Abstract
Integrins are α/β heterodimers, but recent in vitro and in vivo experiments also suggest an ability to associate through their transmembrane domains to form homomeric interactions. While the results of some in vitro experiments are consistent with an interaction mediated by a GxxxG-like motif, homo-oligomers observed after in vivo cross-linking are consistent with an almost opposite helix–helix interface. We have shown recently that both models of interaction are compatible with evolutionary conservation data, and we predicted that the α-helices in both models would have a similar rotational orientation. Herein, we have tested our prediction using in vitro asparagine scan of five consecutive residues along the GxxxG-like motif of the transmembrane domain of α and β integrins, αM and β2. We show that Asn-mediated dimerization occurs twice for every turn of the helix, consistent with two almost opposite forms of interaction as suggested previously for αIIb and β3 transmembrane domains. The orientational parameters helix tilt and rotational orientation of each of these two Asn-stabilized dimers were measured by site-specific infrared dichroism (SSID) in model lipid bilayers and were found to be consistent with our predicted computational models. Our results highlight an intrinsic tendency for integrin transmembrane α-helices to form two opposite types of homomeric interaction in addition to their heteromeric interactions and suggest that integrins may form complex and specific networks at the transmembrane domain during function.
Keywords: asparagine scan, integrins, infrared, oligomerization, transmembrane, GxxxG motif
Integrins are heterodimeric type I transmembrane proteins formed by noncovalent association of an α and a β subunit. Each subunit contains a large extracellular domain, a single transmembrane (TM) spanning α-helix, and a short cytoplasmic tail (Carman and Springer 2003). Different types of α integrins can combine with different β counterparts, forming a variety of heterodimers. In humans, 18 α-chains can interact with eight different β-chains to form 24 different α/β heterodimers with varied functions (Hemler 1999). By spanning the membrane, the integrins serve as a dynamic linkage between cytoplasm and extracellular space, transducing signals across the membrane to mediate cell growth, differentiation, gene expression, motility, and apoptosis (Watt 2002). Inside out signal transduction involves integrin cytoplasmic tails separation and subsequent ectodomain conformational changes, which alter the affinity of integrins for extracellular ligands (Hantgan et al. 1999; Kim et al. 2003; Travis et al. 2003).
Several experimental results suggest that the α and β subunits of integrins interact specifically at the membrane domain (Hughes et al. 1996; Xiong et al. 2001; Adair et al. 2005); for example, electron cryomicroscopy and single particle analysis (Adair and Yeager 2002), cysteine scanning mutagenesis (Luo et al. 2004), or the observed activation after disruption of transmembrane interactions in integrin αIIbβ3 (Luo et al. 2005). At present, there is a general consensus on the type of transmembrane interaction in the inactive, low-affinity form of integrin (Gottschalk et al. 2002; Luo et al. 2004; Li et al. 2005).
However, in addition to the above evidence for heteromeric α/β transmembrane interaction, α and β TM chains have been shown to have a strong tendency to form homo-oligomers (αα or ββ) in vitro, both in zwitterionic and acidic micelles (Li et al. 2001) and in biological membranes (Lu et al. 2001; Li et al. 2004; Schneider and Engelman 2004). The functional relevance of these homomeric interactions has been discussed (Luo et al. 2004; Li et al. 2005), with a possible role in integrin clustering when binding to multimeric ligands.
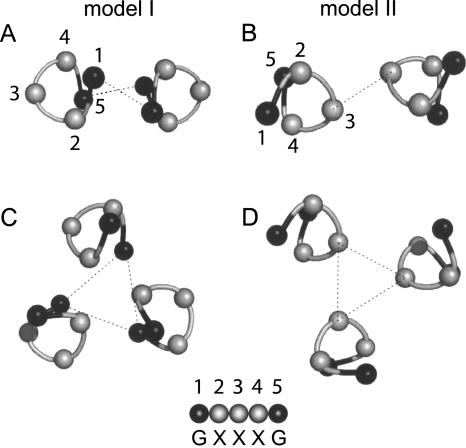
In a recent computational work (Lin et al. 2006), we have tested the stability of integrin homomeric (dimeric and trimeric) transmembrane models, using evolutionary conservation data as a filter (Briggs et al. 2001). We found that two homomeric models, with almost opposite form of interaction (which we referred to as models I and II) (see Fig. 1) were stable; that is, only these two models appeared during the simulation of each of the homologous sequences.
Figure 1.
Schematic representation of the two modes of interaction (I and II). Slices through the α-helical structure of the integrin TM model we predicted previously using a computational method (Lin et al. 2006). Only the α carbons are shown. (A) Homodimer model I; (B) homodimer model II; (C) homotrimer model I; (D) homotrimer model II. In model I, the interaction takes place mediated by residues 1 and 5 (black balls) of the GxxxG-like motif. In model II, the interaction is opposite, through residue 3. Dotted lines are shown only to guide the eye. A more detailed representation of these structures is shown in Lin et al. (2006).
In our model I (Fig. 1), the helix–helix interaction is mediated by a GxxxG-like motif, similarly to the interaction observed for dimeric glycophorin A (GpA) (Lemmon et al. 1992). This is not surprising, because the GxxxG motif is prevalent in TM sequences (Arkin and Brunger 1998) and in integrins in particular. The importance of this and other related motifs in α-helical TM domains has been demonstrated in exhaustive statistical analyses (Senes et al. 2000). In addition, a selection of a random library of TM sequences for homodimerization clearly showed that the GxxxG motif is sufficient for strong helix–helix interactions (Russ and Engelman 2000). The importance of this motif is highlighted by other experimental observations. For example, using the TOXCAT assay (Russ and Engelman 1999), a test that measures the oligomerization in the Escherichia coli inner membrane of a chimeric protein containing a TM helix, a sequence critical for integrin αIIb-TM homodimerization that involved the GxxxG motif was suggested by Li et al. (2004). Also, using a GALLEX assay (Schneider and Engelman 2003), a two-hybrid system that monitors heterodimerization of membrane proteins in the E. coli inner membrane, the GxxxG-like motif was found to have an important role in homomeric transmembrane interactions in α4 and β7 (Schneider and Engelman 2004).
Interactions consistent with model II (see Fig. 1) have also been observed experimentally: For example, in α TM chains of αIIb/β3, an α–α dimer was stabilized by disulfide cross-linking using the cysteine mutant W967C (Luo et al. 2004). In β TMs, in vivo asparagine substitution G708N (Li et al. 2003) led to integrin activation and homotrimeric interactions were observed. In the cases above, the residues involved in the helix–helix interaction are opposite to the “G” residues in the GxxxG motif, hence consistent with our model II. These studies, however, were performed with the system αIIb/β3, and a detailed model for helix–helix interaction was not shown.
Herein, we have conducted an asparagine scan study to test our prediction, using synthetic peptides corresponding to the TM domains of integrin. Asparagine scan mutagenesis relies on the strong hydrogen bonds between asparagine side chains located in neighboring transmembrane domains, and their stabilization of helix–helix interactions (Choma et al. 2000; Zhou et al. 2001; Li et al. 2003; Ruan et al. 2004a,b). Asn residues are not abundant in transmembrane α-helices (Stevens and Arkin 1999), and when present generally point toward other α-helices. The assumption in asparagine scans is that only when the asparagine side chain is in a favorable position will oligomerization be enhanced. However, this method can only be used when TM interactions for the native peptide are weak, that is, when only monomers are present in SDS in the absence of asparagine. This condition is met for αM and β2 TMs, but not for other integrin TMs, for example, αIIb, β3, or β4, which homo-oligomerize in SDS (data not shown). Hence, we have performed our study using αM and β2 TMs. The pair αM/β2, also referred to as Mac-1 or CR-3, is a major surface antigen on human leukocytes. Dysfunction of this integrin is associated with Leukocyte Adhesion Deficiency-1 (LAD1) and Glanzmann thrombasthenia (McDowall et al. 2003; Wehrle-Haller and Imhof 2003).
Because we expected the GxxxG motif to be involved in integrin TM interactions, we sequentially mutated to asparagine a stretch of five consecutive residues encompassing the GxxxG-like motif, both in the α- and β-chain. If only one model of homomeric interaction exists and is mediated by a GxxxG-like motif, oligomerization should be favored when Asn is at positions 1 and 5 of the motif (see Fig. 1). Conversely, if two opposite models exist (Lin et al. 2006), oligomerization should also be observed when Asn is at position 3. Finally, if the Asn scan is not specific, oligomerization should be detected at any position.
Note that although the models I and II represent opposite TM interactions, the rotational orientation of the component α-helices is very similar, according to our previous prediction (Lin et al. 2006). We have tested this by determining the rotational orientation of the α-helices in these oligomers using site-specific infrared dichroism (SSID) (Arkin et al. 1997; Torres et al. 2002) with isotopically labeled synthetic peptides when they are incorporated in model lipid bilayers (DMPC).
Results
Figure 1 shows the two schematic models (I and II) of interaction for homodimers and homotrimers in the transmembrane domain of integrins that we predicted previously using a computational method (Lin et al. 2006). To test the validity of these models, we have used here an asparagine scan strategy, targeting five consecutive transmembrane residues spanning the GxxxG-like motif, labeled 1–5 in Figure 1. We have used the transmembrane domains of αM and β2 integrins, which form a known integrin pair (Hynes 2002). These two transmembrane domains are monomeric in SDS in their native form.
Electrophoreses of αM-TM and β2-TM
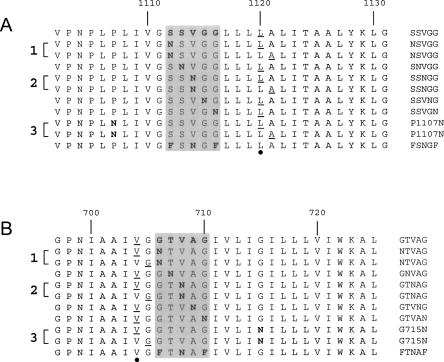
The αM-TM and β2-TM peptides synthesized are shown in Figure 2. In this figure, the location of the GxxxG-like motif is shown by a shaded area (see the figure legend for details).
Figure 2.
Sequences corresponding to the synthesized peptides αM and β2. (A) α-Helical transmembrane (TM) domains of integrin αM. (B) α-Helical TM domains of integrin β2. The five consecutive residues corresponding to the GxxxG-like motif are enclosed in a shaded box, and shown on the right of the sequences. Mutated (Asn) residues are indicated in bold, and the residues isotopically labeled with 13C=18O are underlined. The three pairs of peptides labeled with two consecutive residues, and used to find the orientational parameters with SSID, are indicated by a number on the left (1–3). The residue for which the rotational orientation, ω, was calculated is indicated by a black dot at the bottom of each panel.
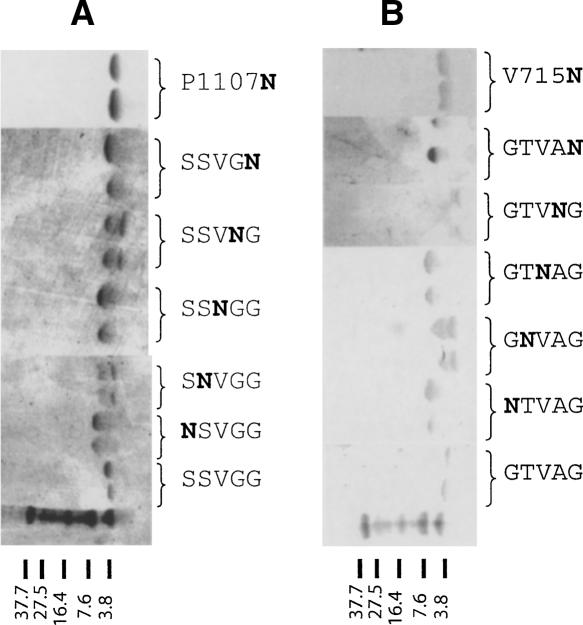
Figure 3 shows the SDS electrophoreses of each one of these peptides. Those corresponding to αM (Fig. 3A) show that the peptide corresponding to the native sequence (labeled SSVGG) is monomeric. In contrast, introduction of asparagine at positions 1 and 5 of the GxxxG-like motif (NSVGG and SSVGN) produced dimers. Because of the position of the asparagine mutation, the helix–helix interaction in the dimer observed in SDS can only be mediated by the GxxxG-like motif, or model I in Figure 1A.
Figure 3.
SDS-PAGE electrophoreses of the synthetic TMs αM and β2. (A) Electrophoreses corresponding to αM. (B) Electrophoreses corresponding to β2. Lanes run from left to right. The bottom lane represents the molecular weight markers. The sequence of letters on the right of each lane indicates the Asn mutation within the GxxxG-like motif.
Crucially, a dimer was also observed when we introduced the asparagine at position 3 of the motif (SSNGG). In this case, the asparagine is located almost opposite (200° in a canonical α-helix) from positions 1 and 5; therefore, in this case, the helix–helix interaction would be consistent with model II in Figure 1B.
As expected, introduction of asparagine at positions 2 and 4 (SNVGG and SSVNG), which are not located on the helix–helix interface in any of the two models (see Fig. 1), produced monomers and some dimers, confirming that these positions only contribute partially to the stabilization of the oligomer. An electrophoretic mobility plot corresponding to these results is shown in the Supplemental material.
In addition to those five residues, we introduced asparagine at an additional position in αM. The rationale is that a recent report (Luo et al. 2004) described an α–α dimer stabilized by a disulfide bond when the mutation W967C was introduced in αIIb. This residue is in phase with position 3 of the GxxxG-like motif, and therefore the interaction described in that dimer must have been consistent with our model II. Residue P1107 in αM is equivalent to W967 in αIIb according to the alignment of α-integrin TM domains (Lin et al. 2006); therefore, we introduced an asparagine residue in αM at P1107. This peptide is labeled P1107N in Figure 3A. In this case, a clear dimer was also observed, which is consistent with a model II of interaction. The fact that αIIb and αM present this form of interaction suggests that it may be general for the integrin family and related to integrin function.
For β2-TM (Fig. 3B), the SDS electrophoresis results were very similar to those observed for αM-TM. The native sequence (labeled GTVAG) produced only monomers in SDS, whereas introduction of asparagine at positions 1 and 5 of the GxxxG-like motif (NTVAG and GTVAN) produced only dimers. The position of the asparagine residue in each case implies that the interaction is consistent with model I.
Dimers were also observed when we introduced the asparagine at the third position of the motif (GTNAG). In this case, the asparagine is located almost opposite (200° in a canonical α-helix) from positions 1 and 5, and the interaction between β TMs can only be consistent with model II. When asparagine was introduced at positions 2 and 4 (GNVAG and GTVNG), we could observe both monomers and dimers, which suggests that this position only contributes partially to oligomer stabilization because positions 2 and 4 of the motif are not located exactly on the helix–helix interface in either model (see Fig. 1).
As in αM-TM, we introduced an additional mutation in order to confirm the result of a recent experiment that found that mutation G708N in β3 induced trimerization and activation in vivo (Li et al. 2003). From the alignment of β TM sequences (Lin et al. 2006), we see that the equivalent residue of G708N in the sequence β2 is V715, which we therefore mutated to Asn. Indeed, a dimer was observed (Fig. 3B, V715N). This result is consistent with a model II type of interaction because this position is seven residues apart (almost in phase) with position 3 of the GxxxG-like motif (see Fig. 2).
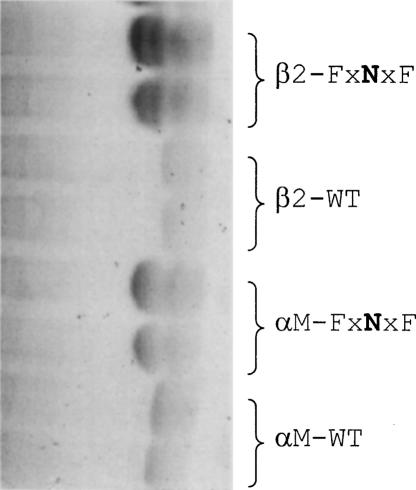
To further confirm that the helix–helix interactions observed after introducing Asn at position 3 (xxNxx) correspond to model II and are not mediated by the “G” residues in the GxxxG-like motif, we repeated the experiment with two additional mutations where positions 1 and 5 were changed to Phe (FxNxF). If a model I interaction was responsible for the dimers observed, the bulky Phe side chains would destabilize it, producing only monomers. Figure 4 shows that, even in the presence of Phe at positions 1 and 5 (FxNxF), dimers are still present, confirming that the dimerization observed is mediated by Asn at position 3, that is, consistent with model II.
Figure 4.
SDS-PAGE electrophoreses of synthetic TMs of integrin containing Phe mutations in the GxxxG-like motif. Lanes run from left to right, and the typed sequence on the right indicates the integrin type and the position of Asn and Phe mutations in the GxxxG-like motif. The mobility is compared to the wild-type (monomeric) sequences of αM and β2.
Orientation of the TM α-helices in a lipid bilayer
Using site-specific infrared dichroism, SSID (Arkin et al. 1997; Torres et al. 2000), it is possible to determine the tilt and rotational orientation for transmembrane α-helices in hydrated lipid bilayers. Therefore, we calculated these orientational values experimentally for αM-TM and β2-TM, using the pairs of labeled sequences (numbered 1–3) shown in Figure 2. Each pair contains two consecutive isotopic labels (underlined) that allow calculation of the orientational parameters (see legend in Fig. 2). In this case, the rotational orientation of the α-helices in models I and II turned out to be very similar (Lin et al. 2006). The reason is that they have opposite interacting faces and also opposite handedness (left or right). Therefore, although it is not possible to know which model (I or II) is present in the lipid bilayer when no asparagine is present, the TM interactions are restrained in the presence of asparagine and the model is known in the mutant sequences. Thus, we only need to confirm that the orientation in each sample (model I or II) is the same, and consistent with that predicted computationally (Lin et al. 2006).
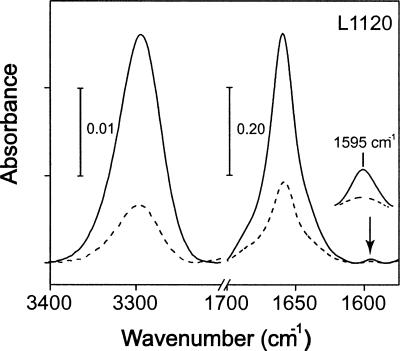
Figure 5 shows the infrared spectra of a labeled αM-TM synthetic peptide in the regions amide A and I, collected at parallel and perpendicular polarizations. Spectra for other samples were similar and are not shown. The frequency of the bands amide A and amide I, centered at ∼3295 and 1655 cm−1, respectively, were consistent with the peptides being completely α-helical. The band corresponding to the isotopic label, 13C=18O (see arrow), is centered at 1595 cm−1 as expected (Torres et al. 2000). Dichroic ratios (see Materials and Methods) were obtained for the amide A band and for the label, and are shown in Supplemental Table 1.
Figure 5.
Representative infrared spectra in the amide A and I regions. The spectrum shown corresponds to αM-TM (NSVGG) reconstituted in DMPC bilayers, collected at parallel (solid line) and perpendicular (broken line) polarization. The isotopically labeled residue (L1120) is indicated. The band corresponding to the 13C=18O label is indicated with an arrow, and displayed as an enlarged view.
SSID analysis of the raw dichroic data (see Materials and Methods), shown in Supplemental Table 1, produced the helix tilt, β, and rotational orientation. We stress that although a difference in ω between experimental and computational results (∼40°) may seem large, we have found for some systems that when analyzing essentially identical models (RMSD < 0.7 Å), the difference in ω may be up to 50°. Therefore, two models cannot be distinguished with confidence if their difference in ω at a particular residue is <50°.
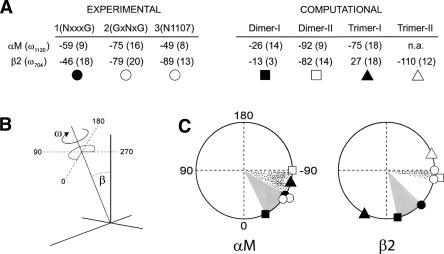
Figure 6 shows that the rotational orientation for residue 1120, ω1120, (calculated using the pair of sequences labeled “1” in Fig. 2, top panel) in the αM mutant NSVGG was −59° ± 20°. The difference in ω with any of the computational models (dimeric or trimeric) of αM was always smaller than 50°; therefore, the orientation of the α-helices in our sample is compatible with both models I and II. However, given that N is located at position 1 of the motif and found to be a dimer in SDS, the structure should correspond only to a dimeric model I (predicted to have ω = −26°).
Figure 6.
Comparison between experimental (this study) and computationally predicted orientational parameters. (A) The residue used to calculate ω (see black dot in Fig. 2) using SSID is indicated in the first column. Experimental values of ω (and helix tilt, shown within parentheses) were obtained using pairs 1, 2, and 3 in Figure 2. Computationally predicted values (Lin et al. 2006) are shown for model I (dimer and trimer) and model II (dimer and trimer). The symbols below the table are used in the lower panel to represent the location of the labeled residue around the helix (ω). (B) Schematic representation of ω and helix tilt, β, in an α-helix. (C) Location of L1120 (for αM) and V704 (for β2), according to experimental (this study) or computational predictions (Lin et al. 2006). The shaded sector (gray or dotted) represents the angular distance between experimental and predicted value in each case.
When Asn is at position 3 (GSNGG), a dimeric model II was found in SDS. For this sample, ω1120 (calculated with the pair labeled “2” in Fig. 2, top panel) was −75° ± 7°, which is consistent with the predicted orientation of a dimer model II (ω = −92°). When Asn was introduced at N1107, ω1120 (calculated with the pair labeled “3” in Fig. 2, top panel) was −49° ± 3°, which is similar to the orientation of the other two mutants, although from the position of the mutation, we know that this must correspond to model II.
To confirm that the introduction of an asparagine residue at these positions did not affect the stability or orientation of the predicted structure, we repeated our molecular dynamics simulations (Lin et al. 2006) when Asn mutations were present. We found that the ω angle (rotational orientation) of the conserved models did not change significantly (data not shown).
In β2, when Asn was at position 1 (NxxxG), ω704 was −46° (calculated using the pair labeled “1” in Fig. 2, lower panel). This is compatible with the predicted dimeric models I and II (−13° and −82°, respectively), but because of the location of the Asn residue, the orientation must correspond to model I. For position 3 (GxNxG), ω704 was −79° (calculated using the pair labeled “2” in Fig. 2, lower panel). This value is only compatible with computational models II (dimer and trimer, with −82° and −110°, respectively), although almost identical to the dimeric form. This is supported by the fact that only dimers were observed in SDS (Fig. 3). When Asn was introduced at G715N, ω704 was −89° (calculated with the pair of sequences labeled “3” in Fig. 2, lower panel), again compatible only with computational model II (either dimer or trimer), but we only observed dimers in SDS, which suggests that the structure in lipid bilayers is dimeric, although we cannot eliminate the possibility that a mixture of dimers and trimers (model II) exists in lipid bilayers.
The rotational orientation of residue 704 in β2 (ω704) was also measured in POPC lipid bilayers, which are in a more fluid, liquid crystal, phase at room temperature. For the pair labeled “1” in Figure 2, lower panel, the result was ω704 = −49° ± 2°, and 17° ± 1° for the helix tilt, entirely consistent with the results obtained in DMPC.
Discussion
Our results clearly show that introduction of Asn at consecutive positions in the membrane domain of αM and β2 integrins, within their five-residue GxxxG-like motif, leads to dimerization. Helix–helix interaction occurs at two opposite helix faces, which is expected from our models I and II described in a previous computational work.
The validity of our results can only be contested if Asn mutations could produce an artifactual dimerization, that is, forcing the α-helices to rotate so that the Asn side chains in opposite helices are always in contact. This is very unlikely for three reasons. First, it would be difficult to explain why this would happen only for alternate residues. Second, equally difficult would be to explain why precisely these two opposite orientations are also those compatible with two evolutionarily conserved models of interaction. Third, if such artifactual reorientation were to take place, the rotational orientation of the helices would deviate significantly from that predicted for models I and II; we show this is not the case using SSID, a technique that we have developed to determine orientational data for many different types of transmembrane α-helical homo-oligomers (Torres et al. 2000, 2001, 2002, 2006). Computational models I and II have approximately the same α-helical rotational orientation (Lin et al. 2006), and this value is entirely consistent with the orientational parameters obtained when the Asn-mutated peptides were incorporated to model lipid bilayers.
We have also shown that these two modes of homomeric interaction in integrin are likely to represent a general theme in integrin function and are not specific to any particular type. Anecdotal evidence, e.g., in α–α interactions (Luo et al. 2004), for model II has been found in the pair αIIb/β3, but we show that these interactions are also present in a completely different integrin pair, αM/β2. Furthermore, the fact that these computational models for integrin homo-oligomeric interaction were previously arrived at using all known α and β integrin TMs (Lin et al. 2006) indicates this is a general feature shared by all integrin TMs, and therefore likely to be associated with a certain common regulatory function.
Clearly, our in vitro results cannot be directly extrapolated to the context of the full-length integrin, but this cannot be used to question the relevance of our findings. First, we have shown that these results are consistent with experiments performed in vivo in other integrins. Second, compared to asparagine scans performed with the full-length integrin, this minimal system has the virtue of simplicity; in in vivo studies, each one of the mutants must express and fold properly, and this may lead to loss of information for various, or many, mutated positions (Li et al. 2003). As a consequence, asparagine scans performed with full-length integrin are difficult to rationalize in terms of a precise TM structure. Third, many in vivo assays use chimeric integrins, for example, the GALLEX assay (Schneider and Engelman 2003), where only a small region of the integrin is present. Although this approach can potentially be the target of similar objections, it has nevertheless contributed to our understanding of transmembrane interactions.
The function of these proposed dual homomeric interactions in integrins is at present only speculative, but it is likely related to the clustering process that takes place after the transition from the resting heterodimeric α/β state to the active state, where α-helices are separated. There are indications, however, that homomerization can take place even when the heterodimer is still in place. For example, in the α/β heterodimer (αIIb/β3), residues at the “G” position in the GxxxG-like motif of αIIb TM participate in helix–helix contacts in the resting state (Luo et al. 2004). However, αIIb mutation W967C, which points away from the α/β interface, was found to be involved in the formation of the species (αIIb/β3)2, a dimer of dimers, through formation of a disulfide bond (Luo et al. 2004). The latter result suggests that αIIb chains can form homodimers through a GxxxG-like motif-independent interaction (similar to our model II; Fig. 1) in the resting state. The potential coexistence of heterodimer and α homodimers has also been suggested earlier (Gottschalk and Kessler 2004).
For the β subunit, it has been observed that an activating mutation G708N in β3 led to homotrimerization, and this was postulated to drive the equilibrium toward an activated state. As noted previously (Gottschalk and Kessler 2004), however, in the resting state residue G708 is hidden at the α/β interface, which prevents homotrimerization. The conclusion is that the resting state and β homotrimerization via model II of interaction cannot occur simultaneously, although β homo-oligomerization via model I is still possible.
Another model for α/β transmembrane interaction, GpA-like (Schneider and Engelman 2004; Li et al. 2005), where both α and β interact via their GxxxG-like motif, has been proposed to be an intermediate state during the activation process (Schneider and Engelman 2004; Gottschalk 2005). Transition from the resting state to this intermediate state necessitates rotation of the β subunit, and this rotation would expose G708 in αIIb to the lipid environment, making possible the coexistence of α/β heterodimer and β homomerization via model II. The possibility that the α or β homo-oligomerization and α/β heterodimer can be simultaneously present, suggests that homo-oligomerization could provide the energy requirement for α- and β-chain separation (Gottschalk 2005) in certain instances. In any case, the complexity of these interactions makes it difficult at present to understand the role of the GxxxG-mediated form of interaction in the β or α subunits.
The putative coexistence of integrin hetero and homo-oligomers in the cell has been rationalized in a context where heteromeric interactions would stabilize the transmembrane region in a low-affinity and/or intermediate-affinity state (Carman and Springer 2003), whereas homo-oligomers would be present in the active state, cross-linking individual molecules (Gottschalk and Kessler 2004). Our results show that the transmembrane domains of integrins have an intrinsic tendency to form two types of homo-oligomers, with almost opposite orientation, and add considerable complexity in present homo-oligomerization models.
Finally, while the results presented pertain to integrin transmembrane domains, it would be interesting to find such dual homomeric interactions in other membrane proteins. Although the polyvalence of transmembrane α-helices in mediating homo- and heteromeric interactions has been reported previously (Sal-Man et al. 2005; Kroch and Fleming 2006), to our knowledge the integrin example we report here is the first of a dual homo-oligomeric interaction. Similar examples could be found in proteins that experience clustering as part of their function.
Materials and Methods
Synthesis of peptides and labeling
Amino acids with an isotopically labeled carbonyl 13C=18O (Torres et al. 2000, 2001) were obtained by incubating the corresponding 13C=16O carbonyl-containing amino acids (Cambridge Isotopes Laboratories), with a mixture of H2 18O (94.4% Promochem GmbH) and dioxane (3:1, v/v) for 1 h at 100°C at acidic pH (∼1). The amino acids were derivatized with FMOC as described (Wellings and Atherton 1997).
All peptides were synthesized using standard solid-phase FMOC chemistry (Intavis Multipep peptide synthesizer). The peptides were cleaved from the resin with trifluoroacetic acid (TFA) and lyophilized. Lyophilization was performed always in the presence of HCl (typically at an approximate molar ratio 20:1, HCl/peptide) in order to avoid the formation of peptide-TFA adducts that result in a typical TFA band at ∼1685 cm−1 in the infrared amide I region. The lyophilized peptides were dissolved in trifluoroethanol (TFE), TFA, and acetonitrile (1:1:4, v/v/v) (final peptide concentration ∼5 mg/mL) and immediately injected to a 20-mL Jupiter 5 C4-300 column (Phenomenex) equilibrated with H2O. Peptide elution was achieved with a linear gradient to a final solvent composition of 10% H2O, 90% acetonitrile, using a Waters 600 HPLC system. All solvents contained 0.1% (v/v) TFA. The resulting fractions were pooled and lyophilized. Peptide purity was confirmed by mass spectrometry. For α-helix orientational measurements (SSID), an isotopic label, i.e., a residue with a 13C=18O carbonyl group, was introduced during the synthesis of the peptides, at two consecutive positions, in different peptides.
Electrophoresis
The electrophoretic mobility of the peptides was assessed using SDS-PAGE. SDS-containing Tris-Tricine sample solubilizing buffer was added to the lyophilized peptides to a final concentration of 2 mg/mL. After vortexing for 1 min, the sample was heated for 5 min at 70°C and loaded on a Tris-Tricine 10%–20% gel (Bio-Rad). The loading volume was 2 and 5 μL (4 μg and 10 μg of peptide, respectively). The sample was electrophoresed at room temperature at a constant low voltage of 50 V for 6 h. After completion, the SDS-PAGE gel was first stained with Coomassie blue, followed by silver staining with the Silver Stain-Plus kit (Bio-Rad).
Sample preparation for ATR-FTIR
Infrared spectra were recorded on a Nicolet Nexus spectrometer purged with N2 and equipped with a MCT/A detector, cooled with liquid nitrogen. Attenuated total reflection (ATR) spectra were measured with a 25-reflections ATR accessory from Graseby Specac and a wire grid polarizer (0.25 μm; Graseby Specac). A total of 200 interferograms collected at a resolution of 4 cm−1 were averaged for every sample and processed with 1 point zero filling and Happ-Genzel apodization. Preparation of the sample was performed as previously described (Torres et al. 2002, 2006). Briefly, the sample was diluted to a final lipid concentration of 10 mg/mL (>15:1 DMPC/peptide molar ratio), and ∼100 μL of this solution was deposited, in successive aliquots of 10 μL, onto a trapezoidal (50 mm × 2 mm × 20 mm) Ge internal reflection element (IRE). After hydration, bulk water was removed using a dry N2 stream through the ATR compartment, and spectra were collected. The intensity corresponding to the 13C=18O carbonyl stretching vibration was obtained integrating the bands centered at ∼1590 cm−1. The area of the amide A (NH stretching) was calculated by integrating the band centered at 3300 cm−1 between 3200 and 3400 cm−1.
Data analysis
The data were analyzed according to the theory of site-specific dichroism presented in detail elsewhere (Arkin et al. 1997). By measuring the orientation of the amide I transition dipole moment, one can determine the helix tilt angle β and the rotational pitch angle ω of a specific dipole moment about the helix axis. The angle α between the transition dipole moment of the vibrational transition and the Z-axis was taken as 39° for the peptidic C=O bond and 29° for the N–H bond (Marsh et al. 2000). The parameters εx, εy, and εz, the electric-field components, were used according to a thick film approximation (Harrick 1979). Dichroic ratios were calculated as the ratio between the integrated absorptions of the spectra collected with parallel and perpendicular polarized light. The rotational pitch angle ω is defined arbitrarily as 0° when the transition dipole moment, the helix director, and the Z-axis all reside in a single plane. The difference of ω between two consecutive residues was assumed to be 100°, as in a canonical α-helix. The rotational orientation and tilt for each labeled residue were calculated as before (Torres et al. 2000, 2002).
Global Search Molecular Dynamics (GSMD) protocol
The simulations were performed using a Compaq Alpha Cluster SC45, which contains 44 nodes. All calculations were carried out using the parallel version of the Crystallography and NMR System (CNS Version 0.3), the Parallel Crystallography and NMR System (PCNS) (Brunger et al. 1998). The global search was carried out in vacuo as described elsewhere (Adams et al. 1995) using CHI 1.1 (CNS Helical Interactions). The interaction between the helices was assumed to be symmetrical. Clustering, averaging of the final structures, RMSD comparisons, and calculation of helix tilt β, and rotational orientation ω were performed as previously described (Arkin et al. 1995; Lin et al. 2006).
To test the effect of Asn substitution on our computational models, homodimeric models were simulated with the α-helices containing Asn at the same position as in the synthetic peptide. The helix tilt, β, was restrained to the angle obtained previously (Lin et al. 2006). The helices were rotated about their long helical axes in 10° increments until the rotation angle reached 350°. Three trials were carried out for each starting configuration using different initial random velocities. As during the global search (Lin et al. 2006), the simulations were performed for all integrin types.
Acknowledgments
J.T. thanks the financial support of the Biomedical Research Council (BMRC) of Singapore grants (03/1/22/19/238) and (04/1/22/19/361), and the facilities at the Bioinformatics Research Center (BIRC) of Nanyang Technological University. We are also grateful to Paul D. Adams for kindly providing CHI.
Footnotes
Supplemental material: see www.proteinscience.org
Reprint requests to: Jaume Torres, School of Biological Sciences, Nanyang Technological University, 60 Nanyang Drive, 637551 Singapore; e-mail: jtorres@ntu.edu.sg; fax: 65-6791-3856.
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.073234208.
References
- Adair, B.D., Yeager, M. Three-dimensional model of the human platelet integrin αIIbβ3 based on electron cryomicroscopy and X-ray crystallography. Proc. Natl. Acad. Sci. 2002;99:14059–14064. doi: 10.1073/pnas.212498199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adair, B.D., Xiong, J.P., Maddock, C., Goodman, S.L., Arnaout, M.A., Yeager, M. Three-dimensional EM structure of the ectodomain of integrin αVβ3 in a complex with fibronectin. J. Cell Biol. 2005;168:1109–1118. doi: 10.1083/jcb.200410068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams, P.D., Arkin, I.T., Engelman, D.M., Brunger, A.T. Computational searching and mutagenesis suggest a structure for the pentameric transmembrane domain of phospholamban. Nat. Struct. Biol. 1995;2:154–162. doi: 10.1038/nsb0295-154. [DOI] [PubMed] [Google Scholar]
- Arkin, I.T., Brunger, A.T. Statistical analysis of predicted transmembrane α-helices. Biochim. Biophys. Acta. 1998;1429:113–128. doi: 10.1016/s0167-4838(98)00225-8. [DOI] [PubMed] [Google Scholar]
- Arkin, I.T., Rothman, M., Ludlam, C.F., Aimoto, S., Engelman, D.M., Rothschild, K.J., Smith, S.O. Structural model of the phospholamban ion channel complex in phospholipid membranes. J. Mol. Biol. 1995;248:824–834. doi: 10.1006/jmbi.1995.0263. [DOI] [PubMed] [Google Scholar]
- Arkin, I.T., MacKenzie, K.R., Brunger, A.T. Site-directed dichroism as a method for obtaining rotational and orientational constraints for oriented polymers. J. Am. Chem. Soc. 1997;119:8973–8980. [Google Scholar]
- Briggs, J.A.G., Torres, J., Arkin, I.T. A new method to model membrane protein structure based on silent amino acid substitutions. Proteins. 2001;44:370–375. doi: 10.1002/prot.1102. [DOI] [PubMed] [Google Scholar]
- Brunger, A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.S., Kuszewski, J., Nilges, M., Pannu, N.S., et al. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. Sect. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Carman, C.V., Springer, T.A. Integrin avidity regulation: Are changes in affinity and conformation underemphasized? Curr. Opin. Cell Biol. 2003;15:547–556. doi: 10.1016/j.ceb.2003.08.003. [DOI] [PubMed] [Google Scholar]
- Choma, C., Gratkowski, H., Lear, J.D., DeGrado, W.F. Asparagine-mediated self-association of a model transmembrane helix. Nat. Struct. Biol. 2000;7:161–166. doi: 10.1038/72440. [DOI] [PubMed] [Google Scholar]
- Gottschalk, K.E. A coiled-coil structure of the αIIbβ3 integrin transmembrane and cytoplasmic domains in its resting state. Structure. 2005;13:703–712. doi: 10.1016/j.str.2005.02.014. [DOI] [PubMed] [Google Scholar]
- Gottschalk, K.E., Kessler, H. A computational model of transmembrane integrin clustering. Structure. 2004;12:1109–1116. doi: 10.1016/j.str.2004.02.039. [DOI] [PubMed] [Google Scholar]
- Gottschalk, K.E., Adams, P.D., Brunger, A.T., Kessler, H. Transmembrane signal transduction of the αIIbβ3 integrin. Protein Sci. 2002;11:1800–1812. doi: 10.1110/ps.4120102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hantgan, R.R., Paumi, C., Rocco, M., Weisel, J.W. Effects of ligand-mimetic peptides Arg-Gly-Asp-X (X = Phe, Trp, Ser) on αIIbβ3 integrin conformation and oligomerization. Biochemistry. 1999;38:14461–14474. doi: 10.1021/bi9907680. [DOI] [PubMed] [Google Scholar]
- Harrick, N.J. Internal reflection spectroscopy. Wiley; NY: 1979. Principles of internal reflection spectroscopy; pp. 13–65. [Google Scholar]
- Hemler, M. Extracellular matrix, anchor, and adhesion proteins. Oxford University Press; Oxford, UK: 1999. [Google Scholar]
- Hughes, P.E., Diaz-Gonzalez, F., Leong, L., Wu, C., McDonald, J.A., Shattil, S.J., Ginsberg, M.H. Breaking the integrin hinge. A defined structural constraint regulates integrin signaling. J. Biol. Chem. 1996;271:6571–6574. doi: 10.1074/jbc.271.12.6571. [DOI] [PubMed] [Google Scholar]
- Hynes, R.O. Integrins: Bidirectional, allosteric signaling machines. Cell. 2002;110:673–687. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- Kim, M., Carman, C.V., Springer, T.A. Bidirectional transmembrane signaling by cytoplasmic domain separation in integrins. Science. 2003;301:1720–1725. doi: 10.1126/science.1084174. [DOI] [PubMed] [Google Scholar]
- Kroch, A.E., Fleming, K.G. Alternate interfaces may mediate homomeric and heteromeric assembly in the transmembrane domains of SNARE proteins. J. Mol. Biol. 2006;357:184–194. doi: 10.1016/j.jmb.2005.12.060. [DOI] [PubMed] [Google Scholar]
- Lemmon, M.A., Flanagan, J.M., Hunt, J.F., Adair, B.D., Bormann, B.J., Dempsey, C.E., Engelman, D.M. Glycophorin A dimerization is driven by specific interactions between transmembrane α-helices. J. Biol. Chem. 1992;267:7683–7689. [PubMed] [Google Scholar]
- Li, R., Babu, C.R., Lear, J.D., Wand, A.J., Bennett, J.S., DeGrado, W.F. Oligomerization of the integrin αIIbβ3: Roles of the transmembrane and cytoplasmic domains. Proc. Natl. Acad. Sci. 2001;98:12462–12467. doi: 10.1073/pnas.221463098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, R.H., Mitra, N., Gratkowski, H., Vilaire, G., Litvinov, R., Nagasami, C., Weisel, J.W., Lear, J.D., DeGrado, W.F., Bennett, J.S. Activation of integrin αIIbβ3 by modulation of transmembrane helix associations. Science. 2003;300:795–798. doi: 10.1126/science.1079441. [DOI] [PubMed] [Google Scholar]
- Li, R.H., Gorelik, R., Nanda, V., Law, P.B., Lear, J.D., DeGrado, W.F., Bennett, J.S. Dimerization of the transmembrane domain of integrin αIIb subunit in cell membranes. J. Biol. Chem. 2004;279:26666–26673. doi: 10.1074/jbc.M314168200. [DOI] [PubMed] [Google Scholar]
- Li, W., Metcalf, D.G., Gorelik, R., Li, R., Mitra, N., Nanda, V., Law, P.B., Lear, J.D., Degrado, W.F., Bennett, J.S. A push-pull mechanism for regulating integrin function. Proc. Natl. Acad. Sci. 2005;102:1424–1429. doi: 10.1073/pnas.0409334102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, X., Tan, S.M., Law, S.K.A., Torres, J. Two types of transmembrane homomeric interactions in the integrin receptor family are evolutionarily conserved. Proteins. 2006;63:16–23. doi: 10.1002/prot.20882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, C.F., Takagi, J., Springer, T.A. Association of the membrane proximal regions of the α and β subunit cytoplasmic domains constrains an integrin in the inactive state. J. Biol. Chem. 2001;276:14642–14648. doi: 10.1074/jbc.M100600200. [DOI] [PubMed] [Google Scholar]
- Luo, B.H., Springer, T.A., Takagi, J. A specific interface between integrin transmembrane helices and affinity for ligand. PLoS Biol. 2004;2:776–786. doi: 10.1371/journal.pbio.0020153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, B.H., Carman, C.V., Takagi, J., Springer, T.A. Disrupting integrin transmembrane domain heterodimerization increases ligand binding affinity, not valency or clustering. Proc. Natl. Acad. Sci. 2005;102:3679–3684. doi: 10.1073/pnas.0409440102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh, D., Muller, M., Schmitt, F.J. Orientation of the infrared transition moments for an α-helix. Biophys. J. 2000;78:2499–2510. doi: 10.1016/S0006-3495(00)76795-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowall, A., Inwald, D., Leitinger, B., Jones, A., Liesner, R., Klein, N., Hogg, N. A novel form of integrin dysfunction involving β1, β2, and β3 integrins. J. Clin. Invest. 2003;111:51–60. doi: 10.1172/JCI14076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan, W., Becker, V., Klingmuller, U., Langosch, D. The interface between self-assembling erythropoietin receptor transmembrane segments corresponds to a membrane-spanning leucine zipper. J. Biol. Chem. 2004a;279:3273–3279. doi: 10.1074/jbc.M309311200. [DOI] [PubMed] [Google Scholar]
- Ruan, W., Lindner, E., Langosch, D. The interface of a membrane-spanning leucine zipper mapped by asparagine-scanning mutagenesis. Protein Sci. 2004b;13:555–559. doi: 10.1110/ps.03357404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russ, W.P., Engelman, D.M. TOXCAT: A measure of transmembrane helix association in a biological membrane. Proc. Natl. Acad. Sci. 1999;96:863–868. doi: 10.1073/pnas.96.3.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russ, W.P., Engelman, D.M. The GxxxG motif: A framework for transmembrane helix–helix association. J. Mol. Biol. 2000;296:911–919. doi: 10.1006/jmbi.1999.3489. [DOI] [PubMed] [Google Scholar]
- Sal-Man, N., Gerber, D., Shai, Y. The identification of a minimal dimerization motif QXXS that enables homo- and hetero-association of transmembrane helices in vivo. J. Biol. Chem. 2005;280:27449–27457. doi: 10.1074/jbc.M503095200. [DOI] [PubMed] [Google Scholar]
- Schneider, D., Engelman, D.M. GALLEX, a measurement of heterologous association of transmembrane helices in a biological membrane. J. Biol. Chem. 2003;278:3105–3111. doi: 10.1074/jbc.M206287200. [DOI] [PubMed] [Google Scholar]
- Schneider, D., Engelman, D.M. Involvement of transmembrane domain interactions in signal transduction by α/β integrins. J. Biol. Chem. 2004;279:9840–9846. doi: 10.1074/jbc.M312749200. [DOI] [PubMed] [Google Scholar]
- Senes, A., Gerstein, M., Engelman, D.M. Statistical analysis of amino acid patterns in transmembrane helices: The GxxxG motif occurs frequently and in association with β-branched residues at neighboring positions. J. Mol. Biol. 2000;296:921–936. doi: 10.1006/jmbi.1999.3488. [DOI] [PubMed] [Google Scholar]
- Stevens, T.J., Arkin, I.T. Are membrane proteins “inside-out” proteins? Proteins. 1999;36:135–143. doi: 10.1002/(sici)1097-0134(19990701)36:1<135::aid-prot11>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- Torres, J., Adams, P.D., Arkin, I.T. Use of a new label, 13C=18O, in the determination of a structural model of phospholamban in a lipid bilayer. Spatial restraints resolve the ambiguity arising from interpretations of mutagenesis data. J. Mol. Biol. 2000;300:677–685. doi: 10.1006/jmbi.2000.3885. [DOI] [PubMed] [Google Scholar]
- Torres, J., Kukol, A., Goodman, J.M., Arkin, I.T. Site-specific examination of secondary structure and orientation determination in membrane proteins: The peptidic 13C=18O group as a novel infrared probe. Biopolymers. 2001;59:396–401. doi: 10.1002/1097-0282(200111)59:6<396::AID-BIP1044>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Torres, J., Briggs, J.A., Arkin, I.T. Multiple site-specific infrared dichroism of CD3-z, a transmembrane helix bundle. J. Mol. Biol. 2002;316:365–374. doi: 10.1006/jmbi.2001.5267. [DOI] [PubMed] [Google Scholar]
- Torres, J., Parthasarathy, K., Lin, X., Saravanan, R., Kukol, A., Liu, D.X. Model of a putative pore: The pentameric α-helical bundle of SARS coronavirus E protein in lipid bilayers. Biophys. J. 2006;91:938–947. doi: 10.1529/biophysj.105.080119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travis, M.A., Humphries, J.D., Humphries, M.J. An unraveling tale of how integrins are activated from within. Trends Pharmacol. Sci. 2003;24:192–197. doi: 10.1016/S0165-6147(03)00069-5. [DOI] [PubMed] [Google Scholar]
- Watt, F.M. Role of integrins in regulating epidermal adhesion, growth and differentiation. EMBO J. 2002;21:3919–3926. doi: 10.1093/emboj/cdf399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehrle-Haller, B., Imhof, B.A. Integrin-dependent pathologies. J. Pathol. 2003;200:481–487. doi: 10.1002/path.1399. [DOI] [PubMed] [Google Scholar]
- Wellings, D.A., Atherton, E. Standard Fmoc protocols. Methods Enzymol. 1997;289:44–67. doi: 10.1016/s0076-6879(97)89043-x. [DOI] [PubMed] [Google Scholar]
- Xiong, J.P., Stehle, T., Diefenbach, B., Zhang, R.G., Dunker, R., Scott, D.L., Joachimiak, A., Goodman, S.L., Arnaout, M.A. Crystal structure of the extracellular segment of integrin αVβ3. Science. 2001;294:339–345. doi: 10.1126/science.1064535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, F.X., Merianos, H.J., Brunger, A.T., Engelman, D.M. Polar residues drive association of polyleucine transmembrane helices. Proc. Natl. Acad. Sci. 2001;98:2250–2255. doi: 10.1073/pnas.041593698. [DOI] [PMC free article] [PubMed] [Google Scholar]