Abstract
Bacteriophage Cro proteins bind to target DNA as dimers but do not all dimerize with equal strength, and differ in fold in the region of the dimer interface. We report the structure of the Cro protein from Enterobacteria phage N15 at 1.05 Å resolution. The subunit fold contains five α-helices and is closely similar to the structure of P22 Cro (1.3 Å backbone room mean square difference over 52 residues), but quite different from that of λ Cro, a structurally diverged member of this family with a mixed α-helix/β-sheet fold. N15 Cro crystallizes as a biological dimer with an extensive interface (1303 Å2 change in accessible surface area per dimer) and also dimerizes in solution with a Kd of 5.1 ± 1.5 μM. Its dimerization is much stronger than that of its structural homolog P22 Cro, which does not self-associate detectably in solution. Instead, the level of self-association and interfacial area for N15 Cro is similar to that of λ Cro, even though these two orthologs do not share the same fold and have dimer interfaces that are qualitatively different in structure. The common Cro ancestor is thought to be an all-helical monomer similar to P22 Cro. We propose that two Cro descendants independently developed stronger dimerization by entirely different mechanisms.
Keywords: evolution of oligomerization, transcription factor, helix-turn-helix, structural evolution
The bacteriophage Cro proteins are a highly diverse family of transcription factors and a model system for protein evolution. Comparisons of Cro orthologs from bacteriophages P22 and λ illustrate the major commonalities and differences among members of this family. Similarities are as follows: P22 and λ cro genes are topologically equivalent within the immunity regions of their respective genomes (Sanger et al. 1982; Vander Byl and Kropinski 2000; Pedulla et al. 2003); both Cro proteins bind, almost certainly as dimers, to three pseudopalindromic DNA sequences in topologically equivalent OR regulatory regions (Maniatis et al. 1975; Johnson et al. 1979; Poteete et al. 1980, 1986); and both proteins contain a similar helix-turn-helix motif for major groove DNA recognition (Sauer et al. 1982; Albright and Matthews 1998; Ohlendorf et al. 1998; Newlove et al. 2004). Major differences are as follows: The two protein sequences show 25% identity or less and no significant similarity in direct BLAST comparisons (Sauer et al. 1982; Newlove et al. 2004); P22 Cro has an all α-helical fold, while λ Cro has a mixed α+β fold (Ohlendorf et al. 1998; Newlove et al. 2004); P22 Cro exists as a monomer in solution even up to low millimolar concentrations (Newlove et al. 2004), while λ Cro dimerizes in solution at very low micromolar concentrations (Jana et al. 1997; Darling et al. 2000b; LeFevre and Cordes 2003; Jia et al. 2005); P22 Cro binds to sites that have an even number of base pairs, with a twofold axis of pseudosymmetry centered between two base pairs, while λ Cro binds to sites with an odd number of base pairs and a symmetry axis that coincides with a central base pair (Maniatis et al. 1975; Poteete et al. 1980); and the specific sequences of the cognate consensus OR half-sites have no clear similarity (Hall et al. 2005). In sum, these two proteins appear to conserve a broadly similar biological role while evolving major differences in sequence, structure, and specific function.
We wish to understand the evolution of different folds and oligomeric states in the Cro family, in terms of both the range of differences and the underlying mutational mechanisms. Toward this end, we have assembled databases of Cro sequences and selected particular family members for in-depth structural characterization. As part of one study, we grouped a database of 56 apparent Cro homologs into linkage clusters in which every member was connected to at least one other member by direct sequence similarity, and to every other member by either direct or transitive sequence similarity (Newlove et al. 2004). Seventy-five percent of the database sequences belonged to a single major cluster, which included P22 Cro and λ Cro. These two homologs were on opposite sides of the cluster and were connected indirectly by a transitive homology pathway involving three “intermediate” Cro sequences. In a subsequent study, reported elsewhere (Roessler et al. 2008), we targeted these three sequences for structural studies as a means of investigating how structural properties of Cro proteins change as one traverses the sequence space connecting two very structurally different family members. One major outcome of this “stepping stone” study was that subunit structures determined for the intermediate Cro sequences clearly fell into either the all-α fold class represented by P22 Cro or the α+β fold class represented by λ Cro. In addition, the all-α Cro proteins were effectively monomeric in solution, while the α+β Cro proteins showed varying levels of solution dimerization in the low to high micromolar range. These results suggested that two approximate categories, all-α monomers and α+β dimers, might be sufficient to describe Cro structural diversity, at least within the main cluster of sequences.
As an alternative, complementary approach, we are targeting “outlier” Cro proteins not within the main sequence similarity cluster, motivated by the supposition that more unique Cro sequences might be more likely to encode unique and as yet undiscovered structural properties. The putative Cro protein from bacteriophage N15 (Lobocka et al. 1996; Ravin et al. 2000) is such an outlier. Not only is N15 Cro outside the main Cro cluster, it has almost no directly recognizable sequence homologs in the nonredundant protein database. The lone hits in a recent BLAST search included a nearly identical sequence from bacteriophage phi-K02 and a borderline similarity to the putative Cro protein of Yersinia pestis phage PY54. The N15 Cro sequence is not directly similar to those of either P22 Cro or λ Cro, but certain comparisons suggest closer homology with the former. For example, standard PSI-BLAST analysis initiated from P22 Cro identifies N15 Cro as a hit in the second round, and the two sequences share 30% identity across 55 residues. N15 Cro and λ Cro show 22%–25% identity across a comparable region of sequence, and no connection in standard PSI-BLAST searches. Like P22 Cro and unlike λ Cro, N15 Cro has putative OR binding sites with an even number of base pairs (Lobocka et al. 1996).
Here we report the crystal structure of N15 Cro at 1.05 Å resolution and characterize its dimerization by sedimentation equilibrium. The subunit structure of N15 Cro proved quite similar to that of P22 Cro, consistent with the hints of sequence and functional similarity outlined above. Surprisingly, however, N15 Cro dimerizes in solution with a strength equal to that of λ Cro. N15 Cro can thus be regarded as representing a new category of Cro protein, the all-α helical dimer. We discuss the evolutionary implications of this finding.
Results
Subunit structure of N15 Cro resembles the all-α fold of P22 Cro
We cloned, expressed, and purified N15 Cro and determined its crystal structure at 1.05 Å resolution in space group P21 (see Materials and Methods). Statistics are summarized in Table 1, and initial unrefined electron density is shown in Figure 1. There are two molecules (chains A and B) in the asymmetric unit, related by a noncrystallographic pseudo-twofold axis that runs nearly parallel to the crystallographic screw axis. Chains A and B are similar in structure (0.5 Å backbone root mean square difference [rmsd] for residues 1–62). Sixty-six of 71 residues were modeled for chain A, while 64 were modeled for chain B. Five to seven residues at the C-terminal end of each chain appear to be disordered.
Table 1.
Crystallographic data for N15 Cro (PDB ID 2HIN)
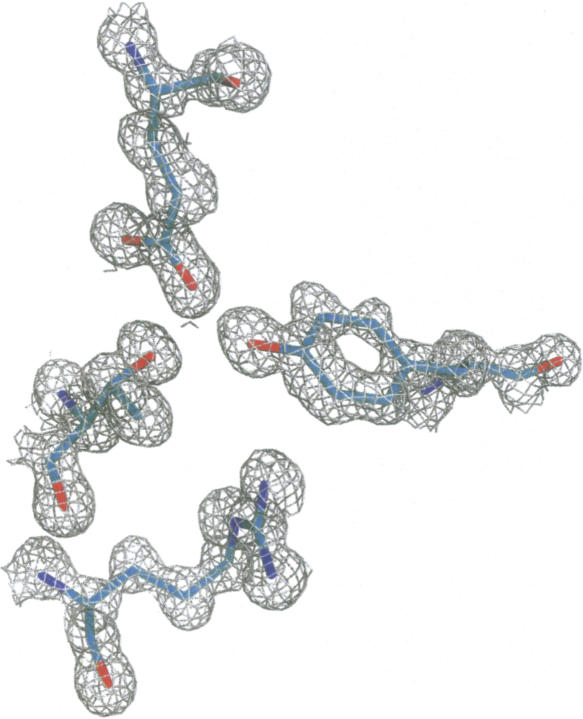
Figure 1.
Electron density in the phased map from ACORN contoured at 1.2 σ and final refined coordinates for residues Tyr 50 and Glu 45 from chain A, and Thr 57 and Arg 60 from chain B. This figure was prepared using PyMOL (DeLano Scientific).
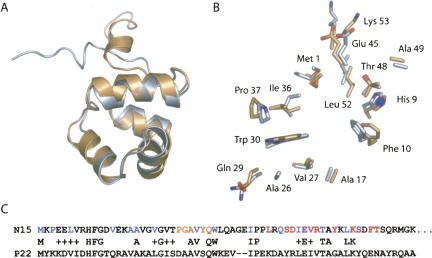
Each subunit has a fold consisting of five α-helices (Fig. 2A), clearly recognizable as the repressor fold (CATH 1.10.260) common to a variety of prokaryotic transcription factors. In terms of fold, N15 Cro clearly must be grouped with all-α Cro proteins like P22 Cro rather than with mixed α+β proteins like λ Cro. In a DALI structural similarity search of the Protein Data Bank (PDB) with chain A of N15 Cro as a query, the closest structural relative was, in fact, P22 Cro (Z-score of 7.5, 2.1 Å rmsd over 56 residues, with 30% sequence identity). Superposition of P22 Cro and chain A of N15 Cro (Fig. 2A) yielded a backbone rmsd of 1.3 Å over 52 residues. The sole qualitative differences between the two backbone structures are a different orientation for the fifth helix and a shorter third helix in N15 Cro relative to P22 Cro. The 17 residues that are identical in the two sequences (Fig. 2C) all occupy comparable locations in the structure, and most have similar side-chain conformations (Fig. 2B). This group includes eight of the buried residues in N15 Cro, three glycine/proline residues at the N or C termini of helices, two residues likely to make direct contacts to DNA from the third or “recognition” helix, and two residues which are at the homodimer interface of N15 Cro. These similarities reinforce the hypothesis that N15 Cro is a closer relative of P22 Cro than of λ Cro.
Figure 2.
Comparison of N15 Cro and P22 Cro. (A) Ribbon diagram representation of chain A of N15 Cro (gold) superimposed on a minimized average solution structure of P22 Cro derived from the NMR ensemble (1RZS). (B) Locations and conformations of identical side chains in the two proteins (same color-coding and approximate orientation as in part A). (C) Sequence alignment highlighting buried residues (<10% ASA) of N15 Cro (blue), residues in the dimer interface within 4 Å of the other chain (red), residues both buried and within 4 Å of the other chain (purple), and putative base-contacting residues in the recognition helix (orange). P22 Cro and N15 Cro have 32% sequence identity over 53 residues. Parts of this figure were prepared using PyMOL (DeLano Scientific).
N15 Cro crystallizes as the biological dimer
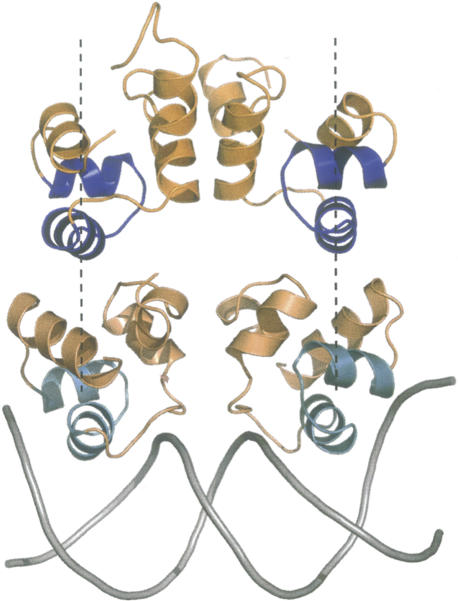
The dimer formed by chains A and B of N15 Cro has imperfect twofold symmetry and an overall conformation consistent with an ability to bind two adjacent, reasonably spaced DNA half-sites. This is nicely illustrated by comparison of the N15 Cro dimer with the structure of the CI N-terminal domain from bacteriophage 434 bound to cognate OR DNA (Fig. 3; Aggarwal et al. 1988). CI proteins bind phage OR sites in competition with Cro proteins from the same phage and are related to Cro by ancient gene duplication, so their DNA complexes can provide informative qualitative models. In particular, the OR sites from phage 434, like those from phages P22 and N15, have even numbers of base pairs and an approximate twofold axis between two base pairs (Wharton et al. 1984). One might thus reasonably suppose that a biological dimer of free N15 Cro should approximately superimpose onto the 434 CI–DNA complex. Indeed, the two helix-turn-helix motifs in N15 Cro have the correct spatial separation and orientation for recognition of the OR half-sites of phage 434. In sum, N15 Cro crystallizes as a biological dimer despite the absence of DNA.
Figure 3.
N15 Cro crystallizes as the biological dimer. The N15 Cro dimer (chains A and B in gold with helix-turn-helix motif highlighted in blue) is shown above the crystal structure of 434 CI repressor complexed with operator DNA (PDB ID 2OR1; protein: wheat with helix-turn-helix motif highlighted in cyan; DNA: gray). Note the approximately equivalent spacing and orientation of the recognition helices in the two dimers. This figure was prepared using PyMOL (DeLano Scientific).
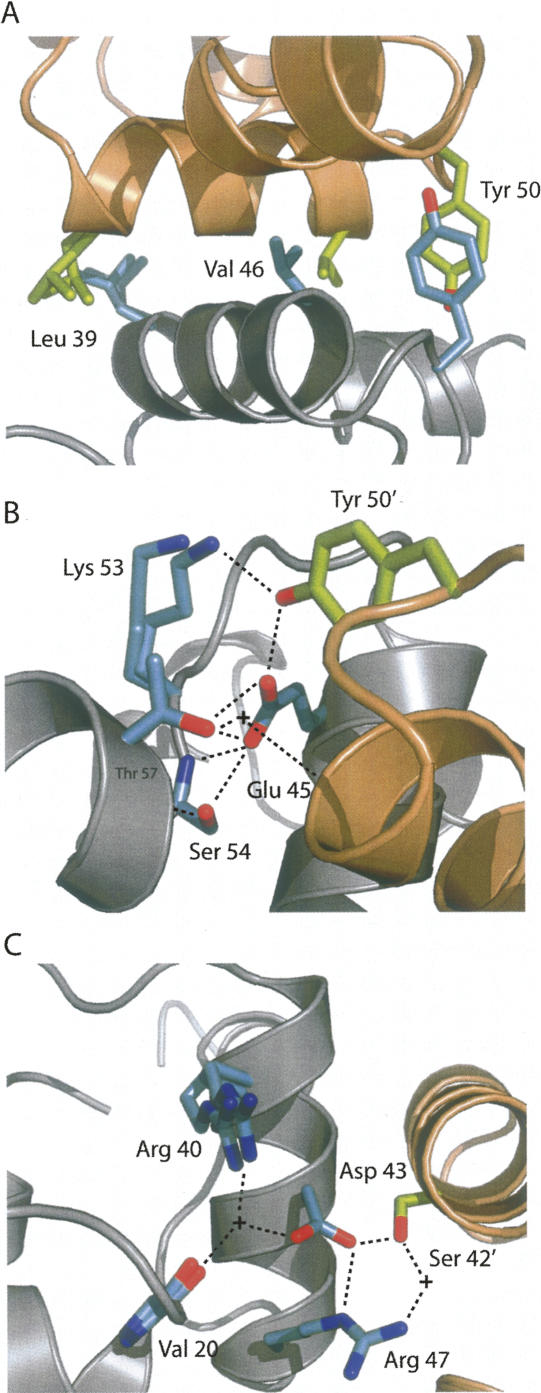
The dimer interface of N15 Cro is tightly packed and exhibits three extensive networks of interacting residues, both polar and nonpolar. First, a triad of homotypic hydrophobic interactions, involving Leu 39, Val 46, and Tyr 50, occurs across the tight interface made between the fourth helices from each subunit (Fig. 4A). Second, a polar network is formed by Glu 45, Lys 53, Ser 54, and Thr 57 within subunit A, and both Glu 45 and Lys 53 within this network interact with Tyr 50 in subunit B across the dimer interface (Fig. 4B). The Lys 53–Tyr 50′ contact only occurs from subunit A to B and does not appear in the reciprocal interaction network. Thr 57 also makes a water-mediated contact to the backbone of the other subunit at Val 46′. Third, a second polar network is formed by Val 20, Arg 40, Asp 43, and Arg 47 of subunit A, with an ordered water forming a bridge between the first three residues (Fig. 4C). Asp 43 and Arg 47 in turn hydrogen bond to Ser 42 in subunit B across the interface, either directly in the case of Asp 43 or through a water molecule in the case of Arg 47. In the reciprocal interaction network from subunit B to A, Arg 47 does not participate.
Figure 4.
Interactions in the dimer interface of N15 Cro. (A) Homotypic hydrophobic “zipper” contacts. (B,C) Two different polar networks involving ordered water molecules, shown as plus signs. Hydrogen bonds are indicated by dashed lines. Ribbon and side chains from chain A are shown in gray and cyan, respectively, while the ribbon and side chains from chain B are shown in wheat and green, respectively. Multiple conformations were modeled for several residues. This figure was prepared using PyMOL (DeLano Scientific).
N15 Cro dimerizes in solution like λ Cro but unlike P22 Cro
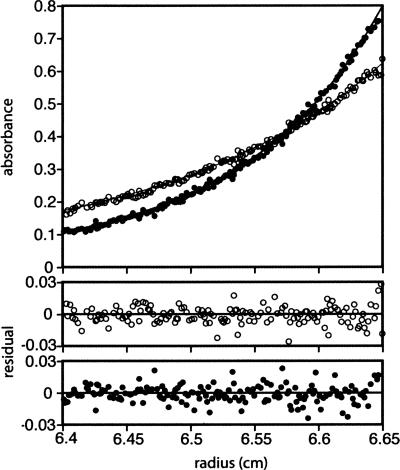
Since N15 Cro forms a biological dimer in the crystal, we wondered whether it would dimerize detectably in solution. P22 Cro, the closest structural relative of N15 Cro, shows no significant dimerization in sedimentation equilibrium experiments, nor do other Cro proteins with all-α folds characterized to date (Newlove et al. 2004; Roessler et al. 2008). λ Cro, by contrast, does dimerize significantly with a K d of ∼3 μM in both sedimentation equilibrium and FRET experiments (LeFevre and Cordes 2003; Jia et al. 2005). However, λ Cro has an α+β fold, with the dimer interface formed by a C-terminal β-hairpin that replaces the fourth and fifth helices observed in P22 Cro and N15 Cro. Since N15 Cro shares the same fold as the weakly dimerizing P22 Cro, one might also naively predict that it would dimerize weakly. Surprisingly, sedimentation equilibrium experiments (Fig. 5) show that N15 Cro dimerizes with nearly the same equilibrium constant as λ Cro. Global monomer-dimer fits of six radial distribution curves, including three concentrations ranging from 5–20 μM and two rotor speeds of 23,000 and 30,000 rpm, yielded a K d of 5.1 ± 1.5 μM.
Figure 5.
Dimerization of N15 Cro characterized by sedimentation equilibrium. Two radial absorbance curves are shown for a loading concentration of 10 μM of N15 Cro and rotor speeds of 23,000 (open circles) and 30,000 rpm (filled circles), monitored at a wavelength of 235 nm. The curve fits shown are based on parameters from global monomer-dimer fitting of three concentrations and two rotor speeds. Residuals for each of the two curves shown are at the bottom of the figure.
This result suggests that the dimer interfaces of N15 Cro and P22 Cro must be rather different despite their shared overall fold. Indeed, most key interfacial residues in N15 Cro are not conserved in P22 Cro (see alignment in Fig. 2C). Only Glu 45 and Lys 53 are identical, and the nonidentical residues differ by nonconservative mutations, with the exception of the replacement of Val 46 in N15 Cro by Ile in P22 Cro. Most striking is the absence of the Tyr 50 side chain in P22, being mutated to Gly. In N15 Cro, Tyr 50 makes both polar and nonpolar interactions in the interface, and its absence might be expected to weaken dimerization. The P22 Cro dimer structure is not known, but based on the sequence alignment (Fig. 2C), one expects very different interactions in the dimer interfaces of the two proteins, and the dimerization of P22 Cro could be weaker by virtue of these differences, even if the overall conformation of the dimer and relative disposition of the subunits are similar.
To begin exploring the origin of N15 Cro's stronger than expected dimerization, we performed a brief characterization of Y. pestis phage PY54 Cro (Hertwig et al. 2003), the closest nonredundant (less than 90% identity) sequence relative of N15 Cro. PY54 Cro is 33% identical in sequence to N15 Cro over 60 residues, and the two proteins have four of 11 identical interfacial residues (Glu 45, Lys 53, Ser 54, and Thr 57) with three additional interfacial positions differing by conservative mutations (Asp 43 to Glu, Arg 47 to Lys, and Phe 56 to Tyr). Four of the putative interfacial residues in PY54 Cro are also identical with aligned residues in P22 Cro (Tyr 42, Glu 45, Gly 50, and Lys 53). By contrast, N15 Cro and P22 Cro share only two identical interfacial residues (Glu 45 and Lys 53) (see also Fig. 2C). The dimer interface sequence of PY54 thus has an intermediate quality between those of P22 Cro and N15 Cro.
Sedimentation equilibrium analysis of a C-terminally histidine-tagged version of PY54 Cro (data not shown) suggested dimerization at least as strong as, if not stronger than, that of N15 Cro. Previous experiments on histidine-tagged N15 Cro had yielded K d values of 5.7 ± 1.0 μM, almost exactly the same as the untagged version, suggesting that the histidine tag was unlikely to affect sedimentation analysis for these proteins. Ideal single-species fits of radial distribution curves for tagged PY54 Cro, at loading concentrations of 5–20 μM and rotor speeds of 23,000 and 30,000 rpm, yielded apparent molecular weights within 15% of the expected dimer molecular weight. Global monomer-dimer fits of these data yielded K d < 1 μM. Because these values were at the edge of accurately measurable ranges for absorbance-monitored sedimentation experiments on these proteins, we report only an approximate maximum K d value. Presuming that PY54 has a helical fold, these preliminary data suggest that significant solution dimerization among all-α Cro proteins is not unique to N15 Cro, nor does it depend critically on conservation of one specific pattern of interfacial residues. A complete mutagenesis study of sequence determinants of dimerization in these proteins is underway in our laboratory.
The N15 Cro dimer interface is completely different from that of λ Cro
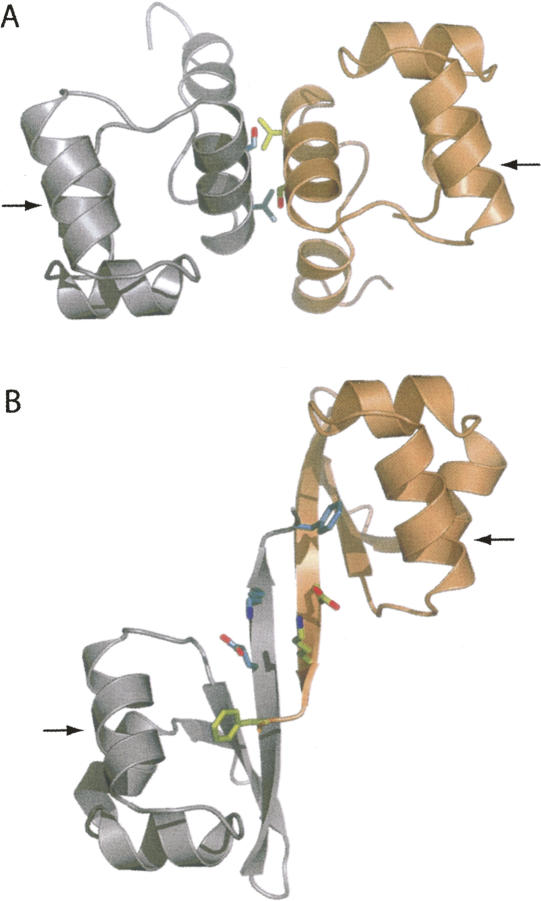
We now compare the dimer interfaces of N15 Cro and λ Cro. In accord with their similar dimerization strength, the dimer interfaces of N15 Cro and λ Cro (Fig. 6) have almost exactly the same amount of buried surface area. The total accessible surface area for the N15 Cro (Fig. 6A) subunits considered separately is 9046 Å2, while the accessible surface of the dimer is 7743 Å2. A total of 1303 Å2 is buried in the subunit interface. The total accessible surface area for the λ Cro subunits (from the complex of λ Cro with cognate DNA) (Fig. 6B; Albright and Matthews 1998) considered separately is 9459 Å2, while the accessible surface of the dimer is 8105 Å2. A total of 1354 Å2 is buried in the subunit interface. To the extent that buried surface area correlates with interfacial strength, it makes sense that the two proteins dimerize at the same concentration.
Figure 6.
Comparison of N15 Cro (A) and λ Cro (B) (PDB ID 6CRO) dimers, with the two chains colored differently in each case. For N15 Cro, close contacts between helix 4 from each subunit are illustrated by showing interactions between Ser 42 and Val 46. For λ Cro, ionic interactions among Glu 54 and Lys 56 are shown, as is Phe 58, which extends from the end of strand 3 in one monomer into the hydrophobic core of the other, forming a “ball-and-socket.” Note the similar orientation but different spacing of the two recognition helices in each dimer, which are the helices nearest the viewer in this perspective, indicated by arrows. This figure was prepared using PyMOL (DeLano Scientific).
In other respects, the dimer interfaces of N15 Cro and λ Cro (Fig. 6) could hardly be more different. The dimer interface of λ Cro (Fig. 6B) involves an extension of the third strand (β3) of a three-stranded antiparallel β-sheet. β3 extensions from each subunit interact in antiparallel fashion to form an intermolecular β-sheet. Apart from the attendant main-chain hydrogen bonds, there are ionic and nonpolar cross-strand interactions in the region of residues 54–56, and a phenylalanine residue at the end of β3 that inserts into a cavity in the hydrophobic core of the other subunit, forming a flexible “ball-and-socket” interaction (Albright and Matthews 1998). N15 Cro, in contrast, uses an entirely helical interface that features a mutual close approach of the fourth helix from each subunit (Fig. 6A). The two subunits are less intertwined and have independent hydrophobic cores, but the interface is nonetheless very tight. A notable difference in the two dimers is the relative position of helix 3 from each subunit, which acts as a DNA “reading head” in the major groove at each half-site. In both N15 Cro and λ Cro, the two helices have an antiparallel disposition, but they are considerably farther away in λ Cro. The difference in inter-helical distance could be explained by the fact that N15 Cro has one fewer base pairs separating its two putative half-sites.
Discussion
Cro proteins differ widely in fold and dimerization strength as a result of divergent evolution (Newlove et al. 2004). Given that differences in Cro fold map to the dimer interface in the C-terminal half of the domain, one might predict that the two properties would show evolutionary linkage. Indeed, work reported elsewhere (Roessler et al. 2008) supports a rough division of Cro protein structures in solution into all-α monomers and α+β dimers. In this work, however, we reported the structure of the biological dimer of Enterobacteria phage N15 Cro, which shows an all-α subunit structure similar to the monomeric, all-α P22 Cro, but low micromolar dimerization similar to the α+β λ Cro protein. These results show that detectable solution dimerization of Cro proteins is not limited to family members with the α+β subunit fold. Moreover, as we discuss below, the ancestral Cro structure was likely to have been an all-α monomer. Thus, low micromolar solution dimerization appears to have evolved independently at least twice in the Cro lineage, by very different mechanisms: once within the framework of the ancestral fold, and once concurrent with or following evolution of a novel α+β fold.
We first need to address whether it is reasonable to discuss dimer evolution in terms of a coherent Cro lineage that includes P22 Cro, λ Cro, and N15 Cro. Because Cro proteins are so variable in sequence, structure, and function, there could be some doubt as to whether they are true orthologs (homologs which are related by speciation rather than gene duplication) despite their conserved gene position in the immunity region of lambdoid bacteriophages. Indeed, phage 434 Cro is likely the product of in situ paralogous gene displacement by a CI repressor N-terminal domain (Sauer et al. 1982; Newlove et al. 2004), the gene for which is adjacent to cro and related to it by an ancient gene duplication. The proteins being compared in this study, however, are likely to be orthologs and to represent linear descent from a common ancestor. First, the observation that the closest known structural relative of N15 Cro is P22 Cro, along with fairly close PSI-BLAST connections between these two proteins, reinforces the impression that they are true orthologs. Second, previously established sequence similarity networks that include P22 Cro and λ Cro, but exclude other proteins such as members of the CI family, suggest that these two proteins are true orthologs as well (Newlove et al. 2004).
Within the Cro lineage, the all-α subunit fold observed in N15 Cro and P22 Cro is almost certainly ancestral to the α+β fold observed in λ Cro. The all-α fold is observed in many prokaryotic transcription factors, including the phage CI repressors to which Cro proteins are related by ancient gene duplication. The α+β topology of λ Cro, on the other hand, is so far unique to some subset of the Cro family. The all-α ancestor also probably exhibited little to no solution dimerization. Other all-α Cros investigated in a separate study show essentially indetectable solution dimerization (Roessler et al. 2008), as do the DNA-binding domains of the CI repressors (Mondragon et al. 1989; Neri et al. 1992; Huang and Oas 1995), though λ CI N-terminal domain exhibits dimerization as strong as 0.3 mM depending upon where the domain boundary is defined (Weiss et al. 1987). In sum, the ancient Cro protein was probably effectively an all-α monomer.
From these origins, N15 Cro and λ Cro represent distinct modes of evolving a stronger oligomeric interface within a family of orthologous proteins. N15 Cro appears to employ what has been termed a “sticky billiard ball” mechanism (Bennett et al. 1995), in which simple variation in surface side chains is used to build the interface. λ Cro, by contrast, has undergone a change in subunit fold, followed or accompanied by evolution of an intertwined interface involving β-sheet backbone interactions. This interface gives the appearance of domain or segment swapping (Bennett et al. 1995; Xu et al. 1998), and reversal of this apparent swapping has been achieved by insertion mutations in protein engineering experiments (Mossing and Sauer 1990; Albright et al. 1996; Mossing 1998). From an evolutionary perspective, however, experiments in our laboratory have suggested that λ Cro's dimer interface strength could derive instead from a “surrogacy” mechanism, in which mutational removal of a critical ancestral hydrophobic core residue in a monomer was compensated by insertion of a similar but nonequivalent residue from a second subunit (LeFevre and Cordes 2003; Newlove et al. 2006). The dimer resulting from this replacement features a ball-and-socket in which a phenylalanine side chain from one subunit inserts into the core of the other (see Fig. 6B; Albright and Matthews 1998).
The functional significance of differences in Cro dimerization bears some discussion. Even Cro proteins with low micromolar dimer dissociation constants will be largely monomeric in solution at the low nanomolar concentrations typically required for operator binding (Jana et al. 1997; Darling et al. 2000a,b). Differences in Cro solution dimerization are probably a matter of degree rather than a fundamental change in the predominant oligomeric state of the free protein under binding conditions. Other things being equal, stronger solution dimerization will simply increase the affinity of the free protein for operator DNA by stabilizing the functionally competent form of the protein.
Solution dimerization varies over at least three to four orders of magnitude for known Cro proteins, amounting to a putative effect on binding strength of 4–6 kcal/mol or more. That orthologous proteins tolerate this level of evolutionary variation in a functionally important property is noteworthy. However, the lowest energy solution dimer may not be exactly the same as the DNA-bound dimer (Kyogoku et al. 1995; Matsuo et al. 1995; Albright and Matthews 1998; Ohlendorf et al. 1998), and energy might be required to adjust the structure to the form observed in sequence-specific protein–DNA complexes. Thus, the contribution of differences in dimerization to DNA-binding affinity is difficult to assess from solution dimerization studies. If the effects are as substantial as they appear, variations in dimerization strength must be offset by changes in direct protein–DNA interactions if comparable operator DNA affinities are to be conserved. How well conserved they actually are is not yet known, but preliminary experiments in our laboratory indicate that P22 Cro can bind its cognate consensus DNA at a half-maximal protein concentration of ∼25 nM (A.J. Miller and M.H.J. Cordes, unpubl.) down between one and two orders of magnitude from the corresponding binding strength of λ Cro under comparable conditions. The affinity of N15 Cro for cognate DNA is unknown but is the subject of current investigation.
Materials and Methods
Cloning, expression, and purification
Genomic DNA for bacteriophage N15 was obtained as a gift from Dr. Sherwood Casjens at the University of Utah. The cro gene was amplified by PCR with introduction of flanking NdeI and PaeR7I restriction sites at the 5′ and 3′ ends of the coding sequence, respectively, with a stop codon immediately preceding the PaeR7I site. The PCR product was digested with NdeI and PaeR7I and ligated with an NdeI–PaeR7I fragment of the expression plasmid pET21b, yielding the expression construct pMD104. N15 Cro protein was overexpressed from plasmid pMD104 in Escherichia coli strain BL21(λDE3) in a 15-L fermentor vessel at 37°C, with a stir rate of 350 rpm, an airflow rate of 15 L/min, and a small amount of antifoam added. Overxpression was induced with 100 μg/mL IPTG when the culture reached an A600 of 0.55–0.6. Cells were harvested by centrifugation of 1 L aliquots of culture at 5000g for 10 min and resuspended in a total of 200 mL cold lysis buffer (100 mM Tris [pH 8.0], 500 mM KCl, 1 mM EDTA, 10 mM MgCl2). Four hundred microliters of 100 mM phenylmethylsulfonyl fluoride (PMSF) were then added, and the cells were lysed by sonication in a stainless steel beaker on ice, using 8 × 1 min bursts. An additional 400 μL of 100 mM PMSF were then added to the lysate, which was then diluted with 1.4 L cold lysis buffer. Sixteen microliters of 10% (v/v) polyethyleneimine (PEI) were added with stirring at 4°C, and stirring continued for 15 min. The lysate was then centrifuged at 14,000g for 30 min at 4°C. The supernatant was collected, ammonium sulfate (502 g; 50% saturation) was added with stirring at 4°C, and the stirring was continued for 1 h. The mixture was centrifuged at 20,000g for 30 min at 4°C, the pellet was discarded, and additional ammonium sulfate (549 g; 90% saturation) was added with stirring at 4°C to precipitate the remaining proteins. Stirring was continued overnight, and precipitated proteins were pelleted by centrifugation at 20,000g for 90 min at 4°C. The pellet was resuspended in ∼20 mL PC buffer (20 mM Tris [pH 8.0]), 0.1 mM EDTA, 5% glycerol, and 1.4 mM β-mercaptoethanol) and dialyzed extensively against the same. The dialysate was centrifuged to remove precipitates and loaded in 5 mL aliquots onto a Mono S HR 10/10 column equilibrated in PC buffer. Consistent with its mildly alkaline pI of 8.1, N15 Cro did not flow through immediately but also did not bind strongly, eluting at the very beginning of a 0–1 M sodium chloride gradient. Mono S fractions containing N15 Cro were identified using SDS-PAGE, combined, dialyzed into PC buffer plus 200 mM sodium chloride, and loaded at a flow rate of 1.3 mL/min onto a Sephacryl S-100 HiPrep 26/60 size-exclusion column equilibrated in the same buffer. Under these conditions, N15 Cro exhibited an elution volume of ∼190 mL. On the basis of a calibration curve for the Sephacryl S-100 column, we predicted a molecular weight of 21 kD, more consistent with a dimeric or trimeric species than with a monomer. Appropriate fractions from the size exclusion step were again identified using SDS-PAGE, combined, and reduced to a volume of 1.3 mL and a protein concentration of 35 mg/mL using Centricon YM-3 centrifugal concentrators. The final yield of N15 Cro was 3 mg/L of culture.
A synthetic gene encoding PY54 Cro was constructed from two pairs of mutually priming oligonucleotides and cloned into a pET21b expression vector as described elsewhere for prophage Afe01 Cro (Roessler et al. 2008), yielding an expression construct for PY54 with a C-terminal LEHHHHHH sequence tag; the comparable construct for tagged N15 Cro was generated by deletion of the stop codon from pMD104 using QuikChange mutagenesis (Stratagene), leaving an in-frame LEHHHHHH tag deriving from the vector. Tagged N15 Cro was purified by denaturing Ni-NTA agarose affinity chromatography, followed by dialysis into SB250 buffer (50 mM Tris [pH 7.5], 250 mM KCl, 0.2 mM EDTA) and size-exclusion chromatography on a Sephacryl S-100 26/60 column. Purification of tagged PY54 Cro was performed similarly but included modifications to maintain a reduced state for two cysteine residues present at positions 17 and 22 in the sequence. Specifically, nickel affinity chromatography included 15 mM β-mercaptoethanol (BME) in lysis and affinity column buffers, and 3 mM BME was included in the SB250 buffer during refolding dialysis and for the size-exclusion column. Following the size-exclusion step, an extra reduction protocol was performed involving dialysis into reducing buffer (100 mM sodium phosphate at pH 6.0, 5 mM EDTA), mixing with an equal volume of reducing agent (50 mM dithiothreitol [DTT], 100 mM BME), and incubation for 1 h at 37°C. The purified reduced protein was then dialyzed into SB250 plus 1 mM DTT.
Sedimentation equilibrium
Sedimentation equilibrium experiments were performed on a Beckman Optima XL-I analytical ultracentrifuge. Protein concentrations of 5, 10, and 20 μM in SB250 buffer (plus 1 mM DTT in the case of PY54 Cro) were scanned at wavelengths between 228 and 280 nm. Samples were dialyzed extensively prior to centrifugation and referenced directly against dialysis buffer. All experiments were performed at 20°C and rotor speeds of 23,000 and 30,000 rpm. After establishment of sedimentation equilibrium at each speed, each radial absorbance curve was measured as an average of 15 replicate scans with a radial spacing of 0.001 cm. To extract dimer dissociation constants, six curves (three concentrations and two speeds) were globally fitted to monomer-dimer models using the nonlinear least-squares analysis program WinNonLin (David Yphantis, University of Connecticut; Michael Johnson, University of Virginia; Jeff Lary, National Analytical Ultracentrifuge Center, Storrs, Connecticut). Relevant parameters including molecular masses, solvent densities, and partial specific volumes were calculated using the program SEDNTERP (John Philo, Amgen, Thousand Oaks, California and RASMB). Absorbance offsets were constrained to be zero. Errors are reported as 95% confidence intervals.
Crystal structure of N15 Cro
N15 Cro was crystallized by the hanging-drop method. A protein solution having a concentration of 35 mg/mL N15 Cro was mixed with an equal volume of 0.1 M Tris (pH 8.5) and 1.8 M ammonium sulfate and equilibrated over the precipitant solution. Visible crystals quickly (<24 h) formed that had a cubic space group (F23) and diffracted poorly (6–7 Å). Over several days to weeks, and especially upon disturbing of the drops, these crystals were superseded by monoclinic crystals (space group P21) that diffracted extremely well, typically better than 1.5 Å. Several crystals were flash frozen in liquid nitrogen, and diffraction data were collected at 100 K at the Stanford Synchrotron Radiation Laboratory, Beamline 9–2. The crystal used for structure solution and refinement diffracted to 1.05 Å. The data were reduced and scaled using CrystalClear (d*trek) (Pflugrath 1999). Due to the high resolution of the data, the phase problem was solvable by the program ACORN (Yao et al. 2006) using as a seed structure an idealized 10-residue polyalanine α-helix. The seed structure was placed in the cell using rotation and translation functions; the top solution had a correlation coefficient (CC) of 0.054. Thirty-one cycles of density modification raised CC to 0.39 for all reflections and 0.73 for strong reflections (E > 1). An electron density map calculated with phases from ACORN contained easily interpreted electron density for two molecules in the asymmetric unit. An initial model was built into this electron density map using ARP/WARP (Perrakis et al. 1999) and completed using COOT (Emsley and Cowtan 2004). Cycles of refinement and model building were performed using Refmac5 (Murshudov et al. 1997) and COOT. The asymmetric unit includes two molecules of N15 Cro (chains A and B) related by a noncrystallographic pseudo-twofold axis. The final refinement cycles included anisotropic thermal parameters for all non-hydrogen atoms and had hydrogen atoms added at calculated positions. Residues 67–71 of chain A and residues 65–71 of chain B could not be located in the density maps and were not included in the final model. Residues 63–66 of chain A were visible at some level but were difficult to model. We therefore set residues 63–66 of chain A to an occupancy of 0.5, although no second conformer was modeled. Alternate conformations were included for 30 of 130 modeled residues in the protein. Thirty-one of 154 water molecules were set at an occupancy of 0.5 due to proximity to other water molecules or to atoms in the protein involved in alternate conformations. Occupancies of two sulfate ions were also set to 0.5 for similar reasons. Data measurement, phasing, and refinement statistics are shown in Table 1. Other computations were performed using the CCP4 suite. Coordinates and structure factors have been deposited with the PDB under the identifier 2HIN.
ACKNOWLEDGMENTS
We thank Sherwood Casjens for a gift of bacteriophage N15 genomic DNA. This research was supported by NIH grants GM066806 (M.H.JC.) and HL62969 (W.R.M.).
Footnotes
Reprint requests to: Matthew H.J. Cordes, Department of Biochemistry and Molecular Biophysics, University of Arizona, Tucson, AZ 85721, USA; e-mail: cordes@email.arizona.edu; fax: (520) 621-9288.
Abbreviations: PCR, polymerase chain reaction; BLAST, basic local alignment search tool; IPTG, isopropyl β-D-thiogalactoside; PMSF, phenylmethanesulfonyl fluoride; PEI, polyethyleneimine; EDTA, ethylenediamine tetraacetic acid; SDS-PAGE, sodium dodecyl sulfate–polyacrylamide gel electrophoresis.
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.073330808.
REFERENCES
- Aggarwal, A.K., Rodgers, D.W., Drottar, M., Ptashne, M., Harrison, S.C. Recognition of a DNA operator by the repressor of phage 434: A view at high resolution. Science. 1988;242:899–907. doi: 10.1126/science.3187531. [DOI] [PubMed] [Google Scholar]
- Albright, R.A., Matthews, B.W. Crystal structure of λ-Cro bound to a consensus operator at 3.0 Å resolution. J. Mol. Biol. 1998;280:137–151. doi: 10.1006/jmbi.1998.1848. [DOI] [PubMed] [Google Scholar]
- Albright, R.A., Mossing, M.C., Matthews, B.W. High-resolution structure of an engineered Cro monomer shows changes in conformation relative to the native dimer. Biochemistry. 1996;35:735–742. doi: 10.1021/bi951958n. [DOI] [PubMed] [Google Scholar]
- Bennett, M.J., Schlunegger, M.P., Eisenberg, D. 3D domain swapping: A mechanism for oligomer assembly. Protein Sci. 1995;4:2455–2468. doi: 10.1002/pro.5560041202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling, P.J., Holt, J.M., Ackers, G.K. Coupled energetics of λ cro repressor self-assembly and site- specific DNA operator binding I: Analysis of Cro dimerization from nanomolar to micromolar concentrations. Biochemistry. 2000a;39:11500–11507. doi: 10.1021/bi000935s. [DOI] [PubMed] [Google Scholar]
- Darling, P.J., Holt, J.M., Ackers, G.K. Coupled energetics of λ cro repressor self-assembly and site-specific DNA operator binding II: Cooperative interactions of cro dimers. J. Mol. Biol. 2000b;302:625–638. doi: 10.1006/jmbi.2000.4050. [DOI] [PubMed] [Google Scholar]
- Emsley, P., Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Hall, B.M., Lefevre, K.R., Cordes, M.H. Sequence correlations between Cro recognition helices and cognate O(R) consensus half-sites suggest conserved rules of protein-DNA recognition. J. Mol. Biol. 2005;350:667–681. doi: 10.1016/j.jmb.2005.05.025. [DOI] [PubMed] [Google Scholar]
- Hertwig, S., Klein, I., Schmidt, V., Beck, S., Hammerl, J.A., Appel, B. Sequence analysis of the genome of the temperate Yersinia enterocolitica phage PY54. J. Mol. Biol. 2003;331:605–622. doi: 10.1016/s0022-2836(03)00763-0. [DOI] [PubMed] [Google Scholar]
- Huang, G.S., Oas, T.G. Structure and stability of monomeric λ repressor: NMR evidence for two-state folding. Biochemistry. 1995;34:3884–3892. doi: 10.1021/bi00012a003. [DOI] [PubMed] [Google Scholar]
- Jana, R., Hazbun, T.R., Mollah, A.K., Mossing, M.C. A folded monomeric intermediate in the formation of λ Cro dimer-DNA complexes. J. Mol. Biol. 1997;273:402–416. doi: 10.1006/jmbi.1997.1256. [DOI] [PubMed] [Google Scholar]
- Jia, H., Satumba, W.J., Bidwell G.L., 3rd, Mossing, M.C. Slow assembly and disassembly of λ Cro repressor dimers. J. Mol. Biol. 2005;350:919–929. doi: 10.1016/j.jmb.2005.05.054. [DOI] [PubMed] [Google Scholar]
- Johnson, A.D., Meyer, B.J., Ptashne, M. Interactions between DNA-bound repressors govern regulation by the λ phage repressor. Proc. Natl. Acad. Sci. 1979;76:5061–5065. doi: 10.1073/pnas.76.10.5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyogoku, Y., Kojima, C., Lee, S.J., Tochio, H., Suzuki, N., Matsuo, H., Shirakawa, M. Induced structural changes in protein-DNA complexes. Methods Enzymol. 1995;261:524–541. doi: 10.1016/s0076-6879(95)61023-5. [DOI] [PubMed] [Google Scholar]
- LeFevre, K.R., Cordes, M.H. Retroevolution of λ Cro toward a stable monomer. Proc. Natl. Acad. Sci. 2003;100:2345–2350. doi: 10.1073/pnas.0537925100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobocka, M.B., Svarchevsky, A.N., Rybchin, V.N., Yarmolinsky, M.B. Characterization of the primary immunity region of the Escherichia coli linear plasmid prophage N15. J. Bacteriol. 1996;178:2902–2910. doi: 10.1128/jb.178.10.2902-2910.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis, T., Ptashne, M., Backman, K., Kield, D., Flashman, S., Jeffrey, A., Maurer, R. Recognition sequences of repressor and polymerase in the operators of bacteriophage λ. Cell. 1975;5:109–113. doi: 10.1016/0092-8674(75)90018-5. [DOI] [PubMed] [Google Scholar]
- Matsuo, H., Shirakawa, M., Kyogoku, Y. Three-dimensional dimer structure of the λ-Cro repressor in solution as determined by heteronuclear multidimensional NMR. J. Mol. Biol. 1995;254:668–680. doi: 10.1006/jmbi.1995.0646. [DOI] [PubMed] [Google Scholar]
- Mondragon, A., Subbiah, S., Almo, S.C., Drottar, M., Harrison, S.C. Structure of the amino-terminal domain of phage 434 repressor at 2.0 Å resolution. J. Mol. Biol. 1989;205:189–200. doi: 10.1016/0022-2836(89)90375-6. [DOI] [PubMed] [Google Scholar]
- Mossing, M.C. Solution structure and dynamics of a designed monomeric variant of the λ Cro repressor. Protein Sci. 1998;7:983–993. doi: 10.1002/pro.5560070416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossing, M.C., Sauer, R.T. Stable, monomeric variants of λ Cro obtained by insertion of a designed β-hairpin sequence. Science. 1990;250:1712–1715. doi: 10.1126/science.2148648. [DOI] [PubMed] [Google Scholar]
- Murshudov, G.N., Vagin, A.A., Dodson, E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Neri, D., Billeter, M., Wuthrich, K. Determination of the nuclear magnetic resonance solution structure of the DNA-binding domain (residues 1 to 69) of the 434 repressor and comparison with the X-ray crystal structure. J. Mol. Biol. 1992;223:743–767. doi: 10.1016/0022-2836(92)90987-u. [DOI] [PubMed] [Google Scholar]
- Newlove, T., Konieczka, J.H., Cordes, M.H. Secondary structure switching in Cro protein evolution. Structure. 2004;12:569–581. doi: 10.1016/j.str.2004.02.024. [DOI] [PubMed] [Google Scholar]
- Newlove, T., Atkinson, K.R., Van Dorn, L.O., Cordes, M.H. A trade between similar but nonequivalent intrasubunit and intersubunit contacts in Cro dimer evolution. Biochemistry. 2006;45:6379–6391. doi: 10.1021/bi052541c. [DOI] [PubMed] [Google Scholar]
- Ohlendorf, D.H., Tronrud, D.E., Matthews, B.W. Refined structure of Cro repressor protein from bacteriophage λ suggests both flexibility and plasticity. J. Mol. Biol. 1998;280:129–136. doi: 10.1006/jmbi.1998.1849. [DOI] [PubMed] [Google Scholar]
- Pedulla, M.L., Ford, M.E., Karthikeyan, T., Houtz, J.M., Hendrix, R.W., Hatfull, G.F., Poteete, A.R., Gilcrease, E.B., Winn-Stapley, D.A., Casjens, S.R. Corrected sequence of the bacteriophage p22 genome. J. Bacteriol. 2003;185:1475–1477. doi: 10.1128/JB.185.4.1475-1477.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrakis, A., Morris, R., Lamzin, V.S. Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- Pflugrath, J.W. The finer things in X-ray diffraction data collection. Acta Crystallogr. D Biol. Crystallogr. 1999;55:1718–1725. doi: 10.1107/s090744499900935x. [DOI] [PubMed] [Google Scholar]
- Poteete, A.R., Ptashne, M., Ballivet, M., Eisen, H. Operator sequences of bacteriophages P22 and 21. J. Mol. Biol. 1980;137:81–91. doi: 10.1016/0022-2836(80)90158-8. [DOI] [PubMed] [Google Scholar]
- Poteete, A.R., Hehir, K., Sauer, R.T. Bacteriophage P22 Cro protein: Sequence, purification, and properties. Biochemistry. 1986;25:251–256. doi: 10.1021/bi00349a035. [DOI] [PubMed] [Google Scholar]
- Ravin, V., Ravin, N., Casjens, S., Ford, M.E., Hatfull, G.F., Hendrix, R.W. Genomic sequence and analysis of the atypical temperate bacteriophage N15. J. Mol. Biol. 2000;299:53–73. doi: 10.1006/jmbi.2000.3731. [DOI] [PubMed] [Google Scholar]
- Roessler, C.G., Hall, B.M., Anderson, W.J., Ingram, W.M., Roberts, S.A., Montfort, W.R., Cordes, M.H. Transitive homology-guided structural studies lead to discovery of Cro proteins with 40% sequence identity but different folds. Proc. Natl. Acad. Sci. 2008;105:2343–2348. doi: 10.1073/pnas.0711589105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger, F., Coulson, A.R., Hong, G.F., Hill, D.F., Petersen, G.B. Nucleotide sequence of bacteriophage λ DNA. J. Mol. Biol. 1982;162:729–773. doi: 10.1016/0022-2836(82)90546-0. [DOI] [PubMed] [Google Scholar]
- Sauer, R.T., Yocum, R.R., Doolittle, R.F., Lewis, M., Pabo, C.O. Homology among DNA-binding proteins suggests use of a conserved super-secondary structure. Nature. 1982;298:447–451. doi: 10.1038/298447a0. [DOI] [PubMed] [Google Scholar]
- Vander Byl, C., Kropinski, A.M. Sequence of the genome of Salmonella bacteriophage P22. J. Bacteriol. 2000;182:6472–6481. doi: 10.1128/jb.182.22.6472-6481.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss, M.A., Pabo, C.O., Karplus, M., Sauer, R.T. Dimerization of the operator binding domain of phage λ repressor. Biochemistry. 1987;26:897–904. doi: 10.1021/bi00377a034. [DOI] [PubMed] [Google Scholar]
- Wharton, R.P., Brown, E.L., Ptashne, M. Substituting an α-helix switches the sequence-specific DNA interactions of a repressor. Cell. 1984;38:361–369. doi: 10.1016/0092-8674(84)90491-4. [DOI] [PubMed] [Google Scholar]
- Xu, D., Tsai, C.J., Nussinov, R. Mechanism and evolution of protein dimerization. Protein Sci. 1998;7:533–544. doi: 10.1002/pro.5560070301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, J.X., Dodson, E.J., Wilson, K.S., Woolfson, M.M. ACORN: A review. Acta Crystallogr. D Biol. Crystallogr. 2006;62:901–908. doi: 10.1107/S0907444906008122. [DOI] [PubMed] [Google Scholar]