Abstract
The assembly of FtsZ plays an important role in bacterial cell division. Mycobacterium tuberculosis FtsZ (MtbFtsZ) has a single cysteine residue at position 155. We have investigated the role of the lone cysteine residue in the assembly of MtbFtsZ using different complimentary approaches, namely chemical modification by a thiol-specific reagent 5,5′-dithiobis-(2-nitrobenzoic acid) (DTNB) or a cysteine-chelating agent HgCl2, and site-directed mutagenesis of the cysteine residue. HgCl2 strongly reduced the polymerized mass of MtbFtsZ while it had no detectable effect on the polymerization of Escherichia coli FtsZ, which lacks a cysteine residue. HgCl2 inhibited the protofilamentous assembly of MtbFtsZ and induced the aggregation of the protein. Further, HgCl2 perturbed the secondary structure of MtbFtsZ and increased the binding of a hydrophobic probe 1-anilinonaphthalene-8-sulfonic acid (ANS) with MtbFtsZ, indicating that the binding of HgCl2 altered the conformation of MtbFtsZ. Chemical modification of MtbFtsZ by DTNB also decreased the polymerized mass of MtbFtsZ. Further, the mutagenesis of Cys-155 to alanine caused a strong reduction in the assembly of MtbFtsZ. Under assembly conditions, the mutated protein formed aggregates instead of protofilaments. Far-UV CD spectroscopy and ANS binding suggested that the mutated MtbFtsZ has different conformation than that of the native MtbFtsZ. The effect of the mutation or chemical modification of Cys-155 on the MtbFtsZ assembly has been explained considering its location in the MtbFtsZ crystal structure. The results together suggest that the cysteine residue (Cys-155) of MtbFtsZ plays an important role in the assembly of MtbFtsZ into protofilaments.
Keywords: MtbFtsZ, EcFtsZ, cysteine, DTNB, FtsZ assembly
FtsZ is an essential cell division protein and is highly conserved among prokaryotes (Margolin 2005; Michie and Lowe 2006). FtsZ polymerizes to form protofilaments and bundles in vitro (Mukherjee and Lutkenhaus 1994). These protofilaments and bundles ultimately constitute a ring-like structure at the mid cell, which is called the Z-ring (Errington et al. 2003). The Z-ring is highly dynamic in nature (Stricker et al. 2002) and the constriction of the Z-ring divides cell into two equal daughter cells. A perturbation of FtsZ functions either by mutation (Addinall et al. 1996) or by FtsZ-targeting agents (White et al. 2002; Margalit et al. 2004; Beuria et al. 2005; Jaiswal et al. 2007; Rai et al. 2007) has been shown to cause an abnormal increase in the cell size, ultimately leading to the inhibition of bacterial proliferation.
FtsZ is considered as the bacterial homolog of eukaryotic cytoskeletal protein tubulin, because of their similarity in structural folds and similar GTP binding signature motif (Lowe and Amos 1998). Interestingly, Mycobacterium tuberculosis FtsZ (MtbFtsZ) has a single cysteine residue at position 155, whereas no cysteine is present in its Escherichia coli or Bacillus subtilis counterpart. Cysteine being a free sulfhydryl-containing amino acid plays important roles in the structure and function of various proteins. For example, tubulin has 20 cysteine residues spread throughout the heterodimers; 12 cysteine residues are present in α-tubulin and eight cysteine residues are present in β-tubulin (Krauhs et al. 1981; Ponstingl et al. 1981). One or more of these cysteine residues have been shown to be important for tubulin assembly (Bai et al. 1989; Luduena and Roach 1991; Hosono et al. 2005). Apart from tubulin, cysteine plays crucial roles in functional properties of many other proteins. For example, the mutation of Cys-129 to serine in E. coli K1 CMP-NeuAc synthase caused the enzyme to become more sensitive to heat and chemical denaturation without affecting the structure as such (Zapata et al. 1993). Cysteine residues have also been observed to be very important in case of ion-conducting channels, aquaporins, and amino acid transporters (Kuwahara et al. 2000; Boado et al. 2005; Martial et al. 2007). Three cysteine residues, C462, C583, and C588, were found to be essential for chloride ion conductance of the trout anion exchanger 1 (tAE1), and C462 was found to be important for plasma membrane expression of tAE1 (Martial et al. 2007). Mutation of Cys-88 and Cys-439 of LAT1 light chain altered amino acid transport (Boado et al. 2005). In the case of a water channel AQP-CE2 in Caenorhabditis elegans, Cys-132 was found to be a mercury-sensitive site and was estimated to take part in the formation of the aqueous pore of the channel (Kuwahara et al. 2000).
In the present study, we investigated the role of the single cysteine residue (C155) of MtbFtsZ on its assembly properties using three complimentary approaches, namely site-directed mutagenesis of the cysteine residue, chemical modification of MtbFtsZ by thiol reactive agents 5,5′-dithiobis-(2-nitrobenzoic acid) (DTNB), or by HgCl2. We found that the C155 residue plays an important role in the assembly of MtbFtsZ. Since the perturbation of FtsZ assembly leads to the inhibition of bacterial cell division (Addinall et al. 1996; Margolin 2005), it is tempting to propose that the C155 of MtbFtsZ may be a potential anti-tubercular drug target.
Results
HgCl2 inhibited the assembly of MtbFtsZ
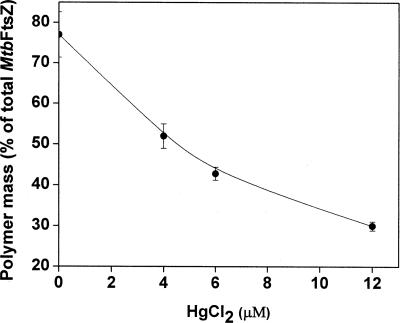
MtbFtsZ has a single cysteine residue at position 155. HgCl2, a known cysteine-chelating agent, was used to examine the role of cysteine residue in the polymerization of MtbFtsZ. HgCl2 decreased the polymerized mass of MtbFtsZ in a concentration-dependent manner (Fig. 1). For example, in the absence and presence of 12 μM HgCl2, 77 ± 5% and 30 ± 1% of the total MtbFtsZ were pelleted as polymers, respectively. The polymerized mass of MtbFtsZ was decreased by 32.8 ± 4%, 44.3 ± 2%, and 60.9 ± 2% in the presence of 4, 6, and 12 μM HgCl2, respectively (Fig. 1). If the lone cysteine residue of MtbFtsZ were the target of HgCl2, then it is logical to think that the presence of excess free sulfhydryl groups will suppress the inhibitory effects of HgCl2 on the assembly of MtbFtsZ. To test the idea, MtbFtsZ (6 μM) was incubated without or with 30 μM 2-mercaptoethanol, and then the protein mixtures were polymerized in the absence or presence of different concentrations of HgCl2. Under the experimental conditions used, 2-mercaptoethanol (30 μM) alone had no detectable effect on the assembly of MtbFtsZ. For example, 79.6 ± 7% and 79.9 ± 6% of MtbFtsZ were pelleted down in the absence and presence of 30 μM 2-mercaptoethanol, respectively. In the absence of 2-mercaptoethanol, HgCl2 (6 μM) decreased the polymer mass of MtbFtsZ by 46.3 ± 4%. However, HgCl2 had a minimal inhibitory effect on the assembly of MtbFtsZ in the presence of 30 μM 2-mercaptoethanol. For example, in the presence of 2-mercaptoethanol, HgCl2 (6 μM) inhibited polymerized mass of MtbFtsZ by ∼5%. The data suggested that HgCl2 inhibited the assembly of MtbFtsZ by chelating the cysteine residue of the protein. To examine further whether Hg2+ exerted its inhibitory effects on the assembly of MtbFtsZ through its cysteine residue, we determined the effects of HgCl2 on the assembly of E. coli FtsZ (EcFtsZ), which does not contain a cysteine residue. EcFtsZ (6 μM) was polymerized in the absence and presence of different concentrations of HgCl2. HgCl2 had no detectable effect on the assembly of EcFtsZ. For example, the polymeric mass of EcFtsZ in the absence and presence of 4, 6, and 12 μM HgCl2 was found to be 38 ± 1.3%, 40 ± 1.0%, 40 ± 1.1%, and 40.1 ± 1.0%, respectively. The results together indicated that HgCl2 inhibited the assembly of MtbFtsZ by chelating its cysteine residue.
Figure 1.
HgCl2 reduced the assembled mass of MtbFtsZ. MtbFtsZ (6 μM) was polymerized in the absence and presence of different concentrations (4, 6, and 12 μM) of HgCl2.
In the absence of HgCl2, MtbFtsZ formed linear protofilaments and bundles (Fig. 2 top panel). In the presence of 6 μM HgCl2, the number of protofilaments per field of view decreased significantly and some aggregates were also observed (Fig. 2 middle panel). In the presence of 12 μM HgCl2, aggregates of MtbFtsZ were predominantly found and no protofilament-like structure was visible (Fig. 2 bottom panel). The results suggested that the cysteine residue of MtbFtsZ is important for the assembly of FtsZ into protofilaments.
Figure 2.
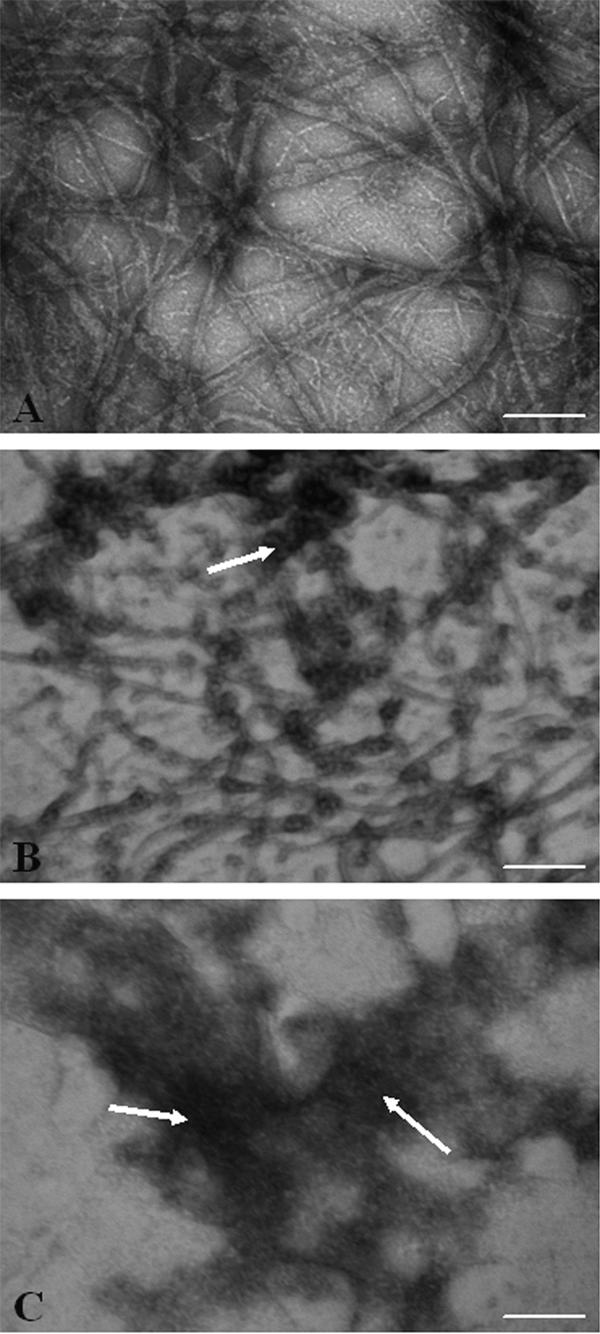
HgCl2 altered morphology of MtbFtsZ polymers. MtbFtsZ was polymerized and negatively stained using 2% uranyl acetate. Shown are MtbFtsZ polymers in the absence (A) and presence of 6 μM (B) and 12 μM (C) HgCl2, respectively. Arrowheads are showing aggregates. Scale bar is 200 nm.
HgCl2 decreased the secondary structural content of MtbFtsZ
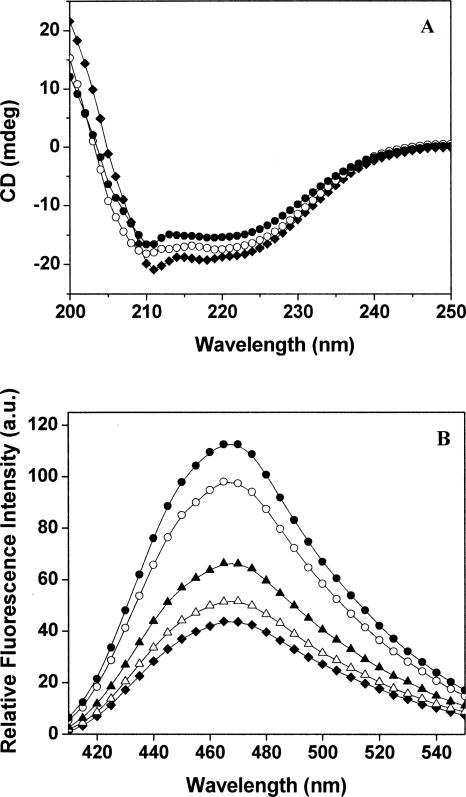
HgCl2 perturbed the far-UV CD spectra of MtbFtsZ (Fig. 3A). For example, the CD signal (222 nm) of MtbFtsZ was decreased by 18 ± 1% in the presence of 6 μM HgCl2 as compared to the control, indicating that HgCl2 altered the helical structure of FtsZ.
Figure 3.
Effect of HgCl2 on the conformation of MtbFtsZ. (A) MtbFtsZ was incubated in the absence and presence of different concentrations of HgCl2 at room temperature. Shown are the far-UV CD spectra of MtbFtsZ in the absence (♦) and presence of 3 μM (○) and 6 μM (●) HgCl2. (B) MtbFtsZ (4 μM) in 25 mM HEPES buffer (pH 6.5) was incubated in the absence and presence of different concentrations of HgCl2 at 25°C for 30 min. Then, the reaction mixtures were incubated with 100 μM ANS for an additional 30 min. The fluorescence spectra of MtbFtsZ–ANS complex in the absence (♦) and presence of 1 μM (△), 2 μM (▲), 4 μM (○), and 6 μM (●) HgCl2, respectively, are shown.
HgCl2 induced conformational change in MtbFtsZ
MtbFtsZ does not contain a tryptophan residue. It has been shown that a hydrophobic fluorescent probe 1-anilinonaphthalene-8-sulfonic acid (ANS) binds to FtsZ (Yu and Margolin 1998). The FtsZ–ANS complex fluorescence has been used to monitor the conformational change in FtsZ (Santra and Panda 2003; Mukherjee et al. 2005). Therefore, conformational change of FtsZ in the presence of HgCl2 was probed by monitoring MtbFtsZ–ANS fluorescence, as HgCl2 had no affect on the fluorescence spectra of free ANS in the absence of FtsZ. The fluorescence intensity of ANS in the presence of MtbFtsZ increased with increasing concentration of HgCl2, indicating that HgCl2 induces conformational change in MtbFtsZ (Fig. 3B). For example, the fluorescence intensity of MtbFtsZ–ANS complex at 470 nm was found to be 43 and 112 (a.u) in the absence and presence of 6 μM HgCl2, respectively.
Chemical modification of cysteine residue of MtbFtsZ by DTNB inhibited MtbFtsZ assembly
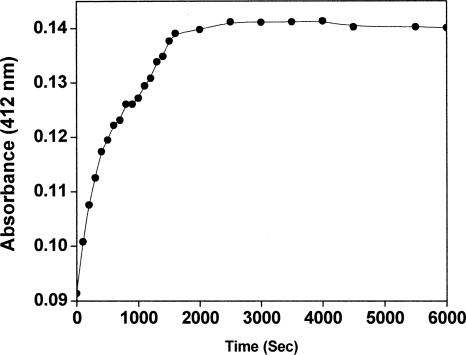
MtbFtsZ (6 μM) was mixed with 200 μM DTNB and the kinetics of the chemical modification of the protein were monitored by measuring the absorbance of the reaction mixture with time at 412 nm (Fig. 4). The stoichiometry of incorporation of TNB per MtbFtsZ molecule after 4 h of incubation was calculated to be 0.71 ± 0.06. DTNB-modified MtbFtsZ was polymerized in the presence of 100 mM KCl, 5 mM MgCl2, and 1 mM GTP. Under the experimental conditions used, the polymerized mass of TNB-MtbFtsZ was reduced by 70 ± 8% compared to that of the native MtbFtsZ. MtbFtsZ was treated similarly in the absence of DTNB and no significant effect of dialysis on the assembly was observed.
Figure 4.
Kinetics of chemical modification of MtbFtsZ with DTNB. The kinetics of labeling of MtbFtsZ with DTNB were monitored by measuring the absorbance at 412 nm.
Effects of C155A–MtbFtsZ mutation on polymer mass
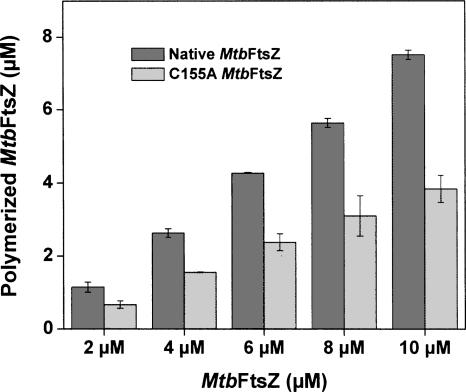
To check whether the cysteine155 has a role in the assembly of MtbFtsZ, a mutant of MtbFtsZ (C155A) was constructed by replacing the cysteine residue of MtbFtsZ with an alanine residue. Increasing concentrations (2–10 μM) of native and mutant FtsZ were polymerized to compare the polymer level of both the proteins. There was a concentration-dependent increase in polymer mass of both the proteins, but the polymer mass of C155A was significantly lower compared to that of the native MtbFtsZ. For example, with 10 μM MtbFtsZ, 7.5 μM of total protein was polymerized, whereas in the case of 10 μM C155A–MtbFtsZ only 3.8 μM of the total protein was sedimented as polymer (Fig. 5).
Figure 5.
C155A mutation inhibited MtbFtsZ assembly. Different concentrations of MtbFtsZ (2–10 μM), both native and C155A, were polymerized in the presence of 100 mM KCl, 5 mM MgCl2, and 1 mM GTP for 30 min at 37°C. Dark gray bars show native MtbFtsZ and light gray bars denote C155A MtbFtsZ.
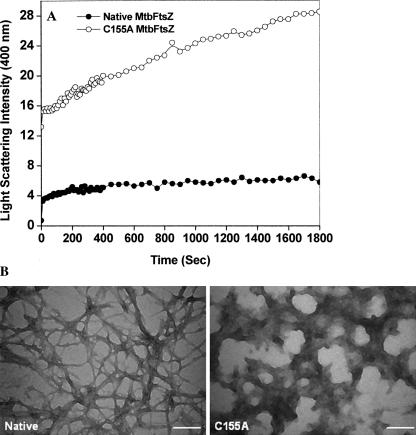
The effect of C155A mutation on polymerization of MtbFtsZ was further examined by monitoring light scattering (Fig. 6A). Surprisingly, the extent of light scattering signal for the mutant MtbFtsZ was fivefold higher than that for the native protein, which was apparently contradictory to the results obtained from the sedimentation assay. This might be either due to the formation of large polymers or due to the formation of aggregates. The polymer morphology of both the native and mutant proteins was analyzed using electron microscopy (Fig. 6B). With native MtbFtsZ, long thin protofilaments as well as bundles of protofilaments were observed, whereas, in the case of C155A, mostly aggregates were observed. No protofilamentous structure was detected with the mutated FtsZ, indicating that the increase in the light scattering signal was due to the formation of aggregates and not due to the polymerization of C155A into protofilaments.
Figure 6.
C155A mutation in MtbFtsZ induced aggregate formation of MtbFtsZ. (A) MtbFtsZ (native and mutant) were polymerized with 100 mM KCl, 5 mM MgCl2, and 1 mM GTP at 37°C. Shown are the traces with native (●) and C155A (○) MtbFtsZ. (B) Both native and mutant MtbFtsZ were polymerized and negatively stained using 2% uranyl acetate. Scale bar is 200 nm.
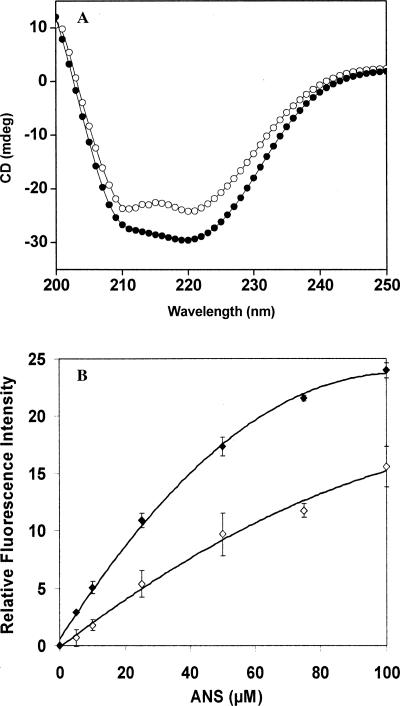
Differences in the far-UV CD spectra of the native and mutant (C155A) MtbFtsZ indicated that the mutation of C155 residue of MtbFtsZ altered the secondary structure of the protein (Fig. 7A). Further, both mutant (C155A) and native MtbFtsZ were incubated with a hydrophobic probe ANS and the fluorescence intensity at 470 nm was monitored. Fluorescence intensity of both the proteins increased with increasing concentrations of ANS (Fig. 7B). However, the fluorescence intensity of C155A–MtbFtsZ was higher than that of the MtbFtsZ, indicating that the mutation caused an alteration in the structure of MtbFtsZ (Fig. 7B).
Figure 7.
C155A mutation induced conformational change in MtbFtsZ. (A) The far-UV CD spectra of 5 μM C155A mutant (○) and native (●) MtbFtsZ in 25 mM sodium phosphate buffer, pH 7.0, are shown. One of the six similar experiments is shown in the figure. (B) The binding of ANS to native (⋄) and C155A (♦) MtbFtsZ. MtbFtsZ (native and mutant) were incubated without and with different concentrations of ANS (5–100 μM) and the fluorescence spectra were recorded using 370 nm as the excitation wavelength. The appropriate blanks were subtracted from the experimental spectra.
Discussion
The present study suggests that the modification of the lone cysteine residue (C155) of MtbFtsZ perturbs the assembly of the protein. Under the assembly conditions used in this work, native MtbFtsZ formed protofilaments and bundles. However, both chemically modified MtbFtsZ and mutant C155A–MtbFtsZ produced large aggregates rather than the typical protofilaments and bundles. There are at least two possible reasons for the impairment of the MtbFtsZ assembly upon cysteine mutation or modification. Cysteine residues are shown to play important roles in the folding of proteins (Branden and Tooze 1999). C155 in MtbFtsZ might be involved in proper folding of protein and the modification or the mutation of the cysteine residue may induce a conformational change in MtbFtsZ (discussed below). A change in conformation of MtbFtsZ upon cysteine modification by HgCl2 was evident from the altered far-UV CD spectra of MtbFtsZ and the increased fluorescence intensity of MtbFtsZ–ANS complex in the presence of HgCl2 (Fig. 3A,B). Similar conformational changes were also observed with C155A–MtbFtsZ.
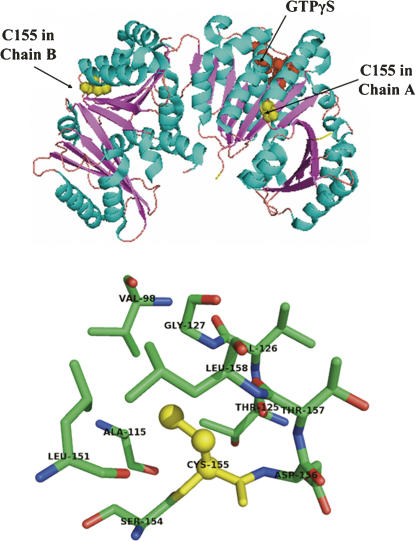
An analysis of the sequence alignment of a number of bacterial FtsZs suggested that C155 is highly conserved among Mycobacterium species and indicated that the cysteine residue is replaced by a valine residue in bacteria other than the Mycobacterium. Further, a careful analysis of the MtbFtsZ crystal structure (Fig. 8 top panel) revealed that the side chain of C155 is pointing toward the hydrophobic pocket and is surrounded by residues like V98, A115, T125, G127, L151, and L158 (Fig. 8 bottom panel). The three-dimensional structure of the protein is thus consistent with the occurrence of valine at position 155 in FtsZ orthologs. In several other nonpathogenic organisms, like Salinispora arenicola and Corynebacterium glutamicum, a cysteine residue is present at this position, indicating that the C155 is well conserved.
Figure 8.
C155 location on the crystal structure of MtbFtsZ dimer. (Top panel) The position of C155 in MtbFtsZ. C155 is located between the helix H8 and strand B6. Figure shows the location of GTPγS bound to MtbFtsZ with respect to C155. Cyan and magenta colors show helices and sheets, respectively. Cysteine is shown as yellow spheres and GTPγS as red spheres. (Bottom panel) The environment around Cys-155. Shown as sticks are residues within a sphere of 5.0 Å around the SG atom of Cys-155. Cys-155 side chain atoms shown as yellow spheres are placed in the hydrophobic pocket defined by V98, A115, T125, G127, L151, and L158 residues. Figures were drawn with PyMOL (Delano Scientific) software using a PDB coordinate file (code 1rlu).
MtbFtsZ has been reported to exist as dimers (Leung et al. 2004). It is apparent from the crystal structure of MtbFtsZ that the C155 is located at the loop between helix H8 and strand β6 (Fig. 8 top panel). The detailed examination of the C155 residue and its surrounding amino acid residues in the reported three-dimensional structure of MtbFtsZ (PDB: 1rlu) suggested that the side chain of C155 is not pointing toward the surface of the molecule; thus, it is possible that the C155 may not participate in the intermolecular contacts. The cysteine side chain is pointing into a hydrophobic pocket constituted by residues V98, A115, T125, G127, L151, and L158 (Fig. 8 bottom panel). The substitution of the C155 by an alanine may not disturb the monomer structure radically as both cysteine and alanine have similar hydrophobicity, except that an alanine residue has a shorter side chain than a cysteine residue, which may cause a destabilizing void or a cavity in the hydrophobic pocket. It is thus possible that C155A mutation may induce a conformational change in the FtsZ monomer. This is consistent with the CD and ANS binding studies reported in this study (Fig. 7). The conformational change may not allow the mutant (C155A) FtsZ to form protofilaments. Alternatively, the cysteine side chain may swing out and get exposed on the surface of MtbFtsZ during the assembly process and actively takes part in protofilament formation. Chemical modifications or the mutation of the C155 could perturb the surface conformation of MtbFtsZ and inhibit its assembly.
The inhibition of MtbFtsZ polymerization due to the mutation or the modification of cysteine residue contradicts a previous report in which MtbFtsZ was labeled using fluorophores (fluorescein-5-maleimide and tetramethylrhodamine-5-maleimide) (Chen et al. 2007). The covalent modification of MtbFtsZ had minimal effects on its assembly (Chen et al. 2007). The discrepancy in the results might be due to several reasons. It has been reported that maleimide groups react primarily with free sulfhydryls at pH range of 6.5–7.5. However, the reactivity of maleimide strongly increases for amines at pH >7.5 (Bigelow and Inesi 1991; Haughland 2002) (http://www.piercenet.com/files/0359dh5.pdf). In the previous study (Chen et al. 2007), MtbFtsZ was labeled at pH 7.9. It is highly plausible that, at this elevated pH, maleimides could modify one of the 12 lysine residues present in MtbFtsZ. We tested whether fluorescein-5-maleimide (FM) at pH 7.9 could modify a residue other than a cysteine residue in FtsZ using EcFtsZ. If FM reacts only with the cysteine residue, it should not chemically modify EcFtsZ because the protein does not have a cysteine residue. We found that EcFtsZ could be covalently modified with FM at pH 7.9. Under the experimental conditions used, the stoichiometry of FM incorporated per EcFtsZ was determined to be 0.3–0.4. In addition, the polymerization ability of the FM-modified EcFtsZ was found to be similar to that of the native protein (data not shown). Therefore, it might be possible that the lysine groups of MtbFtsZ were covalently modified by the maleimide group instead of the cysteine at pH 7.9 (reaction conditions used by Chen et al. [2007]). Also the labeling efficiency reported in the previous study was 30%–40%. Considering that both lysines and cysteine were modified, the net effect of modification on the assembly is expected to be minimal. Though the cysteine-modified FtsZ predominantly forms aggregates, the possibility that the cysteine-modified FtsZ monomers can add to the protofilaments in low numbers is difficult to rule out. We used the site-directed mutagenesis approach to demonstrate that the cysteine residue of MtbFtsZ plays an important role in the assembly of MtbFtsZ. We found that, with C155A substitution, there was a decrease in polymer mass and the mutant FtsZ predominantly formed aggregates. Similar results were obtained when HgCl2 and DTNB were used to modify the cysteine residue.
Although it is tempting to think that the cysteine-targeted agents may not have target specificity, several studies suggested that cysteine residues may be considered as important drug targets for cancer and viral diseases (Rice et al. 1995; Huang et al. 1998; Ramboarina et al. 1999; Shan et al. 1999; Scozzafava et al. 2000; Casini et al. 2002; Hosono et al. 2005). For example, several cysteine-targeting agents such as 3 nitrosobenzamide derivatives, disulfide benzamide, and dithianes are considered as potential drug candidates for viral diseases (Rice et al. 1995; Huang et al. 1998; Ramboarina et al. 1999; Casini et al. 2002). Similarly, T138067 (Shan et al. 1999) and arylsulfonyl-N,N-diethyldithiocarbamates (Scozzafava et al. 2000), which specifically bind to the cysteine residues of β-tubulin, have been projected as potent antitumor agents. In addition, an antimitotic drug 2,4-dichlorobenzyl thiocyanate, was shown to inhibit tubulin assembly at substoichiometric concentrations by specifically targeting Cys-239 of β-tubulin (Bai et al. 1989).
Recent evidences suggest that the perturbation of FtsZ assembly by natural or synthetic small molecules inhibits bacterial proliferation, suggesting that FtsZ can be considered as an attractive antibacterial target (Margalit et al. 2004; Beuria et al. 2005; Jaiswal et al. 2007; Rai et al. 2007). To our knowledge, this is the first report suggesting that the cysteine 155 plays an important role in the assembly of MtbFtsZ. On the basis of findings of the present study, one can speculate that the region around cysteine residue in MtbFtsZ may play a critical role in regulating FtsZ assembly and the Z-ring formation in vivo in M. tuberculosis. This introduces an interesting possibility of using sulfhydryl-directed agents to specifically inhibit the assembly of FtsZ in M. tuberculosis.
Materials and Methods
Reagents
HEPES (N-[2-Hydroxyethyl]piperazine-N′-[2-ethanesulfonic acid]), isopropyl-β-D-thiogalactopyranoside (IPTG), monosodium glutamate, guanosine-5′-triphosphate (GTP), 5,5′-dithiobis-(2-nitrobenzoic acid) (DTNB), 2-mercaptoethanol were obtained from Sigma. HgCl2 was purchased from Merck. Fluorescein-5-maleimide and 1-Anilinonaphthalene-8-sulfonic acid (ANS) were purchased from Molecular Probes. All other chemicals used were of analytical grade.
Expression and purification of MtbFtsZ and C155A–MtbFtsZ
Site-directed mutagenesis and sequencing of the mutant C155A–MtbFtsZ were performed commercially by Bangalore Genei. Nucleotide sequence of the mutant plasmid was confirmed with the help of Macrogen. Both native and mutant proteins were overexpressed and purified as described earlier (Jaiswal et al. 2007). FtsZ concentration was measured by the Bradford method using bovine serum albumin as a standard and stored at −80°C. Protein was thawed and spun at high speed to remove aggregates prior to use. The purity of FtsZ was estimated to be ∼98% from a Coomassie stained sodium dodecyl sulfate polyacrylamide gel electrophoresis.
Chemical modification of MtbFtsZ with DTNB
DTNB was prepared in 25 mM phosphate buffer (pH 7.0). MtbFtsZ (20 μM) in 25 mM HEPES buffer, pH 7.0, was incubated with 200 μM DTNB at 4°C for 4 h. Incorporation ratio of DTNB to MtbFtsZ was determined by dividing the bound TNB concentration by the concentration of MtbFtsZ. The bound TNB concentration was determined by measuring the absorbance of free TNB− at 412 nm using molar extinction coefficient of 12,800 M−1cm−1. Then, the protein mixture was dialyzed in 25 mM HEPES buffer, pH 6.5 for 7 h to remove the free TNB−. Protein concentration was measured by the Bradford method (Bradford 1976) and the labeled protein was used for the polymerization reaction as described earlier.
Chemical modification of EcFtsZ with fluorescein-5-maliemide
Fluorescein-5-maliemide (FM) was prepared in dimethylformamide (DMF). EcFtsZ (15 μM) in 25 mM Tris buffer, pH 7.9, was incubated with 10-fold higher concentration (150 μM) of FM for 4 h on ice. Reaction mixture was passed through a pre-equilibrated Biogel P-6 column and then dialyzed extensively for 24 h in 25 mM HEPES buffer, pH 7.9, containing 100 mM KCl and 5 mM MgCl2 to remove the free probe. EcFtsZ concentration was measured by Bradford assay and FM concentration was measured by taking absorbance at 492 nm using 83,000 M−1cm−1 as molar absorbance coefficient. Incorporation stoichiometry was calculated by dividing the FM concentration in the labeled EcFtsZ by the protein concentration. Incorporation ratio (FM per EcFtsZ molecule) was found to be 0.3–0.4.
Sedimentation assay
Native and mutant MtbFtsZ (6 μM) were polymerized in 25 mM HEPES buffer, pH 6.5, containing 100 mM KCl, 5 mM MgCl2, and 1 mM GTP at 37°C for 30 min. Reaction mixture was then centrifuged at 227,000g for 30 min at 30°C. The protein concentration in the supernatant was measured by the Bradford method. Protein concentration in the pellet was calculated by subtracting the supernatant protein concentration from the total protein concentration.
MtbFtsZ (6 μM) was incubated in the absence and presence of 4, 6, and 12 μM HgCl2 on ice for 10 min in 25 mM HEPES buffer, pH 6.5, and polymerized as described earlier.
Effect of HgCl2 on the assembly of MtbFtsZ in the presence of 2-mercaptoethanol
FtsZ was first incubated without or with 30 μM 2-mercaptoethanol for 30 min on ice. Then, 6 μM HgCl2 was added to the reaction milieu and incubated for an additional 30 min. After the incubation, the reaction mixture was polymerized by adding 100 mM KCl, 5 mM MgCl2, and 1 mM GTP at 37°C for 30 min. Reaction mixtures were centrifuged at 227,000g for 30 min at 30°C. Protein concentration in the pellet was determined.
Light scattering assay
Native and mutant MtbFtsZ (6 μM) were prepared in 25 mM HEPES buffer, pH 6.5, 100 mM KCl, and 5 mM MgCl2. After the addition of 1 mM GTP, the sample was immediately placed in a cuvette at 37°C and the polymerization reaction was followed by monitoring 90° light scattering at 400 nm using a JASCO 6500 spectrofluorometer.
Electron microscopy
MtbFtsZ (6 μM) in 25 mM HEPES buffer containing 100 mM KCl and 5 mM MgCl2 was polymerized in the presence of GTP at 37°C for 30 min. The FtsZ polymers in the samples were fixed with prewarmed 0.5% glutaraldehyde for 5 min. FtsZ polymer solution (50 μL) was placed on the carbon-coated copper grids (300 mesh size) and then blotted dry. The grids were subsequently subjected to negative staining by 1% uranyl acetate solution and air-dried. The samples were examined using a FEI Tecnai-G2 12 electron microscope (Jaiswal et al. 2007). In the case of HgCl2-treated samples, MtbFtsZ was incubated without and with different concentrations of HgCl2 prior to polymerization.
Secondary structure studies with HgCl2
Sodium phosphate buffer (25 mM, pH 7.0) was degassed for optimum transparency and was further cleared by passing it through 0.2 μm syringe filter (Millipore). MtbFtsZ was incubated in the absence and presence of 3 and 6 μM HgCl2 for 10 min at room temperature. The secondary structure was monitored over the wavelength range of 200–250 nm using a 0.1-cm path length cuvette and the ellipticity was determined at 222 nm. A spectral bandwidth of 10 nm and time constant of 1 s were used for all measurements. Each spectrum was recorded using an average of five scans. The CD spectra of 5 μM C155A–MtbFtsZ were also recorded in the similar way.
ANS fluorescence measurement
Native MtbFtsZ (4 μM) in 25 mM HEPES buffer (pH 6.5) was incubated in the absence and presence of increasing concentration (1, 2, 4, and 6 μM) of HgCl2 at 25°C for 30 min. Further, 100 μM ANS was added to all the reaction mixtures and incubated for additional 30 min. Fluorescence spectra were recorded using JASCO FP-6500 fluorescence spectrophotometer. An appropriate blank spectrum of free ANS was subtracted from the respective experimental spectrum. A quartz cuvette of 0.3-cm path length was used for all experiments, and 5 nm and 10 nm were used as excitation and emission bandwidths, respectively. MtbFtsZ–ANS fluorescence was monitored using 370 nm as the excitation wavelength and recording the emission spectrum over the range of 410–550 nm.
Native or mutated MtbFtsZ (6 μM) in 25 mM HEPES buffer (pH 6.5) was incubated with different concentrations (5, 10, 25, 50, 75, 100 μM) of ANS at room temperature for 30 min. Fluorescence spectra were recorded using 370 nm as an excitation wavelength.
Acknowledgments
The work is supported by a grant from the Department of Science and Technology, Government of India. D.P. is a Swarnajayanti fellow. R.J. thanks CSIR for fellowship. We thank SAIF, IIT Bombay for providing the electron microscopy facility. We would like to thank Dr. Vinay Kumar for insightful discussion on this manuscript.
Footnotes
Reprint requests to: Dulal Panda, School of Biosciences and Bioengineering, Indian Institute of Technology Bombay, Powai, Mumbai 400076, India; e-mail: panda@iitb.ac.in; fax: 91-22-2576-4770.
Abbreviations: MtbFtsZ, Mycobacterium tuberculosis FtsZ; EcFtsZ, Escherichia coli FtsZ; C155A–MtbFtsZ, MtbFtsZ in which cysteine 155 is replaced with alanine; IPTG, isopropyl-β-D-thiogalactopyranoside; DTNB, 5,5′-dithiobis-(2-nitrobenzoic acid); ANS, 1-anilinonaphthalene-8-sulfonic acid; FM, fluorescein-5-maliemide.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.083452008.
References
- Addinall, S.G., Bi, E., Lutkenhaus, J. FtsZ ring formation in fts mutants. J. Bacteriol. 1996;178:3877–3884. doi: 10.1128/jb.178.13.3877-3884.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai, R.L., Lin, C.M., Nguyen, N.Y., Liu, T.Y., Hamel, E. Identification of the cysteine residue of β-tubulin alkylated by the antimitotic agent 2,4-dichlorobenzyl thiocyanate, facilitated by separation of the protein subunits of tubulin by hydrophobic column chromatography. Biochemistry. 1989;28:5606–5612. doi: 10.1021/bi00439a040. [DOI] [PubMed] [Google Scholar]
- Beuria, T.K., Santra, M.K., Panda, D. Sanguinarine blocks cytokinesis in bacteria by inhibiting FtsZ assembly and bundling. Biochemistry. 2005;44:16584–16593. doi: 10.1021/bi050767+. [DOI] [PubMed] [Google Scholar]
- Bigelow, D.J., Inesi, G. Frequency-domain fluorescence spectroscopy resolves the location of maleimide-directed spectroscopic probes within the tertiary structure of the Ca-ATPase of sarcoplasmic reticulum. Biochemistry. 1991;30:2113–2125. doi: 10.1021/bi00222a016. [DOI] [PubMed] [Google Scholar]
- Boado, R.J., Li, J.Y., Chu, C., Ogoshi, F., Wise, P., Pardridge, W.M. Site-directed mutagenesis of cysteine residues of large neutral amino acid transporter LAT1. Biochim. Biophys. Acta. 2005;1715:104–110. doi: 10.1016/j.bbamem.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Branden, C., Tooze, J. Introduction to protein structure. 2d ed. Garland Publishing; New York: 1999. Enzymes assist formation of proper disulfide bonds during folding; pp. 96–98. [Google Scholar]
- Casini, A., Scozzafava, A., Supuran, C.T. Cysteine-modifying agents: A possible approach for effective anticancer and antiviral drugs. Environ. Health Perspect. 2002;110:801–806. doi: 10.1289/ehp.02110s5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y., Anderson, D.E., Rajagopalan, M., Erickson, H.P. Assembly dynamics of Mycobacterium tuberculosis FtsZ. J. Biol. Chem. 2007;282:27736–27743. doi: 10.1074/jbc.M703788200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errington, J., Daniel, R.A., Scheffers, D.J. Cytokinesis in bacteria. Microbiol. Mol. Biol. Rev. 2003;67:52–65. doi: 10.1128/MMBR.67.1.52-65.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haughland, R.P. Handbook of fluorescent probes and research products. 9th ed. Molecular Probes; New York: 2002. [Google Scholar]
- Hosono, T., Fukao, T., Ogihara, J., Ito, Y., Shiba, H., Seki, T., Ariga, T. Diallyl trisulfide suppresses the proliferation and induces apoptosis of human colon cancer cells through oxidative modification of β-tubulin. J. Biol. Chem. 2005;280:41487–41493. doi: 10.1074/jbc.M507127200. [DOI] [PubMed] [Google Scholar]
- Huang, M., Maynard, A., Turpin, J.A., Graham, L., Janini, G.M., Covell, D.G., Rice, W.G. Anti-HIV agents that selectively target retroviral nucleocapsid protein zinc fingers without affecting cellular zinc fingers. J. Med. Chem. 1998;41:1371–1381. doi: 10.1021/jm9708543. [DOI] [PubMed] [Google Scholar]
- Jaiswal, R., Beuria, T.K., Mohan, R., Mahajan, S.K., Panda, D. Totarol inhibits bacterial cytokinesis by perturbing the assembly dynamics of FtsZ. Biochemistry. 2007;46:4211–4220. doi: 10.1021/bi602573e. [DOI] [PubMed] [Google Scholar]
- Krauhs, E., Little, M., Kempf, T., Hofer-Warbinek, R., Ade, W., Ponstingl, H. Complete amino acid sequence of β-tubulin from porcine brain. Proc. Natl. Acad. Sci. 1981;78:4156–4160. doi: 10.1073/pnas.78.7.4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwahara, M., Asai, T., Sato, K., Shinbo, I., Terada, Y., Marumo, F., Sasaki, S. Functional characterization of a water channel of the nematode Caenorhabditis elegans . Biochim. Biophys. Acta. 2000;1517:107–112. doi: 10.1016/s0167-4781(00)00268-2. [DOI] [PubMed] [Google Scholar]
- Leung, A.K., Lucile White, E., Ross, L.J., Reynolds, R.C., DeVito, J.A., Borhani, D.W. Structure of Mycobacterium tuberculosis FtsZ reveals unexpected, G protein-like conformational switches. J. Mol. Biol. 2004;342:953–970. doi: 10.1016/j.jmb.2004.07.061. [DOI] [PubMed] [Google Scholar]
- Lowe, J., Amos, L.A. Crystal structure of the bacterial cell-division protein FtsZ. Nature. 1998;391:203–206. doi: 10.1038/34472. [DOI] [PubMed] [Google Scholar]
- Luduena, R.F., Roach, M.C. Tubulin sulfhydryl groups as probes and targets for antimitotic and antimicrotubule agents. Pharmacol. Ther. 1991;49:133–152. doi: 10.1016/0163-7258(91)90027-j. [DOI] [PubMed] [Google Scholar]
- Margalit, D.N., Romberg, L., Mets, R.B., Hebert, A.M., Mitchison, T.J., Kirschner, M.W., RayChaudhuri, D. Targeting cell division: Small-molecule inhibitors of FtsZ GTPase perturb cytokinetic ring assembly and induce bacterial lethality. Proc. Natl. Acad. Sci. 2004;101:11821–11826. doi: 10.1073/pnas.0404439101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin, W. FtsZ and the division of prokaryotic cells and organelles. Nat. Rev. Mol. Cell Biol. 2005;6:862–871. doi: 10.1038/nrm1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martial, S., Guizouarn, H., Gabillat, N., Pellissier, B., Borgese, F. Importance of several cysteine residues for the chloride conductance of trout anion exchanger 1 (tAE1) J. Cell. Physiol. 2007;213:70–78. doi: 10.1002/jcp.21093. [DOI] [PubMed] [Google Scholar]
- Michie, K.A., Lowe, J. Dynamic filaments of the bacterial cytoskeleton. Annu. Rev. Biochem. 2006;75:467–492. doi: 10.1146/annurev.biochem.75.103004.142452. [DOI] [PubMed] [Google Scholar]
- Mukherjee, A., Lutkenhaus, J. Guanine nucleotide-dependent assembly of FtsZ into filaments. J. Bacteriol. 1994;176:2754–2758. doi: 10.1128/jb.176.9.2754-2758.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee, A., Santra, M.K., Beuria, T.K., Panda, D. A natural osmolyte trimethylamine N-oxide promotes assembly and bundling of the bacterial cell division protein, FtsZ and counteracts the denaturing effects of urea. FEBS J. 2005;272:2760–2772. doi: 10.1111/j.1742-4658.2005.04696.x. [DOI] [PubMed] [Google Scholar]
- Ponstingl, H., Krauhs, E., Little, M., Kempf, T. Complete amino acid sequence of α-tubulin from porcine brain. Proc. Natl. Acad. Sci. 1981;78:2757–2761. doi: 10.1073/pnas.78.5.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai, D., Singh, J.K., Roy, N., Panda, D. Curcumin inhibits FtsZ assembly: An attractive mechanism for its antibacterial activity. Biochem. J. 2007;410:147–155. doi: 10.1042/BJ20070891. [DOI] [PubMed] [Google Scholar]
- Ramboarina, S., Moreller, N., Fournie-Zaluski, M.C., Roques, B.P. Structural investigation on the requirement of CCHH zinc finger type nucleocapsid protein of human immunodeficiency virus 1. Biochemistry. 1999;38:9600–9607. doi: 10.1021/bi9905258. [DOI] [PubMed] [Google Scholar]
- Rice, W.G., Supko, J.G., Malspeis, L., Buckheit R.W., Jr, Clanton, D., Bu, M., Graham, L., Schaeffer, C.A., Turpin, J.A., Domagala, J., et al. Inhibitors of HIV nucleocapsid protein zinc fingers as candidates for the treatment of AIDS. Science. 1995;270:1194–1197. doi: 10.1126/science.270.5239.1194. [DOI] [PubMed] [Google Scholar]
- Santra, M.K., Panda, D. Detection of an intermediate during unfolding of bacterial cell division protein FtsZ: Loss of functional properties precedes the global unfolding of FtsZ. J. Biol. Chem. 2003;278:21336–21343. doi: 10.1074/jbc.M301303200. [DOI] [PubMed] [Google Scholar]
- Scozzafava, A., Mastrolorenzo, A., Supuran, C.T. Arylsulfonyl-N,N-diethyl-dithiocarbamates: A novel class of antitumor agents. Bioorg. Med. Chem. Lett. 2000;10:1887–1891. doi: 10.1016/s0960-894x(00)00375-9. [DOI] [PubMed] [Google Scholar]
- Shan, B., Medina, J.C., Santha, E., Frankmoelle, W.P., Chou, T.C., Learned, R.M. Selective, covalent modification of β-tubulin residue Cys-239 by T138067, an antitumor agent with in vivo efficacy against multidrug-resistant tumors. Proc. Natl. Acad. Sci. 1999;96:5686–5691. doi: 10.1073/pnas.96.10.5686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stricker, J., Maddox, P., Salmon, E.D., Erickson, H.P. Rapid assembly dynamics of the Escherichia coli FtsZ-ring demonstrated by fluorescence recovery after photobleaching. Proc. Natl. Acad. Sci. 2002;99:3171–3175. doi: 10.1073/pnas.052595099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, E.L., Suling, W.J., Ross, L.J., Seitz, L.E., Reynolds, R.C. 2-alkoxycarbonyl aminopyridines: Inhibitors of Mycobacterium tuberculosis FtsZ. J. Antimicrob. Chemother. 2002;50:111–114. doi: 10.1093/jac/dkf075. [DOI] [PubMed] [Google Scholar]
- Yu, X.C., Margolin, W. Inhibition of assembly of bacterial cell division protein FtsZ by the hydrophobic dye 5,5′-bis-(8-anilino-1-naphthalenesulfonate) J. Biol. Chem. 1998;273:10216–10222. doi: 10.1074/jbc.273.17.10216. [DOI] [PubMed] [Google Scholar]
- Zapata, G., Roller, P.P., Crowley, J., Vann, W.F. The role of cysteine residues 129 and 329 in Escherichia coli K1 CMP-NeuAc synthase. Biochem. J. 1993;295:485–491. doi: 10.1042/bj2950485. [DOI] [PMC free article] [PubMed] [Google Scholar]