a specific asparagine residue of Rho proteins by a SN2 mechanism (Sekine et al. 1989). This modification has no major effect on nucleotide binding, intrinsic and protein-stimulated GTPase activity, or effector coupling of Rho proteins (Sehr et al. 1998; Genth et al. 2003b). However, the ADP-ribosylated Rho proteins are trapped in the cytoplasm by its guanine-dissociation inhibitor (GDI) regulator inhibiting its GEF-driven GTP-loading. Thus, ADP-ribosylation prevents Rho from performing its cellular functions at the membrane surface (Genth et al. 2003a; Aktories et al. 2004).
C3-like exoenzymes are 24-kDa enzymes that are produced by various Gram-positive pathogens. So far, seven C3-like isoforms have been described: Two are produced by Clostridium botulinum types C and D (C3bot1 and C3bot2) (Aktories and Frevert 1987; Aktories et al. 1988; Rubin et al. 1988; Nemoto et al. 1991), one from Clostridium limosum (C3lim) (Just et al. 1992), one from Bacillus cereus (C3cer) (Just et al. 1995), and three from Staphylococcus aureus (C3stau1, C3stau2, and C3stau3, also known as EDIN-A, EDIN-B, and EDIN-C) (Inoue et al. 1991; Wilde et al. 2001; Yamaguchi et al. 2001). C3bot1 shares 65% and 63% identity sequence with C3bot2 and C3lim, respectively, but is only 30% identical to C3cer. The three isoforms from S. aureus are about 35% identical with C3bot1 and 66%–77% identical between each other. All C3-like exoenzymes modify asparagine 41 on RhoA, RhoB, and RhoC specifically, while the C3stau also acts on RhoE/Rnd3 (Wilde et al. 2001).
The overall structure of the C3bot1 exoenzyme has been described in its free and NAD-bound states (Han et al. 2001; Ménétrey et al. 2002). Comparison of both of these structural states has highlighted three different conformational changes undergone by this toxin. A widespread protein “flexure” movement of the entire protein and two localized conformational changes of the PN (phosphate-picotinamide) and ARTT (ADP-ribosyltransferase turn-turn) loops. The PN loop delimits the NAD binding site and provides two residues important for NAD binding (Ménétrey et al. 2002; Wilde et al. 2003). Facing the PN loop, the ARTT loop contributes to catalysis and protein target recognition and interaction. Three residues are critical for these functions and define the ARTT motif (ΦxxQxE) (Han et al. 2001): (1) a highly conserved catalytic glutamate (Glu214 in C3bot1), which is required for NAD glycohydrolysis (Aktories et al. 1995; Saito et al. 1995; Bohmer et al. 1996; Wilde et al. 2002b, 2003); (2) a phenylalanine/tyrosine residue (Phe209 in C3bot1), which has been shown to be involved in protein target recognition and interaction (Wilde et al. 2002a,b, 2003), and (3) a glutamine (Gln212 in C3bot1), which is critical for ADP-ribose transfer to the target protein (Ménétrey et al. 2002; Wilde et al. 2002b, 2003).
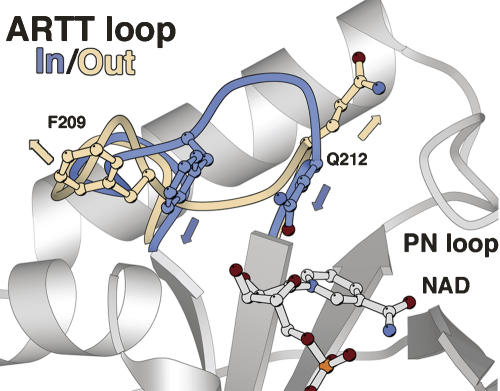
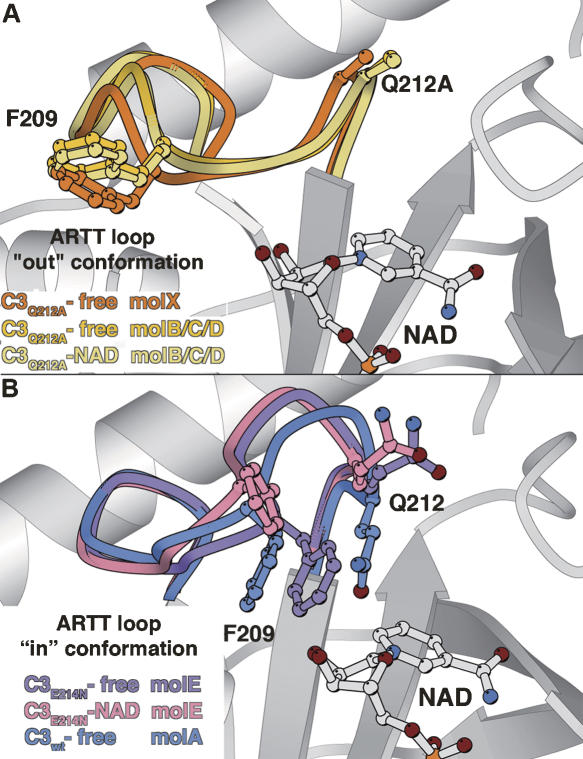
However, the relationship of C3 conformational changes along the catalytic process remains poorly understood. Previously, we had noted that NAD binding to C3 exoenzyme triggered different conformational changes specifically within the ARTT loop (Ménétrey et al. 2002). In this study, we present seven new crystal structures of the wild-type C3bot1 and of two ARTT-loop mutants: C3-Q212A and C3-E214N in the free state, in complex with its true substrate, NAD, and with the NAD-hydrolysis product, nicotinamide. Two distinct conformations have been observed for the ARTT loop with the side chains of Gln212 and Phe209 (residues from C3bot1) pointing either inside or outside of the NAD binding pocket (termed here the “in” and the “out” conformation, respectively) (Fig. 1). Altogether, our data suggest a higher degree of complexity as local protein–protein contacts also affect the ARTT loop conformation of the C3 exoenzyme.
Figure 1.
The ARTT loop conformations. The “in” and “out” conformations of the ARTT loop are shown in blue and beige, respectively. The “in” conformation which is defined by Phe209 and Gln212 from the ARTT motif extended inside the NAD-binding site is shown in blue. The “out” conformation with both residues extended outside is shown in beige. The NAD is shown in ball-and-stick representation.
Results
High-resolution structure of the C3wt exoenzyme
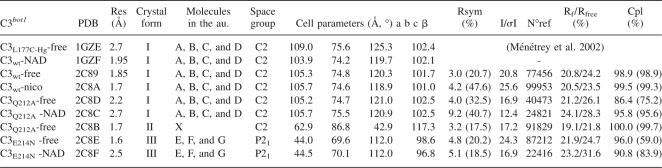
The crystal structure of the free state of the wild-type C3bot1 exoenzyme has been determined to be 1.85 Å resolution (PDB code 2C89) (Table 1), improving on the 2.7 Å resolution of our previous free-state structure obtained with the cysteine-mutant L177C of the C3bot1 exoenzyme with bound mercury (1GZE) (Table 1) (Ménétrey et al. 2002). Both of these structures belong to the same crystal form (called hereafter form I), which has four molecules (A, B, C, and D) in the asymmetric unit. The increase in resolution might be due to tighter packing of the molecules in the crystal lattice with a decrease by 6% in the unit cell volume (Table 1). The four molecules in these structures have an average RMSD <0.58 Å against each other (on 188 Cα atoms consisting of residues 45–244, omitting the ARTT loop residues 202–213), consistent with a well-conserved overall fold, confirming that the L177C mutation and presence of mercury have little effect on the structure of C3bot1 exoenzyme.
Table 1.
Crystal structures of C3 exoenzyme from Clostridium botulinum
However, one difference between these structures concerns the ARTT loop. Indeed, the ARTT loop of molecule A in the C3-L177C-Hg-free structure (1GZE) exhibits the “out” conformation, while that in the C3wt-free structure (2C89) adopts the “in” conformation. Note that the ARTT loop of all of the other three molecules (B, C, and D) in both structures retains the “out” conformation (Table 2). Another difference concerns the overall flexure of the C3 exoenzyme, which is similar for molecules A, B, and C in the C3wt-free and C3-L177C-Hg-free structures, respectively, and together, but not shared by molecule D. Molecule D exhibits a more open flexure in C3wt-free compared with C3-L177C-Hg-free, comparable to that observed in molecule D bound to ADP in the C3wt-NAD structure (1GZF) (Table1). In conclusion, these new results reveal that the ARTT loop can adopt the “in” and “out” conformation irrespective of its ligation state, but influenced by protein–protein interactions. This is also the case for the overall flexure of the C3 exoenzyme.
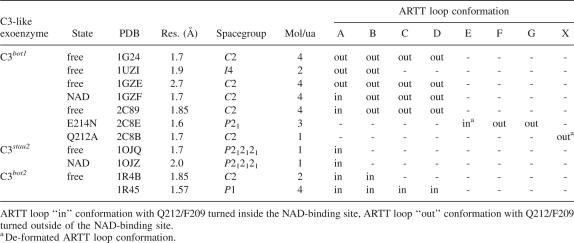
Table 2.
Crystal structures of C3-like exoenzymes from different isoforms, space groups, and states
Structure of the C3wt bound to nicotinamide
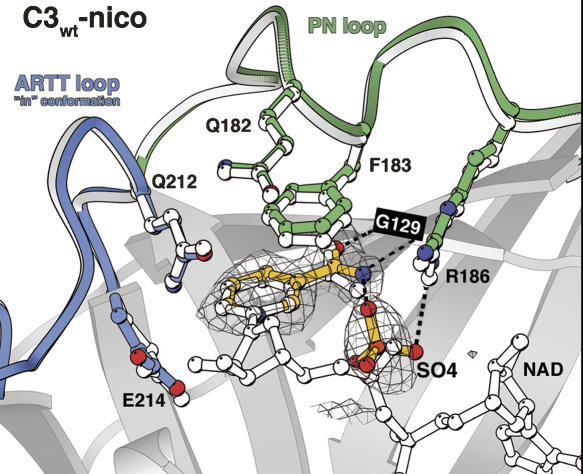
The C3wt-free crystals were soaked with nicotinamide and the structure was solved at 1.7 Å resolution (2C8A) without any significant variation in cell parameters (Table 1). In all four molecules in the asymmetric unit, at the NAD binding site, the electron density shows unambiguously the nicotinamide (Fig. 2). The nicotinamide position and conformation is identical to that of the nicotinamide moiety of NAD in the C3wt-NAD structure (1GZF), with the carboxamide group of nicotinamide making the same bidentate hydrogen bonds with Gly129 (Fig. 2). A sulfate molecule is present inside of the NAD binding site in each of the four C3 molecules to mimic the phosphate from the NMN moiety of NAD. This sulfate makes a hydrogen bond with the carboxamide group from the nicotinamide as the phosphate does in the C3wt–NAD complex (Fig. 2).
Figure 2.
The nicotinamide-binding site. Nicotinamide and sulfate ion inside the NAD binding site of the C3wt-nico structure are represented in ball-and-stick colored in yellow. The electron density grid shows an Fo–Fc omit map contoured at the 2 σ level. The ARTT and the PN loops are indicated in blue and green, respectively. Hydrogen bonds made by the nicotinamide and sulfate ion are shown by a dashed line. For comparison, the NAD and the ARTT and PN loops from the C3wt-NAD structure (1GZF) (Ménétrey et al. 2002) are superimposed and shown in white ball-and-stick representation.
The overall fold of the C3wt-nico (2C8A) is well conserved compared with C3wt-free without any significant variation in flexure (2C89; RMSD <0.45 Å for 188 Cα atoms). The ARTT loop from molecule A adopts the “in” conformation, while that from molecules B, C, and D exhibit the “out” conformation, as in the C3wt-free structure (Table 2). The overall agreement between C3wt-nico and C3wt-free structures suggests that nicotinamide binding does not impose restrictions on the C3 flexure and its ARTT loop conformation. However, the presence of nicotinamide is associated with a defined PN loop (180–187) conformation and the side chain of Phe183 capping the nicotinamide ring (Fig. 2).
Structures of the C3-Q212A mutant free and bound to NAD
We have solved the structures of the C3-Q212A mutant in the free state at 2.2 Å and NAD-bound state at 2.7 Å resolution by soaking C3-Q212A-free crystals with NAD (2C8D and 2C8C, respectively). The crystals are isomorphous with the C3wt crystal form I without significant variations in cell dimensions (Table 1) and with all four molecules superimposing very well on each other and on molecules from the C3wt-free structure (2C89; RMSD <0.50 Å for 188 Cα atoms). In the C3-Q212A-NAD structure, the electron density for NAD is observed in molecules A, B, and C, while for molecule D there is density only for the ADP moiety. Moreover, the hydrolysis state of NAD bound to molecule C differs between C3wt-NAD (1GZF) and C3-Q212A-NAD (2C8C) structures; the wild type is found with NAD hydrolyzed in ADP plus ribose-nicotinamide bound, while the Q212A mutant binds an intact NAD. Such a difference may be due to variations in the duration of the soak and differences in lattice constraints.
In both the free and NAD-bound states of the Q212A mutant, the ARTT loop of molecule A adopts the “in” conformation, while the three other molecules (B, C, and D) are found in the “out” conformation (Table 2), as observed for the wild-type structures (C3wt-free and C3wt-NAD; 2C89 and 1GZF, respectively). The overall flexure between equivalent molecules from C3-Q212A-free (2C8D) and C3wt-free (2C89) is similar to molecule D, slightly more open than molecules A, B, and C. However, in contrast to what has been observed for C3wt (1GZF), the C3-Q212A mutant (2C8C) does not trigger significant movement in the crystal upon NAD binding (or ADP binding for molecule D). In conclusion, no major difference is induced by the Q212A mutation in the free or NAD-bound structures of the C3bot1 exoenzyme.
New crystal form for C3bot1 exoenzyme
Minor changes in crystallization conditions yield a better diffracting monoclinic crystal form (form II) for the C3-Q212A mutant. The structure of C3-Q212A-free (form II) has been solved at 1.7 Å resolution (2C8B) with only one molecule (termed X) in the asymmetric unit (Table 1). Molecule X makes different crystal packing interactions than molecules A–D from the crystal form I, and thus provides supplementary information to understand the variations in the ARTT loop conformation and the C3 flexure. Molecule X of C3-Q212A-free (form II) superimposes well with the molecules A–D of C3-Q212A-free (form I; 2C8D; RMSD <0.69 Å for 188 Cα atoms) with an overall flexure matching that of molecule D.
In form II, the ARTT loop of molecule X from C3-Q212A-free adopts a slightly shifted “out” conformation different from the virtually identical ARTT loop of molecules B, C, or D from C3-Q212A-free in form I (Fig. 3A). The ARTT loop deviation between X (2C8B) and B/C/D (2C8D) is 0.73 Å mean RMSD for residues 203–214 on Cα atoms, while when comparing together molecules B, C, and D in form I, we observed a deviation of only 0.33 Å and 0.35 Å mean RMSD in the free state (2C8D) and against the NAD-bound state (2C8C), respectively. This shift is accompanied by a similar shift of helices α2 and α4 from the N-terminal helical region (Fig. 1), thus preserving the ARTT loop/helix α2 and the helix α2/helix α4 surface contacts. The comparison of the same protein (C3-Q212A-free) in different contact environments reveals that both the ARTT loop and the helical N-terminal region are influenced by protein–protein interactions.
Figure 3.
The ARTT loop deformations. (A) The ARTT loop from the C3-Q212A-free structure (form II, mol X) is superimposed on those of the C3-Q212A-free and C3-Q212A-NAD structures (form I, mol B/C/D). (B) The ARTT loop from the C3E21N-free and C3E21N-NAD structures (form III, mol E) are superimposed on top of each other and on top of the C3wt-free-I structure (mol A). The NAD is shown for clarity.
Structures of the C3-E214N mutant free and bound to NAD
The structures of the C3-E214N mutant in the free state (2C8E) has been determined at 1.6 Å resolution in a new monoclinic crystal form (form III) with three molecules in the asymmetric unit (E, F, and G) (Table 1). Overall, the three molecules from crystal form III superimpose well with each other and with all four molecules from form I of the C3wt-free structure (2C89; RMSD <0.75 Å for 188 Cα atoms). Thus, mutating the catalytic glutamate to asparagine does not affect the overall structure of the C3bot1 exoenzyme. The structure of the NAD-soaked crystal of C3-E214N mutant (2C8F) was refined to 2.5 Å resolution. In the structure, clear electronic density for intact NAD was observed for molecule E, where it adopts the same compact conformation and position inside the active site as in the C3wt–NAD structure (1GZF). However, neither NAD nor NAD hydrolysis products can be seen in molecules F and G, although the NAD binding sites are easily accessible in the crystal.
The overall C3 flexure varies between the three molecules of this new crystal form and against the four molecules from crystal form I. In molecules E from C3-E214N-free (2C8E) and C3-E214N-NAD (2C8F), NAD binding occurs without any change in flexure, while in molecules F a slight change is observed despite no NAD binding. In molecule G, also in the absence of NAD, the flexure matches the closed conformation observed for molecule A with NAD bound (1GZF). Thus, different C3 flexures are observed independently of the NAD-bound state.
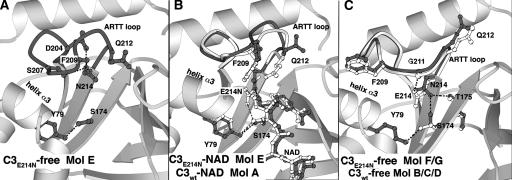
The ARTT loop adopts the “out” conformation in molecules F and G and the “in” conformation in molecule E in both the free and NAD-bound states. The selection of this last conformation is probably because of crystal packing reasons. No major difference is observed in the “out” conformation of the ARTT loop in molecules F and G (2C8E and 2C8F) compared with that in molecules B, C, and D of crystal form I (2C89; 0.35 Å mean RMSD for residues 203–214 on Cα atoms). In contrast, the “in” conformation of molecule E (2C8E) deviates from that of molecule A (2C89) by 1.17 Å RMSD, with the largest deviation observed between positions 208 and 212. This is the largest deviation observed for the ARTT loop “in” conformation to date, but given that the torsion angles of the backbone remain similar to those observed in molecule A, it should be considered a deformed “in” conformation rather than a new ARTT loop conformation (Fig. 3B). The glutamate-to-asparagine mutation and crystal contacts contribute to this deformation to a different degree. The asparagine makes multiple intra-ARTT loop interactions that are not observed with the catalytic glutamate due to different structural and chemical properties. In addition, upon NAD binding, the deformed ARTT loop of molecule E (2C8F) undergoes a further readjustment (RMSD of 0.64 Å, compared with the C3-E214A-free structure; 2C8E), still remaining distinct from the “in” conformation of molecule A (RMSD 1.02 Å; 1GZF; Fig. 3B). Given that both C3-E214N-NAD (2C8F) and C3-E214N-free (2C8E) structures are strictly isomorphous without changes in crystal contacts (Table 1), this readjustment is attributable to NAD binding. Altogether, the ARTT loop deformation observed for the molecule E of the C3-E214N-free structure (2C8E) and its readjustment upon NAD binding (2C8F) induce critical differences in both Phe209 and Gln212 residues compared with molecule A from the C3wt-free structure (2C89) (Fig. 3B). The side chain of Phe209 is closer to the NAD binding site by 2.5 Å, and upon NAD binding, it is pushed further toward the outside to avoid a clash with the side chain of Asn214 and the NAD. The side chain of Gln212 is prevented by Asn214 and Phe209 from positioning itself within the NAD-binding pocket; it points toward the outside.
Additional differences are observed at the nucleotide-binding site of the catalytic-glutamate mutant (E214N). In the absence of NAD, in molecule E, the mutated asparagine side chain points toward the deformed “in” ARTT loop conformation (2C8E) (Fig. 4A), while in the presence of NAD, it extends further toward the NAD (2C8F), as does the catalytic glutamate in the C3wt-NAD molecule A (1GZF) (Fig. 4B). In the C3-E214N-free structure (2C8E), the carboxamide group of the asparagine makes three hydrogen bonds with the carboxylate of Asp204, the hydroxyl group of Ser207, and the carbonyl group of Phe209 (Fig. 4A), while in the C3-E214N-NAD structure (2C8F), this asparagine makes a single hydrogen bond with the hydroxyl group of the NMN-ribose of NAD (Fig. 4B). While the catalytic glutamate can interact with Ser174 (1GZF), the asparagine in this position is too short to do the same (Fig. 4B). The situation is different for the “out” conformation adopted by the ARTT loop in molecules F and G from both C3-E214N structures (2C8E and 2C8F). The “out” conformation of the ARTT loop is characterized by an extra hydrogen bond made by the Gly211 main chain with the catalytic glutamate (2C89) (Fig. 4C). Although the side chain of the mutated asparagine adopts a position inside the NAD binding site, which is similar to that of the catalytic glutamate in molecules B, C, and D, again its side chain is too short to match this interaction (Fig. 4C). Its position is stabilized by a hydrogen bond with the carbonyl group of Thr175 from the STS motif at the bottom of the NAD binding site (Fig. 4C), which cannot be made by the catalytic glutamate due to its longer side chain and its chemical nature (Fig. 4C). Further stabilization of the Asn214 side chain is observed in molecules F and G, where its amide group makes a hydrogen bond with the hydroxyl group of Ser174 from the STS motif (Fig. 4C). This serine side chain shows a high degree of variability adopting various rotamers in different molecules and structures. Certain variations no longer allow it to interact with the hydroxyl group of Tyr79, as this is the case for molecule F/G in the C3-E214N-free structure (2C8) (Fig. 4C).
Figure 4.
The catalytic glutamate mutant (E214N) active site. The ARTT loop and residues from the NAD binding site are shown in gray. Hydrogen bonds are indicated by a dashed line (black). Orientation is conserved between the three panels. (A) C3-E214N-free structure (molE, 2C8E). (B) Comparison of the C3-E214N-NAD structure (molE, 2C8F) with the C3wt-NAD structure (molA, 1GZF; ball-and-sticks in white, and dash line in gray). (C) C3-E214N-free structure (molF/G, 2C8E).
Discussion
Recently, new structural data, in addition to those presented previously and in this study, have enlarged our understanding of the C3 exoenzyme structural properties (Han et al. 2001; Ménétrey et al. 2002; Evans et al. 2003; A. Teplyakov, G. Obmolova, G.L. Gilliland, and S. Narumiya, unpubl.). Thus, altogether, crystal structures for C3 exoenzyme are known (1) in different crystal packing environments, (2) unbound and bound to NAD or NAD-hydrolysis products, (3) with critical residues mutation (Gln212 and Glu214), and (4) from different species (C3bot1, C3bot2, and C3stau2) (see Table 2 for references and PDB codes). Based on these structural data, we present the C3-exoenzyme structural capability, focusing mainly on the ARTT loop and its impact along the catalysis pathway. Note that a C3 flexure reassessment is presented as Supplemental material.
Comparing the structures of the C3bot1 in complex with nicotinamide, the NAD-hydrolysis product of the ADP-ribosyltransferase reaction, and the unbound enzyme, we find that the main difference is localized within the PN loop. As reported previously for C3bot1 (Ménétrey et al. 2002), the PN loop is particularly flexible and poorly defined in the absence of NAD (and/or crystal packing contacts), while in the presence of substrate, it rearranges to interact with the nicotinamide moiety. An aromatic residue from the PN loop changes its conformation to cap the nicotinamide part inside the cleft. We show here that the nicotinamide molecule alone is sufficient to trigger the PN loop and aromatic residue conformational changes observed upon NAD binding. The PN loop appears to be a nicotinamide sensor that plays important roles in locking the NAD or the nicotinamide in position inside the catalytic site during the enzymatic process (Ménétrey et al. 2002).
A conserved glutamate inside the ARTT motif has been identified as a critical residue for the NAD hydrolysis step (Aktories et al. 1995; Saito et al. 1995; Bohmer et al. 1996; Wilde et al. 2002b, 2003). Structural studies have shown that this catalytic glutamate extends toward the NAD binding site irrespective of the ARTT loop conformation and makes direct interactions with NAD (Ménétrey et al. 2002; Evans et al. 2003; this study). The E214N mutation induces changes in the ARTT loop and affects NAD binding. When the ARTT loop adopts the “out” conformation (molecules F and G) (Table 2), the non-catalytic asparagine makes a set of interactions (Fig. 4C) that are only possible due to its shorter side chain and its chemical nature. In this conformation, NAD binding seems to be hindered, since no NAD molecule is found bound to these molecules. This correlates well with biochemical data that show that the catalytic glutamate mutation causes a significant reduction in affinity for NAD (Saito et al. 1995). However, in the “in” conformation (molecule E) (Table 2), NAD can still bind to the C3-E214N (Fig. 4B). The non-catalytic Asn214 makes a hydrogen bond with the O2′ hydroxyl from the NMN ribose as the catalytic glutamate, but it does so with a different orientation due to its shorter side chain (Fig. 4B). Beyond the significant reduction for the NAD hydrolysis step, this catalytic glutamate mutation would also impair other catalytic steps. Indeed, mutation of Glu214 to asparagine triggers the deformation of the “in” conformation of the ARTT loop that displaces the critical Phe209 and Gln212 residues from their respective positions (Fig. 3B). Both Phe209 and Gln212 are pushed away from the catalytic site on NAD binding (Figs. 3B, 4B). This movement is particularly interesting since Phe209 is involved in Rho binding and Gln212 in the ADP-ribose transfer step; thus, such displacements would alter the overall mechanism.
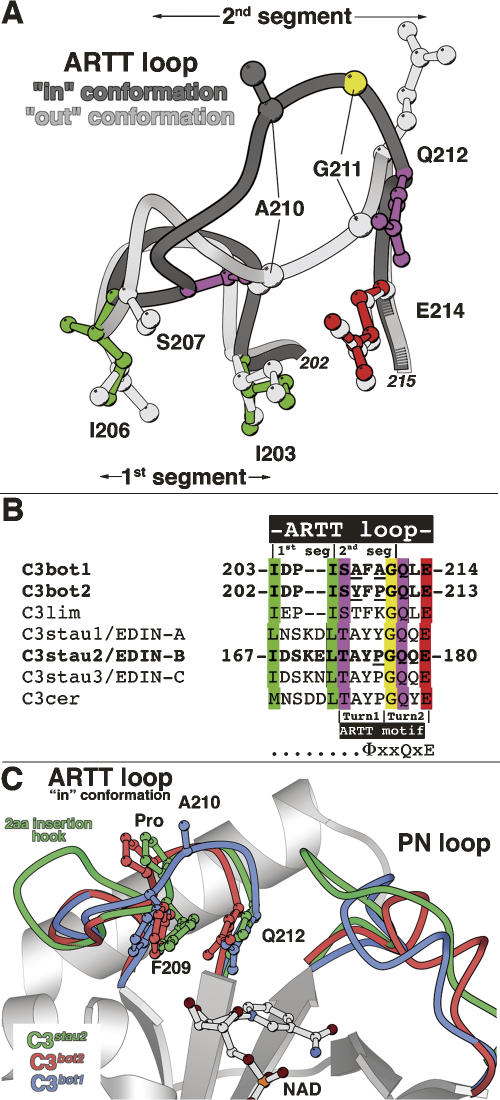
Two conformations have been observed for the ARTT loop so far: the “in” conformation defined by an extended loop with residues making only few self-stabilization interactions, and the “out” conformation defined by a more compact loop with residues making multiple intra-loop interactions with a large contribution from Asp204. The ARTT loop can be defined as two structural segments (Fig. 5A): The first one encompasses residues Ile203–Ile206 with both highly conserved isoleucines as hydrophobic anchors, maintaining the ARTT loop pinched between helices α1 and α2 and probably reducing the flexibility for this segment. The second one comprises residues Ser207–Gln212 with Ser207 and Gln212 being hinge residues that allow this loop to change from the “in” to the “out” conformation and Ala210–Gly211 suffering the greatest displacements between both conformations. The second segment is encompassed by the two-turn ARTT motif (ΦxxQxE) (Fig. 5B) previously defined in the comparison with the C3 exoenzymes and the actin-ADP-ribosyltransferase toxins (Han et al. 2001; Han and Tainer 2002). The last two residues of the ΦxxQxE motif are not included in the second segment, as they are not involved in the movement of the ARTT loop. While the length of the first segment is not conserved among C3 exoenzyme isoforms, that of the second one is (Fig. 5B). Despite differences in sequence, the conformation of the second segment (“in” conformation) between C3bot1, C3bot2 (A. Teplyakov, G. Obmolova, G.L. Gilliland, and S. Narumiya, unpubl.), and C3stau2 (Evans et al. 2003) remains largely unchanged (Fig. 5C). In the C3bot2 (A. Teplyakov, G. Obmolova, G.L. Gilliland, and S. Narumiya, unpubl.), Ala208 and Ala210 from C3bot1 are replaced by tyrosine and proline, respectively, while the C3stau2 (Evans et al. 2003) maintains the Ala208 and shares with C3bot2 the proline at position 210 (Fig. 5B).
Figure 5.
The ARTT loop in C3-like exoenzymes. (A) The two structural segments of the ARTT loop. Superposition of the two distinct conformations of the ARTT loop (dark and light gray) are shown with critical residues for understanding its structural properties. The first segment encompasses two conserved anchor hydrophobic residues (I203 and I206 in green for the “in” conformation). The second segment encompasses two hinge residues, Ser207 and Gln212 (in purple for the “in” conformation), which allow the ARTT loop conformational change. Gly211 (in yellow for the “in” conformation) from this second segment makes the most important deviation between both conformations. Note that the catalytic glutamate is highlighted in red (for the “in” conformation). Equivalent residues from the “out” conformation are shown in light gray for clarity. (B) Sequence alignment of the C3-like exoenzymes. The color code is conserved with the “in” conformation shown in Figure 5A. In bold are the C3 isoforms for which the structure is known. (Underline) Differences in sequence among C3bot1, C3bot2, and C3stau2 mentioned in the text. Note that the position of the two turns of the ARTT motif are indicated (Han and Tainer 2002). (C) Superimposition of the ARTT loop “in” conformation of the C3bot1 (C3wt-NAD; molA), C3bot2, and C3stau2 structure.
Most of the structural differences are seen in the first segment. A two-residue insertion in the first segment of the ARTT loop occurs in C3stau and C3cer isoforms (Fig. 5B). The two-residue insertion protrudes as a hook in C3stau2 compared with the C3bot1 and C3bot2 structures (Fig. 5C; Evans et al. 2003). Evans et al. (2003) proposed that this insertion favors the “in” conformation observed for the ARTT loop. Given that both hydrophobic anchors that delineate the first segment are in the same position in C3bot1 /bot2 and C3stau2 structures, it is more likely that the two-residue insertion in C3stau2 has little effect on the overall ARTT loop conformation. The preference for the “in” conformation is by default, since in both C3stau2 and C3bot2, crystal contacts prevent the ARTT loop from adopting the “out” conformation. The hook protrusion in C3stau that does not exist in C3bot exoenzymes (Fig. 5B,C) could, however, be associated with RhoE/Rnd3 recognition, a feature distinguishing C3stau from the other isoforms.
The numerous crystal structures presented here have allowed us, in most cases, to differentiate between changes as a result of crystal contact interactions from those due to mutations and nucleotide binding. Nonetheless, crystal contacts are not uninteresting, as they could mimic protein–protein interactions. The ARTT loop has a strong tendency to make protein–protein interactions in the crystal lattice, which could reflect the role it has in target recognition. These crystal contacts influence the choice of the “in” or “out” conformation and can also induce loop deformations. We have observed such deformations in two distinct cases; one is the consequence of the crystal-packing environment (C3bot1-Q212A-Free form II, molX; Fig. 3A), while the second is more probably due to the specific mutation within the ARTT loop (C3bot1-E214N, mole; Fig. 3B). These deformations are not specific to a given ARTT loop conformation, since they have been observed in both “in” and “out” conformations. C3stau2 and C3bot2 also have slightly deformed ARTT loops (“in” conformation) compared with that of C3bot1 (Fig. 5C). Superposition of C3bot2 and C3stau2 reveals that the second segment of their ARTT loop adopts virtually the same deformation relative to that of C3bot1 (Fig. 5C), probably caused by the proline, instead of Ala210, shared by both isoforms.
Conclusion
Apart from minor deformations, the ARTT loop has always been observed either in the “in” or the “out” conformation. This emphasizes the fact that the ARTT loop is not a flexible region that can be easily modified by different crystal packing contacts. If this had been the case, more than two distinct conformations would have been observed, but rather, it would appear to be a plastic region able to adopt a limited number of specific conformations associated with a well-defined function. The question regarding the functional relevance of these two ARTT loop conformations is only partially answered. One previously proposed hypothesis suggests that upon NAD binding, the ARTT loop undergoes a conformational change to adopt a distinct conformation that positions Phe209 for Rho binding (Ménétrey et al. 2002). Still compatible with this hypothesis, such a conformational change could also be triggered upon Rho binding and/or during the transfer of the ADP-ribose from the NAD binding site to Asn41 of Rho.
Materials and Methods
Expression and mutation
The mature form of wild-type C. botulinum C3 exoenzyme (residues 41–251) was subcloned into the pET-28a vector, expressed in the BL21 (DE3) strain of Escherichia coli, and purified to homogeneity in one step by cation exchange chromatography (CM Sepharose Fast Flow). Mutants were constructed by site-directed mutagenesis with the pET-28a C3 wild type as templates and the respective oligonucleotides using the Stratagene QuikChange kit according to the manufacturer's instructions. The different C3 mutants (C3-E214N and C3-Q212A) were expressed and purified as the wild type.
Crystallization and NAD soaking
Wild-type and mutant C3bot1 exoenzyme crystals were grown by the sitting-drop vapor diffusion method at 20°C from protein at 12 mg/mL in 15 mM NaCl, 25 mM Tris pH 7.4. To obtain variations in the lattice of wild-type and mutant crystals, the method of reverse screening was used (Stura 1998). The reservoir solutions consisted of a 95% dilution of the same working solution used previously for C3bot1 L177C-Hg-free (Ménétrey et al. 2002) consisting of 22.5% PEG 3350 w/w, 100 mM Li2SO4, 100 mM sodium citrate, pH 3.0, to which a perturbation of 3%–10% MPEG 550 v/v was applied to compensate for variations in protein solubility of the mutants compared with the wild type and to modulate (mainly reduce) the solvent content of the lattice.
Streak seeding was used to define the optimal amount of MPEG 550 to be added and macroseeding was used to obtain large single crystals. A cryoprotectant consisting of 10% propanol, 15% ethylene glycol, 6% xylitol, 22.5% PEG 3350 w/w, and 100 mM citric acid at pH 3 was used to flash-freeze crystals in liquid ethane. Complexes were obtained by adding 10–15 mM of NAD or nicotinamide to the cryoprotectant solution and flash-freezing the crystals in liquid ethane after a 20–30 min soak. The same procedure was used previously (Ménétrey et al. 2002).
X-ray data collection, structure determination, and refinement
X-ray diffraction data set were collected at −170°C on the ESRF beamline ID14-eh1, ID14-eh3, and BM30a. All of the data were processed with DENZO and scaled with SCALEPACK (Otwinowski and Minor 1997) (see Table 1). All structures were solved by molecular replacement with the AmoRe program (Navaza 1994) using the C3bot1 exoenzyme wild-type model (1GZF) lacking the ARTT loop. All C3 molecules from the asymmetric unit were refined independently. All structures were first treated with rigid-body refinements, followed by simulated annealing, energy minimization, and isotropic B-factor refinements through maximum-likelihood as implemented in CNS 1.1 (Brünger et al. 1998). For mutant structures, electronic density was checked before deleted mutated side chain. Then, structures with resolution below 2.2 Å were automatically built (N and C terminus and ARTT loop) and refined with ARP/wARP 5 (Perrakis et al. 1999). Others were manually rectified with Turbo (Roussel and Cambillaud 1989) and refined with CNS 1.1 (Brünger et al. 1998). After protein refinement, NAD, NAD-hydrolyzed products, and nicotinamide were modeled, and water molecules were added using ARP/wARP 5 (Perrakis et al. 1999). The final statistics and PDB accession code (Protein Data Bank) for the models are summarized in Table 1. Note that all figures were computed using Molscript and Bobscript (Kraulis 1991; Esnouf 1999).
Data deposition
The atomic coordinates and structure factors have been deposited at the Protein Data Bank (PDB) under PDB ID, 2C89, 2C8A, 2C8B, 2C8C, 2C8D, 2C8E, and 2C8F.
Acknowledgments
This work was supported by grants from the Programme de Recherche Fondamentale Microbiologie Maladies Infectieuses et Parasitaires. We thank the staff of the European Synchrotron Radiation Facility for assistance during data collection.
Footnotes
Supplemental material: see www.proteinscience.org
Reprint requests to: Julie Ménétrey, Institut Curie, UMR144 26 rue d'Ulm, 75248 Paris Cedex 05, France; e-mail: julie.Menetrey@curie.fr; fax: 33-1-42-34-63-82; or Enrico A. Stura, Service d'Ingénierie Moléculaire des Protéines (SIMOPRO), iBiTec-S, DSV, CEA-Saclay 91191 GIF sur Yvette Cedex, France; e-mail: estura@cea.fr; fax: 33-1-69-08-90-71.
Abbreviations: ARTT, ADP-ribosyl toxin turn-turn; PN, phosphate-nicotinamide; NMN, nicotinamide mono-nucleotide; RMSD, root mean square deviation; GEF, guanine-exchange factor.
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.073398508.
References
- Aktories, K., Frevert, J. ADP-ribosylation of a 21–24 kDa eukaryotic protein(s) by C3, a novel botulinum ADP-ribosyltransferase, is regulated by guanine nucleotide. Biochem. J. 1987;247:363–368. doi: 10.1042/bj2470363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aktories, K., Rosener, S., Blaschke, U., Chhatwal, G.S. Botulinum ADP-ribosyltransferase C3. Purification of the enzyme and characterization of the ADP-ribosylation reaction in platelet membranes. Eur. J. Biochem. 1988;172:445–450. doi: 10.1111/j.1432-1033.1988.tb13908.x. [DOI] [PubMed] [Google Scholar]
- Aktories, K., Jung, M., Bohmer, J., Fritz, G., Vandekerckhove, J., Just, I. Studies on the active-site structure of C3-like exoenzymes: Involvement of glutamic acid in catalysis of ADP-ribosylation. Biochimie. 1995;77:326–332. doi: 10.1016/0300-9084(96)88142-9. [DOI] [PubMed] [Google Scholar]
- Aktories, K., Wilde, C., Vogelsgesang, M. Rho-modifying C3-like ADP-ribosyltransferases. Rev. Physiol. Biochem. Pharmacol. 2004;152:1–22. doi: 10.1007/s10254-004-0034-4. [DOI] [PubMed] [Google Scholar]
- Bohmer, J., Jung, M., Sehr, P., Fritz, G., Popoff, M., Just, I., Aktories, K. Active site mutation of the C3-like ADP-ribosyltransferase from Clostridium limosum–analysis of glutamic acid 174. Biochemistry. 1996;35:282–289. doi: 10.1021/bi951784+. [DOI] [PubMed] [Google Scholar]
- Brünger, A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.S., Kuszewski, J., Nilges, M., Pannu, N.S., et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Esnouf, R.M. Bobscript: Further additions to MolScript version 1.4, including reading and contouring of electron-density maps. Acta Crystallogr. D Biol. Crystallogr. 1999;55:938–940. doi: 10.1107/s0907444998017363. [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville, S., Hall, A. Rho GTPases in cell biology. Nature. 2002;420:629–635. doi: 10.1038/nature01148. [DOI] [PubMed] [Google Scholar]
- Evans, H.R., Sutton, J.M., Holloway, D.E., Ayriss, J., Shone, C.C., Acharya, K.R. The crystal structure of C3stau2 from Staphylococcus aureus and its complex with NAD. J. Biol. Chem. 2003;278:45924–45930. doi: 10.1074/jbc.M307719200. [DOI] [PubMed] [Google Scholar]
- Genth, H., Gerhard, R., Maeda, A., Amano, M., Kaibuchi, K., Aktories, K., Just, I. Entrapment of Rho ADP-ribosylated by Clostridium botulinum C3 exoenzyme in the Rho-guanine nucleotide dissociation inhibitor-1 complex. J. Biol. Chem. 2003a;278:28523–28527. doi: 10.1074/jbc.M301915200. [DOI] [PubMed] [Google Scholar]
- Genth, H., Schmidt, M., Gerhard, R., Aktories, K., Just, I. Activation of phospholipase D1 by ADP-ribosylated RhoA. Biochem. Biophys. Res. Commun. 2003b;302:127–132. doi: 10.1016/s0006-291x(03)00112-8. [DOI] [PubMed] [Google Scholar]
- Han, S., Tainer, J.A. The ARTT motif and a unified structural understanding of substrate recognition in ADP-ribosylating bacterial toxins and eukaryotic ADP-ribosyltransferases. Int. J. Med. Microbiol. 2002;291:523–529. doi: 10.1078/1438-4221-00162. [DOI] [PubMed] [Google Scholar]
- Han, S., Arvai, A.S., Clancy, S.B., Tainer, J.A. Crystal structure and novel recognition motif of Rho ADP-ribosylating C3 exoenzyme from Clostridium botulinum: Structural insights for recognition specificity and catalysis. J. Mol. Biol. 2001;305:95–107. doi: 10.1006/jmbi.2000.4292. [DOI] [PubMed] [Google Scholar]
- Inoue, S., Sugai, M., Murooka, Y., Paik, S.Y., Hong, Y.M., Ohgai, H., Suginaka, H. Molecular cloning and sequencing of the epidermal cell differentiation inhibitor gene from Staphylococcus aureus . Biochem. Biophys. Res. Commun. 1991;174:459–464. doi: 10.1016/0006-291x(91)91438-i. [DOI] [PubMed] [Google Scholar]
- Just, I., Mohr, C., Schallehn, G., Menard, L., Didsbury, J.R., Vandekerckhove, J., van Damme, J., Aktories, K. Purification and characterization of an ADP-ribosyltransferase produced by Clostridium limosum . J. Biol. Chem. 1992;267:10274–10280. [PubMed] [Google Scholar]
- Just, I., Selzer, J., Jung, M., van Damme, J., Vandekerckhove, J., Aktories, K. Rho-ADP-ribosylating exoenzyme from Bacillus cereus. Purification, characterization, and identification of the NAD-binding site. Biochemistry. 1995;34:334–340. doi: 10.1021/bi00001a041. [DOI] [PubMed] [Google Scholar]
- Kraulis, P.J. MOLSCRIPT: A program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 1991;24:946–950. [Google Scholar]
- Ménétrey, J., Ménez, A. Structural features of bacterial ADP-ribosyltransferase toxins. In: Gillet D., Johannes L., editors. Recent Research Developments in Toxins from Bacteria and Other Organisms. Research Signpost; Kerala, India: 2006. pp. 241–263. [Google Scholar]
- Ménétrey, J., Flatau, G., Stura, E.A., Charbonnier, J., Gas, F., Teulon, J., Le Du, M., Boquet, P., Ménez, A. NAD binding induces conformational changes in Rho ADP-ribosylating Clostridium botulinum C3 exoenzyme. J. Biol. Chem. 2002;277:30950–30957. doi: 10.1074/jbc.M201844200. [DOI] [PubMed] [Google Scholar]
- Navaza, J. AMoRe: An automated package for molecular replacement. Acta Crystallogr. A. 1994;50:157–163. [Google Scholar]
- Nemoto, Y., Namba, T., Kozaki, S., Narumiya, S. Clostridium botulinum C3 ADP-ribosyltransferase gene. Cloning, sequencing, and expression of a functional protein in Escherichia coli . J. Biol. Chem. 1991;266:19312–19319. [PubMed] [Google Scholar]
- Otwinowski, Z., Minor, W. HKL package. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Perrakis, A., Morris, R.M., Lamzin, V.S. ARP/wARP can be employed for automatic density interpretation and tracing of a protein model. Resolution around 2.0 Å is the current limit…but we are working on it! Nat. Struct. Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- Roussel, A., Cambillaud, C. Silicon Graphics Geometry Partner Directory. Silicon Graphics; Mountain View, CA: 1989. Turbo; pp. 77–78. [Google Scholar]
- Rubin, E.J., Gill, D.M., Boquet, P., Popoff, M.R. Functional modification of a 21-kilodalton G protein when ADP-ribosylated by exoenzyme C3 of Clostridium botulinum . Mol. Cell. Biol. 1988;8:418–426. doi: 10.1128/mcb.8.1.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito, Y., Nemoto, Y., Ishizaki, T., Watanabe, N., Morii, N., Narumiya, S. Identification of Glu173 as the critical amino acid residue for the ADP- ribosyltransferase activity of Clostridium botulinum C3 exoenzyme. FEBS Lett. 1995;371:105–109. doi: 10.1016/0014-5793(95)00851-y. [DOI] [PubMed] [Google Scholar]
- Sehr, P., Joseph, G., Genth, H., Just, I., Pick, E., Aktories, K. Glucosylation and ADP ribosylation of rho proteins: Effects on nucleotide binding, GTPase activity, and effector coupling. Biochemistry. 1998;37:5296–5304. doi: 10.1021/bi972592c. [DOI] [PubMed] [Google Scholar]
- Sekine, A., Fujiwara, M., Narumiya, S. Asparagine residue in the Rho gene product is the modification site for botulinum ADP-ribosyltransferase. J. Biol. Chem. 1989;264:8602–8605. [PubMed] [Google Scholar]
- Stura, E.A. Strategy 3: Reverse screening. In: Bergfors T.M., editor. Protein crystallization: Techniques, strategies and tips. A laboratory manual. International University Line; La Jolla, CA: 1998. pp. 113–124. [Google Scholar]
- Wilde, C., Chhatwal, G.S., Schmalzing, G., Aktories, K., Just, I. A novel C3-like ADP-ribosyltransferase from Staphylococcus aureus modifying RhoE and Rnd3. J. Biol. Chem. 2001;276:9537–9542. doi: 10.1074/jbc.M011035200. [DOI] [PubMed] [Google Scholar]
- Wilde, C., Barth, H., Sehr, P., Han, L., Schmidt, M., Just, I., Aktories, K. Interaction of the Rho-ADP-ribosylating C3 exoenzyme with RalA. J. Biol. Chem. 2002a;277:14771–14776. doi: 10.1074/jbc.M201072200. [DOI] [PubMed] [Google Scholar]
- Wilde, C., Just, I., Aktories, K. Structure-function analysis of the Rho-ADP-ribosylating exoenzyme C3stau2 from Staphylococcus aureus . Biochemistry. 2002b;41:1539–1544. doi: 10.1021/bi015809i. [DOI] [PubMed] [Google Scholar]
- Wilde, C., Vogelsgesang, M., Aktories, K. Rho-specific Bacillus cereus ADP-ribosyltransferase C3cer cloning and characterization. Biochemistry. 2003;42:9694–9702. doi: 10.1021/bi034583b. [DOI] [PubMed] [Google Scholar]
- Yamaguchi, T., Hayashi, T., Takami, H., Ohnishi, M., Murata, T., Nakayama, K., Asakawa, K., Ohara, M., Komatsuzawa, H., Sugai, M. Complete nucleotide sequence of a Staphylococcus aureus exfoliative toxin B plasmid and identification of a novel ADP-ribosyltransferase, EDIN-C. Infect. Immun. 2001;69:7760–7771. doi: 10.1128/IAI.69.12.7760-7771.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]