Summary
During Drosophila oogenesis, the targeted localization of gurken (grk) mRNA leads to the establishment of the axis polarity of the egg. In early stages of oogenesis, grk mRNA is found at the posterior of the oocyte, whereas in the later stages grk mRNA is positioned at the dorsal anterior corner of the oocyte. In order to visualize real time localization and anchorage of endogenous grk mRNA in living oocytes, we have utilized the MS2-MCP system. We show that MCP-GFP tagged endogenous grk mRNA properly localizes within wild-type oocytes and behaves aberrantly in mutant backgrounds. FRAP experiments of localized grk mRNA in egg chambers reveal a difference in the dynamics of grk mRNA between young and older egg chambers. grk mRNA particles, as a population, are highly dynamic molecules that steadily lose their dynamic nature as oogenesis progresses. This difference in dynamics is attenuated in K10 and sqd1 mutants such that mislocalized grk mRNA in older stages is much more dynamic compared to wild-type. In contrast, in flies with compromised dynein activity, properly localized grk mRNA is much more static. Taken together, we have observed the nature of localized grk mRNA in live oocytes and propose that its maintenance changes from a dynamic to a static process as oogenesis progresses.
Keywords: grk, oogenesis, FRAP, RNA localization, Drosophila
Introduction
Asymmetric messenger RNA localization provides a means to spatially restrict protein synthesis. In many cell types on site translation serves to maintain and elaborate a polarized architecture. However, the fundamental questions of how a transcript moves to its destination, how it remains translationally silent while in transit and how it is anchored at its target site remain unanswered for many mRNAs (reviewed in (St Johnston, 2005). In Drosophila, several asymmetrically localized transcripts have been identified in the developing oocyte. Bicoid mRNA is localized at the anterior of the oocyte, whereas oskar can be found spatially restricted at the posterior of the oocyte (Berleth et al., 1988; Ephrussi et al., 1991; Lehmann and Nusslein-Volhard, 1986). These transcripts along with gurken (grk) mRNA are necessary for the patterning of the Drosophila embryo. gurken encodes a Transforming Growth Factor-α (TGFα) like protein, that is secreted by the oocyte and activates the Epidermal Growth Factor Receptor (EGFR) in the overlying somatic follicle cells. In early oogenesis, grk mRNA accumulates at the posterior of the oocyte. Gurken protein, synthesized from this RNA, induces posterior cell fates in the overlying follicle cells. Soon thereafter, during mid to late oogenesis, grk mRNA and protein are localized to the dorsal anterior corner of the oocyte establishing dorsal cell fates in lateral follicle cells (Neuman-Silberberg and Schupbach, 1996; St Johnston, 2005; Van Buskirk and Schupbach, 1999).
Mutations in several genes have been identified that result in mislocalization of grk mRNA. Some of these genes encode heterogeneous nuclear ribonucleoproteins (hnRNPs) such as squid and hrb27c (Goodrich et al., 2004; Neuman-Silberberg and Schupbach, 1993). Others include genes necessary for cytoskeletal integrity such as cappuccino, spire (actin nucleator) and spindle-F (microtubule organizer) (Abdu et al., 2006; Manseau and Schupbach, 1989; Neuman-Silberberg and Schupbach, 1993). In addition, it has been shown that the motor enzymes, kinesin and dynein, are required for correct grk mRNA localization (Brendza et al., 2002; Clark et al., 2007; Delanoue et al., 2007; Duncan and Warrior, 2002; Januschke et al., 2002). These requirements suggest a model where grk mRNA is assembled into an RNP complex that is transported on filaments by molecular motors. In fact, in vitro synthesized grk mRNA injected during mid-oogenesis assembles into nonmembranous transport particles that first move to the anterior of the oocyte and then dorsally toward the nucleus, and these movements are dependent on dynein. Furthermore, microtubule depolymerization disrupts the directionality of injected grk mRNA implying active transport along microtubules (Clark et al., 2007; Delanoue et al., 2007; MacDougall et al., 2003). However, little is known about how grk mRNA is maintained or anchored at its destination. Recent ultrastructural analysis suggest that grk mRNA is not only transported, but also anchored by dynein to large cytoplasmic structures called sponge bodies at the dorsal-anterior corner (Delanoue et al., 2007).
We have used a system for fluorescent labeling of mRNA in vivo to visualize real time localization and anchoring of endogenous grk mRNA in living oocytes. We show that GFP-labeled endogenous grk mRNA properly localizes within wild-type oocytes and behaves aberrantly in mutant backgrounds. Interestingly, pharmacological studies show that the anchoring of grk mRNA may not be dependent on an intact cytoskeleton, or alternatively, it may utilize a subpopulation of microtubules that are very drug-resistant. Fluorescence Recovery After Photobleaching (FRAP) experiments of grk mRNA particles in live egg chambers reveal a difference in the dynamic state of localized grk mRNA between early and mid-stage egg chambers. grk mRNA particles, as a population, are highly dynamic molecules, exhibiting high fluorescence recovery, during early stages. As oogenesis progresses, grk mRNA steadily loses its dynamic nature. This difference in grk mRNA mobility is attenuated in K10 and sqd1 mutants such that mislocalized grk mRNA in older stages remains much more dynamic. In contrast, in flies with compromised dynein activity, properly localized grk mRNA is more static. Taken together, we have observed the changing nature of localized grk mRNA in live oocytes and have demonstrated that its maintenance can be a dynamic process. We show that localized grk mRNA, as a population, can be highly dynamic or static, depending on the stage of oogenesis. Investigating the changing nature of grk mRNA allows us to gain insight into the mechanisms involved in the maintenance of localized transcripts.
Results
In vivo labeling of gurken mRNA in egg chambers
In order to visualize grk mRNA in live egg chambers we took advantage of a system for fluorescent labeling of mRNA in vivo. We have utilized the MS2-MS2 Coat Protein (MS2-MCP) system pioneered by Singer and colleagues in yeast (Bertrand et al., 1998) and adapted for Drosophila by Gavis and colleagues (Forrest and Gavis, 2003; Weil et al., 2006). Twelve stem-loop binding sites for MCP were inserted into the 3′UTR of grk mRNA. The grk-(MS2)12 transgene rescues the grk2B mutant phenotype indicating that the stem loops do not significantly interfere with the function of the RNA. grk-(MS2)12 was coexpressed with either the hsp83-MCP-GFP or the hsp83-MCP-RFP transgene, which encode MCP fused to either GFP or RFP (Weil et al., 2006). Eggs from flies expressing both transgenes, grk-(MS2)12 and hsp83-MCP-GFP/RFP, (referred to as grk*GFP and grk*RFP) show no obvious alterations in morphology. Moreover, the labeled RNAs, grk*GFP and grk*RFP, have identical localization patterns to wild-type grk mRNA.
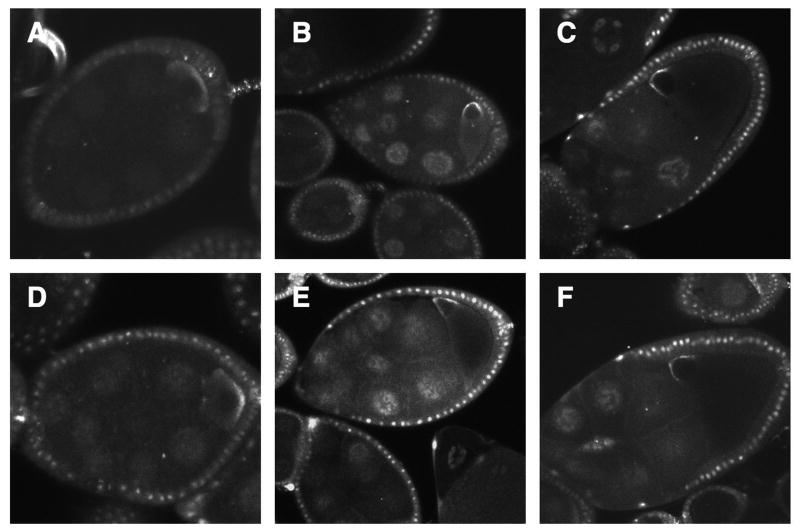
As previously described, grk mRNA is expressed in early egg chambers (stages 1-7) and localized to the posterior end of the oocyte. Starting at stage 8, the oocyte nucleus moves from the center to the anterior edge of the oocyte. As the egg chamber grows, this edge will become the future dorsal anterior corner. During this stage grk mRNA begins to accumulate around the new position of the nucleus as well as the anterior of the oocyte creating a transient cortical ring. From stages 9 to 10B grk mRNA remains in a tight association with the dorsal anterior corner, forming a cap over the nucleus (Neuman-Silberberg and Schupbach, 1993). We used laser scanning confocal microscopy for live imaging of grk*GFP/RFP during oogenesis (Fig. 1). Egg chambers were dissected and kept within a culture dish with nutrient media and then immediately imaged. Throughout early stages 4-7, grk*GFP/RFP can be seen lining the posterior of the oocyte (Fig. 1A). During stage 7, when the oocyte nucleus is centered or shifting to one side, live images of grk*GFP show it to accumulate around the anterior cortex as well as continuing to line the posterior. At stage 8, the oocyte nucleus moves entirely to one side. As soon as the nucleus is asymmetric the amount of grk*GFP at the posterior and ventral lateral sides of the oocyte decreases and instead grk*GFP appears to line the lateral wall posterior to the nucleus (Fig. 1B). At this stage, the dorsal-anterior cap of grk*GFP above the nucleus becomes increasingly prominent. While stage 8 progresses, grk*GFP continues to be visible anteriorly in a prominent ring, meanwhile little to no grk*GFP can be seen at the posterior. As the egg chamber develops through stage 9, less and less grk*RFP accumulates in the ring whereas grk*RFP in the ventral-anterior corner persists a bit longer, also seen by MacDougall et al., 2003 by fluorescent in situ hybridization. During this time, grk*RFP becomes more prominently localized to the dorsal-anterior cap above the nucleus. From late stage 9 to 10B grk*RFP continues to be found above the nucleus and has a fainter appearance.
1. Live imaging of endogenous grk mRNA.
(A-C) grk*GFP expressed in the younger stages of mid-oogenesis. (A) Stage 7 shows posterior accumulation. (B) In early stage 8 there is often a lateral-cortical lining of grk*GFP on the side of the oocyte nucleus (asterisk). (C) During late 8 there is no longer a posterior lining of grk*GFP but instead the beginnings of an anterior ring with significant accumulation at the ventral anterior. (D-E) grk*RFP expressed in older stages of mid-oogenesis. From stages 9 to 10 there is a prominent dorsal-anterior cap and a strong reduction of grk*RFP at the ventral-anterior corner. (F) Control line expressing MCP-RFP alone. The MCP-GFP/RFP protein has a nuclear localization sequence, as a consequence when no grk mRNA with stem loops are present, the protein will enter the follicle cell nuclei (arrowhead) and nurse cell nuclei (arrow). (A-F) White scale bars represent 50 μm.
Actin destabilizing drugs do not disrupt the maintenance of localized grk mRNA in live egg chambers
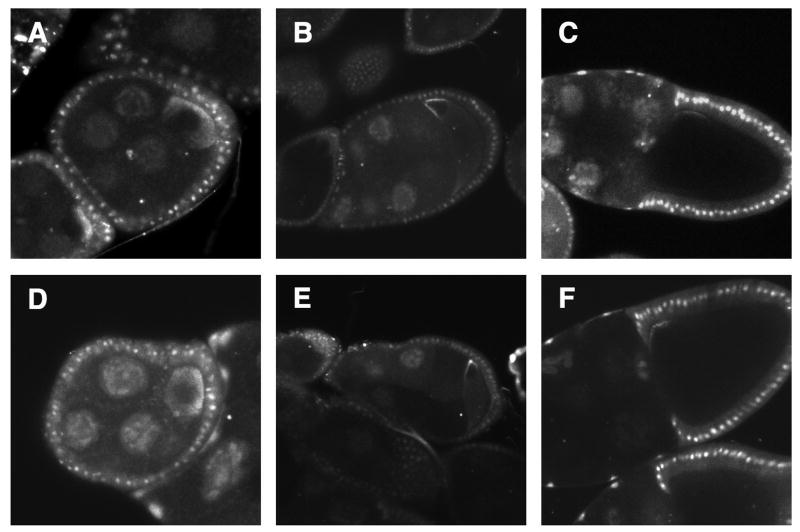
An intact actin cytoskeleton has been shown to be important for asymmetric RNA and protein localization in Drosophila oocytes, reviewed in (Hudson and Cooley, 2002). Treatment with actin destabilizing drugs, like cytochalasin D, disrupts the anchoring of nanos and bicoid mRNA (Forrest and Gavis, 2003; Weil et al., 2006). Also, elimination or reduction of actin binding proteins, such as moesin and tropomyosin, disrupts the posterior maintenance of Osk protein and mRNA (Jankovics et al., 2002; Polesello et al., 2002). In order to investigate whether grk mRNA requires an intact actin framework, we treated live egg chambers expressing grk*RFP with latrunculin A and cytochalasin D, either individually or in combination. To test the effectiveness of these drugs, we stained egg chambers with phalloidin, which revealed very reduced, barely detectable F-actin, except for prominent ring canals. The drug concentrations that we used were capable of blocking nurse cell dumping, which results from actin-dependent contractions of the nurse cells (Gutzeit, 1982). Time lapse imaging of egg chambers from drug treated GFP-actin flies show rapid depolymerization. We were therefore confident that actin was severely disrupted. Interestingly, we found that grk*RFP is well maintained during stages 6-10 (Fig. 2). grk*RFP remains tightly anchored both early at the posterior and later at the dorsal-anterior corner and this anchoring is disrupted only when drug concentrations are high enough to cause the oocyte to collapse. In these cases grk*RFP spreads along the cortex of the oocyte. Additionally, we fed adult female grk*RFP flies latrunculin A and cytochalasin D and observed grk*RFP properly localized (data not shown). Similarly, Saunders and Cohen, 1999 saw no changes in grk transcript localization from cytochalasin D fed flies.
2. Actin is not necessary for anchorage of grk mRNA.
(A-C) Dissected live grk*RFP egg chambers treated with DMSO, the solvent used to dissolve the actin destailizing drugs. grk*RFP is localized properly from young to late stages. (D-F) Egg chambers were treated with the actin destabilizing drugs, cytochalasin D and latrunculin A for 45-60 minutes. There is no obvious disruption of grk*RFP.
Resistant microtubules may explain why grk mRNA remains localized after using microtubule destabilizing drugs
Microtubules are central for the localization of several mRNAs in Drosophila oocytes (St Johnston, 2005). With respect to maintenance, disruption of microtubules has been shown to release bicoid, fs(1)K10, orb and Bicaudal-D from the anterior of the oocyte (Pokrywka and Stephenson, 1995). In order to investigate whether an intact microtubule network is needed to maintain grk mRNA localization, we treated live egg chambers from grk*RFP expressing flies with a combination of colchicine and colcemid, microtubule destabilizing drugs. After drug treatments, grk*RFP localization appeared not to have been disrupted, whether positioned at the posterior, or at the dorsal anterior corner during stages 6-10 (Fig. 3). Time-lapse imaging of drug treated egg chambers over 45-60 minute intervals confirmed these results. These results suggest that anchoring of grk*RFP is not dependent on microtubules, or that the anchoring is meditated by a population of microtubules that are resistant to these inhibitors. To ensure that certain populations of microtubules were indeed disrupted, we monitored microtubule dependent ooplasmic streaming that occurs prior to nurse cell dumping (Gutzeit, 1982). This unidirectional flow ceased within 3 minutes using our treatment of colcemid and colchicine, indicating that the drug treatment was very efficient at disrupting microtubules that promote cytoplasmic streaming. We also imaged drug-treated tau-GFP expressing fly egg chambers. Tau-GFP decorates microtubles and allowed us to follow live depolymerization. Interestingly, while depolymerization of microtubules in the oocyte and nurse cells was plainly evident, the microtubule basket surrounding the oocyte nucleus (previously reported at stage 9 by MacDougall et al., 2003) was clearly resistant to the combined effects of colchicine and colcemid (see movie in Fig. S1). This suggests that there may be different types of microtubules present in the oocyte that have different sensitivities to depolymerizing drugs. Whether grk*RFP is anchored to these resistant microtubules is unclear, but our data clearly shows grk*RFP not disrupted by the treatment.
3. grk*RFP appears to remain unaltered after treatment with MT destabilizing drugs.
(A-C) Dissected live grk*RFP egg chambers treated with ethanol show grk*RFP properly maintained. (D-F) Egg chambers were treated with MT destabilizing drugs, colchicine and colcemid for 45-60 minutes. grk*RFP does not appear to be disrupted in either young and older egg chambers.
We also examined the combined effect of colcemid and colchicine on grk*RFP in egg chambers from females fed with these inhibitors. We observed a range of defects including loss of grk*RFP, mislocalized grk*RFP in the oocyte, and accumulation of grk*RFP in the nurse cells (Fig. S2). In contrast, it has been reported that there is an accumulation of grk transcripts in the oocytes rather than the nurse cells from colchicine-fed flies (Saunders and Cohen, 1999). The combination of two inhibitors used in our experiments may account for the differences. Nevertheless, under these conditions, it is difficult to distinguish between the roles microtubules may play in mRNA transport versus anchoring within the oocyte.
grk mRNA is a dynamic molecule
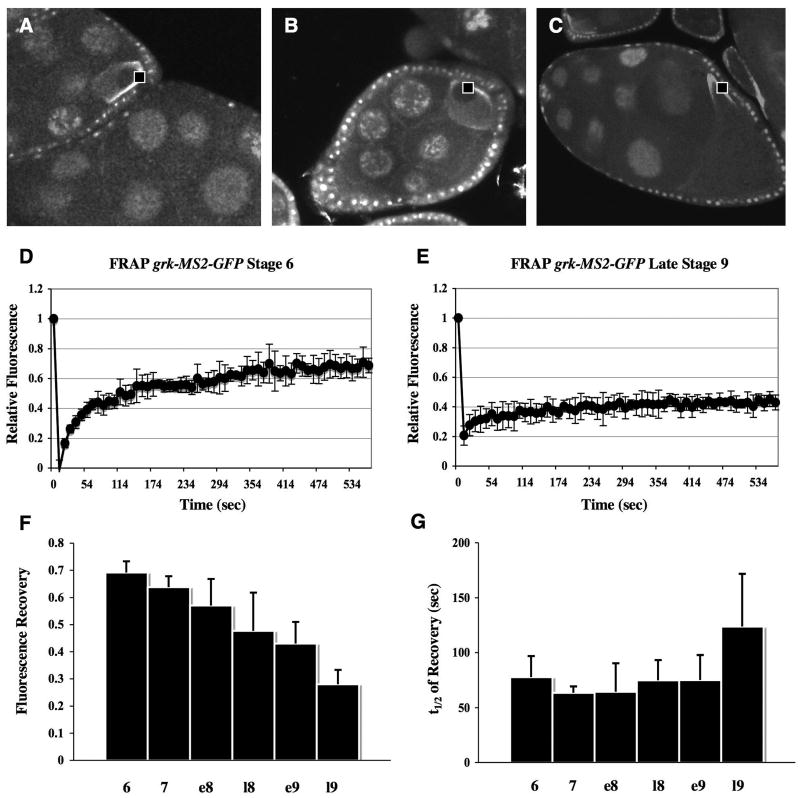
In some cases, mRNAs are stably anchored at their destinations. For example, nanos mRNA diffuses to the posterior of the oocyte and becomes stably anchored in an actin-dependent manner (Forrest and Gavis, 2003). Dynein acts as a static anchor for the apical transcripts, runt and fushi tarazu, in Drosophila blastoderm embryo (Delanoue and Davis, 2005). However, recent work indicates that the maintenance of localized mRNA may also be a dynamic process. bicoid mRNA is maintained at the anterior oocyte cortex by continual active transport on microtubules (Weil et al., 2006). To examine whether grk mRNA is stably anchored during different stages of oogenesis, we performed Fluorescence Recovery After Photobleaching (FRAP) experiments. grk*GFP was photobleached, either in the region posterior to the oocyte nucleus during stages 6 and 7, or on the dorsal-lateral side of the cap above the oocyte nucleus during stages 8 and 9 (see movies in Fig. S4-6). Our FRAP conditions cause photobleaching of a 2.5μm2 area of grk*GFP in the focal plane, as well as grk*GFP as far out of focus as 2-3 μm above and below this plane (Fig. S3D-F). Quantitative analysis shows 70% recovery of fluorescence with a half time of about 80 seconds (t1/2 = τ*ln2, where τ is the time constant of recovery), during stage 6. In contrast, by late stage 9, recovery is reduced to 30% with a t1/2 of over 120 seconds (Fig. 4). Thus, the steady state population of grk*GFP is highly dynamic during early stages of oogenesis, but this dynamic behavior subsides at later stages.
4. grk mRNA is a dynamic molecule.
FRAP experiments were performed on grk*GFP egg chambers stages 6-9. (A-C) Images of grk*GFP egg chambers stages 6-9 with a small box representing the area that is bleached during a FRAP experiment. (D-E) Line graphs representing the average relative fluorescence of grk*GFP during several FRAP experiments. (D) During stage 6, a defined area of fluorescent grk*GFP at the posterior oocyte was bleached and about 70% was recovered. (E) grk*GFP localized at the dorsal-anterior cap during late stage 9 was bleached and had reduced recovery. (F) Bar graph representing the percent fluorescence recovery from stages 6 to late stage 9. There is a steady decrease in the amount of recovery as the oocyte progresses through development. (G) Bar graph representing the half time recovery (t1/2) for stages 6 to late 9. Although recovery steadily decreased, the rate did not, suggesting there could be a similar mechanism occurring during stages 6 to early 9. A minimum of five FRAP experiments were taken for each data point. All error bars represent standard deviations.
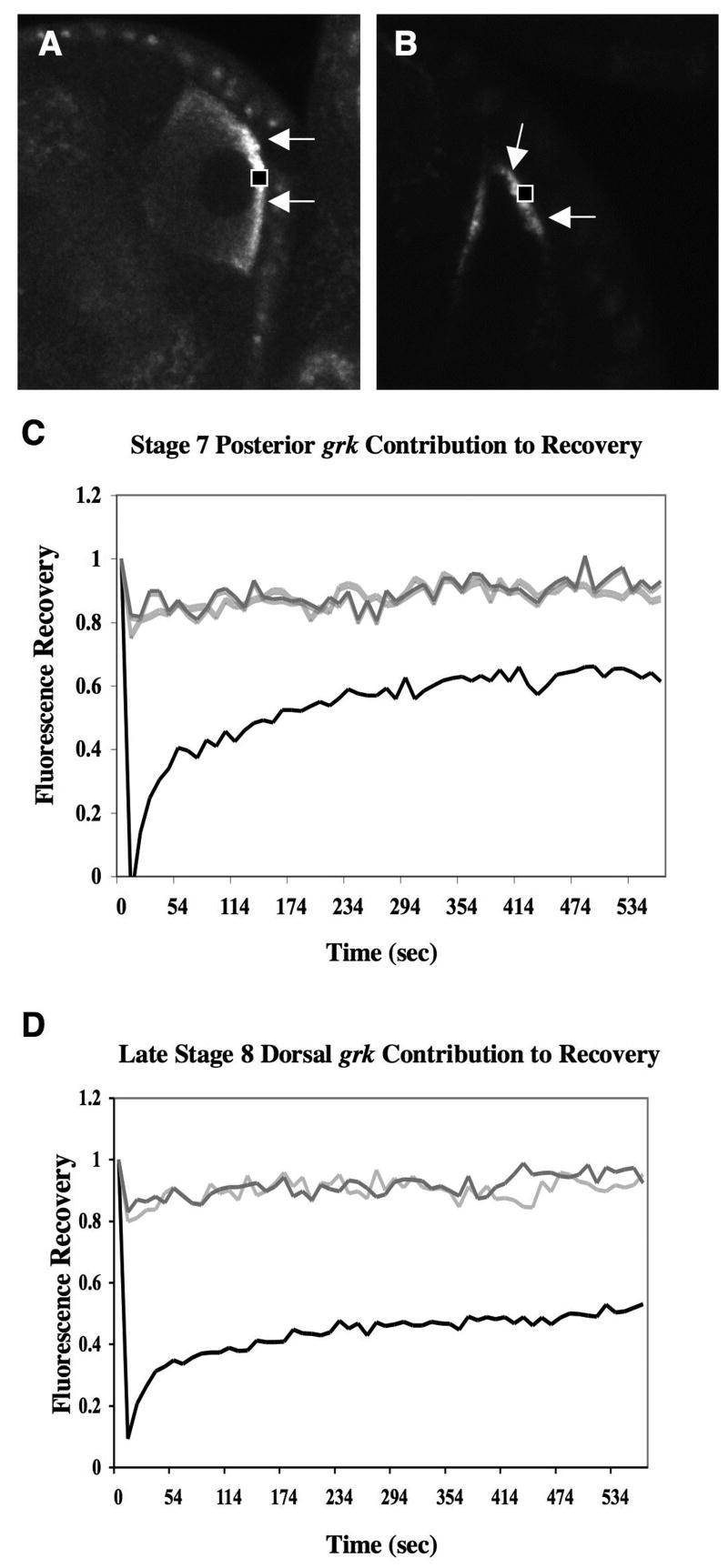
What is the source of fluorescence grk mRNA recovery? Fluorescence recovery could result from lateral movement of grk*GFP that is already localized. To address this possibility, we measured the fluorescence intensity values of the neighboring pools of localized non-bleached grk*GFP that surround the bleached area during a FRAP experiment for a corresponding decrease in fluorescence. Fluorescence of these neighboring regions was not decreased, however, implying that recovery does not primarily involve prelocalized sources of grk*GFP present at the cortex (Fig. 5). In addition, we performed inverse FRAP experiments on posterior localized grk*GFP. We bleached two areas of cortical grk*GFP and measured the fluorescence intensity of the localized, non-bleached grk*GFP in between the two bleached spots. In these cases, there was no appreciable loss of fluorescence to the non-bleached localized grk*GFP, while high fluorescence recovery occurred within the neighboring bleached spots (data not shown). Unless constant and rapid accumulation of new cytoplasmic grk*GFP obscures the fluorescence loss from the cortex, our results indicate that recovery of fluorescence is due to movement of grk*GFP from the cytoplasm to the cortex. To determine whether our fast fluorescent recovery is due to newly synthesized grk*GFP from the nurse cells, we bleached the entire oocyte at stage 7. We observed partial grk*GFP recovery within 20 minutes, more than 10 times longer than recovery observed after bleaching the cortex only (data not shown). Thus, we conclude that at stage 7 mRNA newly transported from the nurse cells cannot be the major source of cortical recovery, instead recovery most likely reflects exchange from the nearby cytoplasm.
5. grk mRNA recovery does not specifically involve depletion of nearby localized sources.
(A-B) Images of stage 7 (A) and late 8 (B) egg chambers from grkHGFP expressing flies illustrating the region bleached during the FRAP experiment (black box), plus arrows pointing to the unbleached areas of grk*GFP monitored for fluorescence loss. (C-D) Line graphs representing average relative fluorescence of grk*GFP and two unbleached neighboring pools of grk*GFP during FRAP experiments. In (C) the two posterior pools that surrounded the bleached region of grk*GFP during stage 7 show no dramatic decrease in fluorescence intensity. The pool on the right of the bleached region is represented by the dark gray line, whereas the pool on the left is the light gray line. (D) Likewise, the surrounding sources of localized grk*GFP found at the dorsal-anterior cap during late stage 8 also had no measurable decrease as recovery occurred (right-dark gray and left-light gray). This suggests that recovery is not due to nearby localized sources of grk*GFP.
Mislocalized grk mRNA in K10 and sqd1 mutant oocytes is dynamic
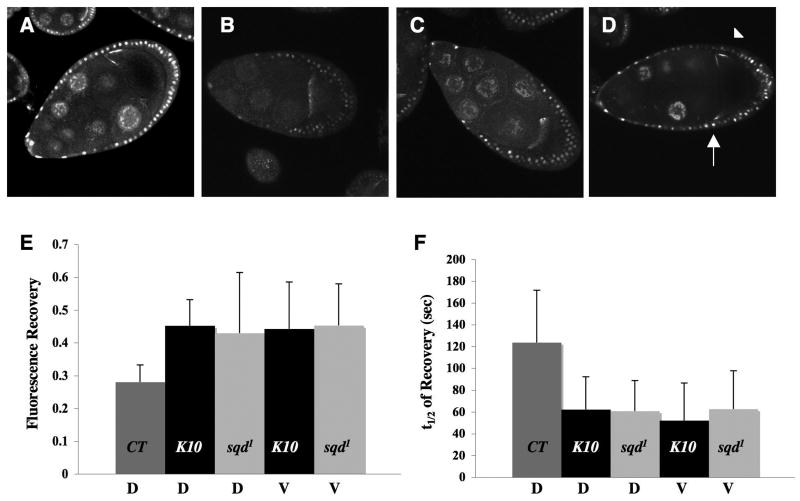
We considered the possibility that mislocalized grkHGFP in mutant backgrounds could manifest altered dynamics. In K10 and sqd1 mutants (Kelley, 1993; Wieschaus, 1978) grk mRNA appears to have normal localization patterns during younger stages of oogenesis, but in mid to late stages grk mRNA is mislocalized along the entire anterior circumference of the oocyte instead of being restricted to the dorsal-anterior corner (Neuman-Silberberg and Schupbach, 1993). In addition, in these mutants grk mRNA is translated along the anterior cortex, resulting in ectopic EGFR activation. Excess receptor activation results in the induction of more dorsal cell fates leading to expanded dorsal appendages on the egg (Kelley, 1993; Neuman-Silberberg and Schupbach, 1993; Neuman-Silberberg and Schupbach, 1994; Norvell et al., 1999). grk*GFP reproducibly shows a similar pattern of localization in either K10 or sqd1 mutant backgrounds (Fig. 6B-D).
6. Mislocalized grk mRNA at the anterior is dynamic.
FRAP experiments were performed on K10; grk*GFP and grk*GFP; sqd1 egg chambers. Images of grk*GFP (A), K10; grk*GFP (B,D) and grk*GFP; sqd1 (C) egg chambers during stage mid-late stage 9. (B-D) Mislocalized grk*GFP forms an anterior ring at the edge of the oocyte. A defined area of fluorescent grk mRNA at the dorsal (arrowhead) and ventral-anterior (arrow) was bleached during FRAP experiments. (E) Previously we noted, grk*GFP localized at the dorsal-anterior cap during mid-late stage 9 had very reduced recovery (∼25% recovery) during FRAP experiments. In contrast, grk*GFP found both at the dorsal and ventral-anterior of K10 and sqd1 mutants are more dynamic (∼45%). (F) Bar graph representing the half time recovery (t1/2) for grk*GFP during mid-late stage 9 from controls and mutant lines. Recovery rates are much faster in the mutants, similar to the younger stages in control line (Figure 4). A minimum of five FRAP experiments were taken for each data point. All error bars represent standard deviations.
To examine the behavior of localized and mislocalized grk mRNA in the K10 and sqd1 mutant oocytes, we performed FRAP experiments. We photobleached the dorsal and ventral anterior corners of K10 and sqd1 (Fig. 6D-F). Interestingly, during mid-late stage 9 egg chambers, we found grk*GFP at both the dorsal and ventral sides to be dynamic. In fact, grk*GFP is more dynamic (∼45% recovery) compared to grk*GFP in a wild-type background (∼25%) at this stage (Fig. 6E and see Fig. S8 for movie). Accordingly, the recovery rates (t1/2) for grk*GFP during mid-late stage 9 egg chambers in the mutant backgrounds are much faster. The nature of grk*GFP dynamics resembles that of a younger stage with faster and higher recovery, significantly different from the control displaying a slow rate with little recovery (Fig. 6F). We also performed FRAP experiments on K10 and sqd1 mutants at stages 6 and 7, when grk mRNA is properly localized at the posterior (see Fig. S9). Surprisingly, we found a substantial amount of variability in K10 mutants and a general increase of recovery in sqd1 mutants.
Compromised dynein activity interferes with the recovery of grk mRNA
Recent evidence for direct binding between grk mRNA and the dynein light chain adds to the mounting support for the potential role of the dynein motor enzyme in grk mRNA localization (Rom et al., 2007). Given the dynamic nature of grk mRNA, we examined the behavior of grk*GFP under conditions that compromise dynein motor activity. First, we overexpressed Dynamitin (Dmn), which is an essential component of the dynactin complex and is necessary for dynein activity (Kamal and Goldstein, 2002). Overexpression of Dmn results in an increase of anteriorly localized grk mRNA during mid-stages of oogenesis, as seen by in situ hybridization, and mispositioning of the oocyte nucleus (Duncan and Warrior, 2002; Januschke et al., 2002). Second, we took advantage of hypomorphic dynein heavy chain alleles (dhc) alleles, which permit oogeneis but have been shown to reduce transport (Clark et al., 2007; Weil et al., 2006).
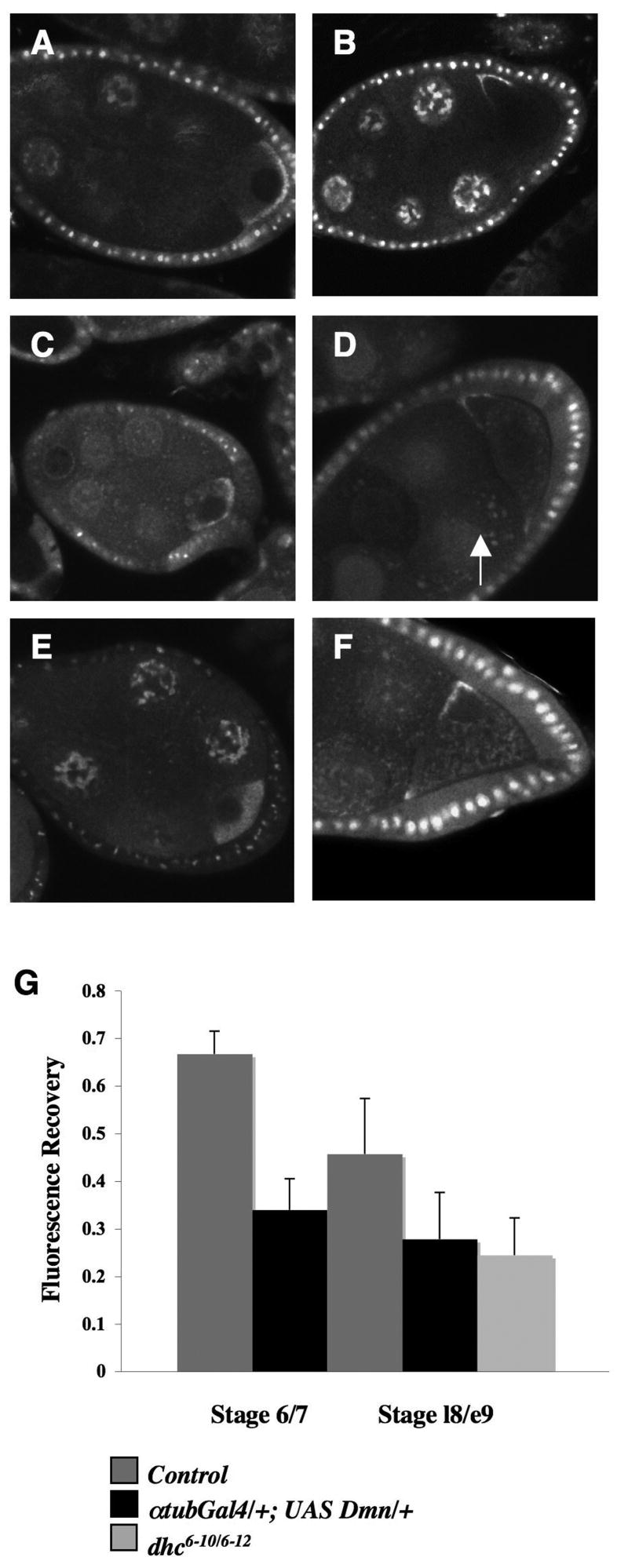
FRAP experiments were performed on localized grk*GFP in oocytes where dynein function was reduced by either of the above methods. In both situations, we observed mislocalized grk*GFP in the oocyte cytoplasm accompanied by large aggregates, from stages 5 to 10. These large aggregates are most clearly visible in the nurse cell cytoplasm and are very remininiscent of the aggregates we observed in colchicine-fed grk*GFP flies (Fig. S2B). Upon Dmn overexpression, a significant portion of grk*GFP is still capable of reaching its proper destination both at the posterior and dorsal-anterior corner (Fig. 7C,D). In Dhc6-10/6-12 egg chambers (Fig. 7E,F), grk*GFP no longer accumulates in a thin crescent along the posterior cortex during stages 6 and 7, and instead is very diffuse throughout the oocyte. During mid-stages of oogenesis there is an accumulation of grk*GFP at the anterior cortex beginning as early as stage 7. A defined area of fluorescent grk mRNA at the posterior and dorsal-anterior was bleached during FRAP experiments. With Dmn overexpression, fluorescence recovery is reduced to 34% in stages 6/7 and 28% in stage late 8/early 9 (l8/e9). Similarly in dynein mutant egg chambers, grk*GFP recovery is reduced to 24% at stage l8/9. This is direct contrast to control fluorescence recovery values of 67% for stage 6/7 and 46% for l8/e9 stages (Fig. 7G). Taken together, our results show that when dynein is compromised by two different methods, we observe a significant reduction in grk mRNA dynamics.
7. Reducing Dynein activity leads to reduced grk mRNA recovery.
FRAP experiments were performed on grk*GFP; αtubGal4/+;UAS-Dmn/+ and grk*GFP; Dhc6-10/6-12 egg chambers. Images of grk*GFP (A,B), grk*GFP; αtubGal4/+;UAS-Dmn/+ egg (C,D) and grk*GFP; Dhc6-10/6-12 (E,F) egg chambers during stages 6/7 (A,C,E) and late 8/early 9 (B,D,F). (A,B) Properly localized grk*GFP lines the posterior cortex of young egg chambers, whereas grk*GFP forms a crescent around the oocyte nucleus at the dorsal anterior corner starting stage 8. In a UAS-Dmn background (C,D), there is mislocalized grk*GFP in the oocyte cytoplasm accompanied by large aggregates that can also be seen in the nurse cells (arrow). Under these conditions, a significant portion of grk*GFP is capable of reaching its destination both at the posterior and dorsal-anterior corner. In hypomorphic alleles (E,F), grk*GFP has a very diffuse localization during young stages and an accumulation at the anterior cortex in later stages. Similar to (C,D) there are aggregates in both the nurse cell and oocyte cytoplasm. A defined area of fluorescent grk at the posterior and dorsal-anterior was bleached during FRAP experiments. (G) In control early stages (6/7) grk*GFP has on average 67% fluorescence recovery and during mid-stages (l8/e9) a 46% recovery during FRAP experiments, in contrast a significant reduction occurs in both young and mid stages when dynein activity is compromised. When Dmn is overexpressed recovery is reduced to 34% in stages 6/7 and 28% in l8/e9. Similarly in the dynein hypomorphs recovery is reduced to 24% in stage l8/e9 egg chambers. A minimum of five FRAP experiments were taken for each data point. All error bars represent standard deviations.
Discussion
In order to visualize grk mRNA in vivo we used the MS2-MCP system. Unlike in situ hybridization or FISH techniques that reveal static images of RNA, the MS2-MCP system has allowed us to observe live samples and film grk mRNA during various stages of oogenesis. Using this new detection method, we have described the live localization patterns of fluorescent grk mRNA. Our description is similar to those originally published (Neuman-Silberberg and Schupbach, 1993) and confirms observations recently published using FISH (MacDougall et al., 2003). We also describe grk mRNA accumulating at the anterior before posterior localization has disappeared. Moreover, in early stage 8 we observe lateral accumulation of grk mRNA that persists on the side of the oocyte where the nucleus is found (the future dorsal side), meanwhile the opposite side (future ventral side) no longer has grk mRNA except for a small accumulation at the anterior cortex. At this transitional stage, large amounts of grk mRNA are lining the entire anterior and dorsal cortex of the oocyte. This configuration eventually resolves to the well defined dorsal-anterior cap, by first restricting grk mRNA on the dorsal side followed by restricting grk mRNA at the anterior. The questions arises as to whether grk mRNA is transported along the lateral wall from the posterior to the dorsal-anterior corner, or whether the lateral RNA decays and new grk mRNA transcripts populate the corner from the nurse cells. Since endogenous grk mRNA does not form large particles that can be resolved and tracked with our imaging system, we are presently not able to address this question. Using photoswitchable fluorescent molecules in combination with the MS2-MCP system could provide some answers to these questions in the future.
The process of RNA localization is likely to involve several of the following steps: RNP assembly, nuclear export, cytoplasmic transport, anchorage, translation and decay (St Johnston, 2005). Stable anchorage versus continuous transport have been proposed as two different mechanisms for the maintenance of mRNAs before translation occurs. Possible anchors can be microfilaments, microtubules, proteins and even noncoding RNAs, like the Xlsirts in Xenopus oocytes (Kloc and Etkin, 2005). Continuous transport can give the appearance of an mRNA having a static anchor, because this dynamic mechanism can result in a net gain of accumulation. In fact, bicoid mRNA has recently been shown to be maintained in late oocytes by continual active transport (Weil et al., 2006). Unexpectedly, it has been demonstrated that Dynein, separate from its motor function, can act as a static anchor for apical transcripts in Drosophila embryos (Delanoue and Davis, 2005). In attempts to study whether the cytoskeleton is a necessary factor for the maintenance of already localized grk transcripts, we used cytoskeletal destabilization drugs. Our results suggest that actin does not play a role in the anchorage. This also corresponds to data obtained by moesin-deficient flies, which have defects in localization of oskar mRNA because of a loose and detached actin network, but in which grk mRNA remains undisrupted (Jankovics et al., 2002; Polesello et al., 2002). Interestingly though, Babu et al., 2004 showed that osk mRNA has two redundant anchoring processes. They noted that latrunculin A treatments did not disrupt the maintenance of osk, unless other proteins, such as Homer, were absent (Babu et al., 2004). It is therefore possible that the actin cytoskeleton is similarly involved in the maintenance of grk mRNA in a redundant manner with other proteins, but so far no actin binding proteins have been found to bind directly to the grk mRNA.
Using microtubule destabilizing drugs, we see no obvious disruption of localized grk mRNA throughout stages 6-10. This agrees with injection studies using in vitro synthesized grk mRNA which show that show localized grk mRNA is not disrupted by the later injection of colcemid (MacDougall et al., 2003). Interestingly, we observed the microtubule network around the oocyte nucleus of stage 9-10 egg chambers to be resistant to our drug treatments. It is not uncommon to observe populations of microtubules to be resistant to depolymerizing drugs. Often it is a characteristic of stable versus dynamic populations of microtubules (Baas et al., 1994; Bannigan et al., 2006; Guillaud et al., 1998; Palazzo et al., 2003). These microtubules persist even when nuclear migration is disrupted in grk mutants, or when the nucleus is displaced in various other mutants (Guichet et al., 2001; Januschke et al., 2002). It is possible that grk mRNA may attach itself to these MTs. Moreover, overexpressing Dmn, a component of the dynein-dynactin complex, disrupts this oocyte nucleus microtubule scaffold resulting in grk mRNA dispersed in the oocyte within stage 10 egg chambers (Januschke et al., 2002), very similar to what we focused in our experiments. During stages 9 and 10, when we observed the resistant microtubule basket, our FRAP studies show little fluorescence recovery. During these later stages, grk mRNA therefore appears to have a more stable anchorage and may no longer be in transit, whereas the high percentage and fast rate of recovery during younger stages suggests a high transport phase. The t1/2 is remarkably similar for stages 6-early 9 suggesting a similar mechanism may be occurring during these stages. However, recovery slows down at mid-late stage 9 and is likely to be indicative of a shift in an equilibrium to a more stably anchored grk mRNA. Recovery does not appear to be from prelocalized sources of grk mRNA. It appears to occur instead from nearby cytoplasm. At least at stage 7, the recovery is not due to nurse cells. At later stages the low amount of recovery could represent grk mRNA in transit from the nurse cells (already close to its destination) (Clark et al., 2007) or this could also be explained by continual active transport. Further studies are needed to distinguish between these possibilities.
The difference in dynamics of grk mRNA between young and old oocytes is altered in K10 and sqd1 mutants. In fact, during the later stages, we found an increase in recovery in the mutants suggesting a more dynamic grk mRNA found at the anterior of the oocytes. The rate of recovery also does not slow down and is more representative of grk mRNA in a younger oocyte. It may be that in addition to a defect in the machinery needed for proper localization (Norvell et al., 1999), the transition to a more stable anchorage is impaired in these mutants. In fact, in a recent publication Delanoue et al., (2007) suggest that Squid is a necessary component for the conversion of grk transport particles to a more stable structure. In addition, we found that compromising the activity of dynein by either overexpressing Dmn or using hypomorphic alleles of the dynein heavy chain led to less fluorescence recovery in both young and mid stages. Although we could not directly distinguish the roles of dynein in either long range transport or anchorage, we have demonstrated that when dynein activity is reduced localized grk mRNA is less dynamic.
Using the MS2-MCP system, we have been able to visualize grk mRNA in live egg chambers and begin to dissect the processes involved in the maintenance of localized transcripts. We show that grk mRNA has a remarkably different set of dynamics between young and older egg chambers, we also measured differences in various mutant backgrounds, which allows us to define various stages involved in the process.
Methods
Drosophila Stocks
hsp83-MCP-RFP and hsp83-MCP-GFP transgenic flies have been described (Forrest and Gavis, 2003; Weil et al., 2006). fs(1)K10, sqd1, dhc6-10, dhc6-12 mutant strains were used (Kelley, 1993; Li et al., 1994; Wieschaus, 1978). Antoine Guichet provided the UAS-Dmn flies (Guichet et al., 2001). Overexpression of Dmn was acheived by using the αtubGal4 driver that begins to be active during stage 4 (matαGal4-67). For marker mutations, Gal4 lines and balancer chromosomes see flybase@indiana.edu.
Transgene Construction
A PmlI site was introduced to 1603 bp of the mRNA, within the 3′UTR of grk mRNA. This site is 343 bp after the stop codon. The mutagenesis was performed on a 5050 bp genomic fragment containing the complete grk locus (Queenan et al., 1999) with adjacent 5′ and 3′ sequences within the pBluescript II SK+ phagemid (Stratagene). The pSL-MS2-12 construct contains 12 MS2 stem loops (gift from K. Forrest). We used a mutated version of the stem loop which results in a very strong affinity (Kd= 10-11) of MS2 Coat Protein to the MS2 stem loop (Johansson et al., 1998; Valegard et al., 1997). The 693 bp BamHI-EcoRV fragment from pSL-MS2-12 was end-filled with Klenow (NE Biolabs) and placed into the PmlI site of grk. A XhoI-EagI grk-MS2-12 fragment was cloned into pCasper4 using XhoI-NotI.
P-Element Mediated Transformation
P-element transformation was performed according to standard procedures (Spradling and Rubin, 1982) using an Eppendorf Transjector 5246 with Eppendorf Femtotips (Eppendorf). Transgene constructs were injected at a concentration of 0.4 μg/μl along with the helper plasmid pTurbo at a concentration of 0.1 μg/μl.
Live Imaging and Inhibitor Treatment
Ovaries were dissected in Schneider's Drosophila Medium (GIBCO). Egg chambers were placed in No. 1.5 glass bottom culture dishes (MatTek) with 200 μl of medium. A No. 1.5 micro cover glass (VWR) was cut with a diamond tip to a coverslip of about 4 mm2 and placed on top of the dissected egg chambers. Egg chambers were treated with either colchicine 200 μg/ml and colcemid 100 μg/ml (Sigma) or latrunculin A 4.2 μg/ml and cytochalasin D 100 μg/ml (Sigma) for 45-65 minutes while in the glass bottom dishes.
FRAP Experiments
FRAP experiments were performed using a Zeiss LSM510 confocal microscope with a 40X 1.2NA water objective lense. Imaging was performed with a 488 nm Argon laser. The following conditions gave minimal photobleaching: 70% laser output, 1.5% AOTF, zoom 4, scan speed 6, 10 seconds per frame. For each FRAP experiment 1-3 prebleach images were taken. A defined region of interest (ROI), 2.25 um2 region, was bleached at 80% power with 15 iterations (∼4 seconds). After bleaching, time series were taken every 10 seconds for a total of 10 minutes. Analysis was done using MATLAB and ImageJ (NIH). In order to test for the survival and transport properties of the egg chamber, we bleached the entire oocyte of stage 8 GFP-Spn-F egg chambers (Abdu et al., 2006) and monitored the recovery of Spn-F protein for 20 minutes. GFP-Spn-F is transported from nurse cells to oocyte and under our experimental conditions significant recovery occurs (data not shown).
Feeding Inhibitor Drugs
Flies were starved for 24 hours. After starvation, the flies were placed in a vial for 24 hours containing fly food media topped with a coated layer of yeast and inhibitor drugs (colchicine 200 μg/ml and colcemid 100 μg/ml or latrunculin A 4.2 μg/ml and cytochalasin D 100 μg/ml). Ovaries were then dissected and fixed with 4% paraformaldehyde.
Supplementary Material
Time-lapse movie of microtubules surrounding the oocyte nucleus of a stage 10 egg chamber from tau-GFP flies. The egg chamber was treated with colchicine and colcemid at time 0′ and subsequently filmed for 40 minutes. There appears to be no obvious depolymerization or breakages of the microtubules.
(A) Adult female grk*RFP flies were fed for 24 hours ethanol mixed in with yeast paste. Stage 9 egg chamber shows properly localized grk*RFP at the dorsal anterior corner. (B-C) grk*RFP flies were fed for 24 hours colchicine and colcemid mixed in with yeast paste. (B) Stage 8 egg chamber shows mislocalized grk*RFP in the oocyte and an accumulation of fluorescence in the nurse cells. (C) Stage 9 egg chamber shows a misplaced oocyte nucleus with no surrounding grk*RFP.
(A-C) Images of egg chambers stage 6 (A), early 8 (B) and mid-late 9 (C) from grkHGFP expressing flies that correspond to the FRAP movies found in supplemental figures 4-6. The black boxes cover the region that is bleached during the FRAP experiments. Panels (D-G) show images of grk*GFP at the dorsal anterior corner (D) pre bleached, (E) post bleached, (F) 2 μm up the plane of focus and (G) 2μm down the plane of focus. A white box outlines the area bleached (OON – oocyte nucleus).
Time-lapse FRAP movie of a stage 6 from grk*GFP flies under the conditions used for quantification found in the Methods section. Movie corresponds to Fig. S3A.
Time-lapse FRAP movie of a early stage 8 egg chamber from grk*GFP expressing flies. Movie corresponds to Fig. S3B.
Time-lapse FRAP movie of a mid-late stage 9 egg chamber from grk*GFP flies. Movie corresponds to Fig. S3C. As oogenesis progresses the amount of fluorescence recovery decreases.
Image of grk*GFP;sqd1 egg chamber with a black box covering the ventral-anterior area photobleached during FRAP experiments.
Time-lapse FRAP movie from the ventral-anterior corner of a grk*GFP;sqd1 egg chamber showing significant fluorescence recovery of the mislocalized mRNA.
FRAP experiments were performed on stage 6 and 7 egg chambers from K10; grkHGFP and grk*GFP;sqd1. The XY scatter plot shows the data point spread of recovery from the different fly lines. Posterior localized grk*GFP in K10 mutants show a high degree of variability, particularly during stage 6, whereas, the sqd1 mutants show a general increase in recovery during both stages (p=0.004 for sqd1 stage 7). A minimum of five FRAP experiments were taken for each data point. All error bars represent standard deviations
Acknowledgments
We thank Kevin Forrest for technical advice and reagents, Thomas Gregor for help with MATLAB analysis and Antoine Guichet for flies. We also thank the members of the Wieschaus and Schupbach labs for their insightful critiques toward the work involved in the manuscript. This work was supported by the Howard Hughes Medical Institute (A.J. and T.S.) and by a grant from the National Institutes of Health (GM067758) to E. Gavis.
Contributor Information
Angela M. Jaramillo, Howard Hughes Medical Institute, Princeton University, NJ 08544, USA
Timothy T. Weil, Department of Molecular Biology, Princeton University, NJ 08544, USA
Joseph Goodhouse, Department of Molecular Biology, Princeton University, NJ 08544, USA.
Elizabeth R. Gavis, Department of Molecular Biology, Princeton University, NJ 08544, USA
Trudi Schupbach, Howard Hughes Medical Institute, Princeton University, NJ 08544, USA.
References
- Abdu U, Bar D, Schupbach T. spn-F encodes a novel protein that affects oocyte patterning and bristle morphology in Drosophila. Development. 2006;133:1477–84. doi: 10.1242/dev.02319. [DOI] [PubMed] [Google Scholar]
- Baas PW, Pienkowski TP, Cimbalnik KA, Toyama K, Bakalis S, Ahmad FJ, Kosik KS. Tau confers drug stability but not cold stability to microtubules in living cells. J Cell Sci. 1994;107(Pt 1):135–43. doi: 10.1242/jcs.107.1.135. [DOI] [PubMed] [Google Scholar]
- Babu K, Cai Y, Bahri S, Yang X, Chia W. Roles of Bifocal, Homer, and F-actin in anchoring Oskar to the posterior cortex of Drosophila oocytes. Genes Dev. 2004;18:138–43. doi: 10.1101/gad.282604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannigan A, Wiedemeier AM, Williamson RE, Overall RL, Baskin TI. Cortical microtubule arrays lose uniform alignment between cells and are oryzalin resistant in the Arabidopsis mutant, radially swollen 6. Plant Cell Physiol. 2006;47:949–58. doi: 10.1093/pcp/pcj067. [DOI] [PubMed] [Google Scholar]
- Berleth T, Burri M, Thoma G, Bopp D, Richstein S, Frigerio G, Noll M, Nusslein-Volhard C. The role of localization of bicoid RNA in organizing the anterior pattern of the Drosophila embryo. Embo J. 1988;7:1749–56. doi: 10.1002/j.1460-2075.1988.tb03004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH, Long RM. Localization of ASH1 mRNA particles in living yeast. Mol Cell. 1998;2:437–45. doi: 10.1016/s1097-2765(00)80143-4. [DOI] [PubMed] [Google Scholar]
- Brendza RP, Serbus LR, Saxton WM, Duffy JB. Posterior localization of dynein and dorsal-ventral axis formation depend on kinesin in Drosophila oocytes. Curr Biol. 2002;12:1541–5. doi: 10.1016/s0960-9822(02)01108-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark A, Meignin C, Davis I. A Dynein-dependent shortcut rapidly delivers axis determination transcripts into the Drosophila oocyte. Development. 2007;134:1955–65. doi: 10.1242/dev.02832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delanoue R, Davis I. Dynein anchors its mRNA cargo after apical transport in the Drosophila blastoderm embryo. Cell. 2005;122:97–106. doi: 10.1016/j.cell.2005.04.033. [DOI] [PubMed] [Google Scholar]
- Delanoue R, Herpers B, Soetaert J, Davis I, Rabouille C. Drosophila Squid/hnRNP helps Dynein switch from a gurken mRNA transport motor to an ultrastructural static anchor in sponge bodies. Dev Cell. 2007;13:523–38. doi: 10.1016/j.devcel.2007.08.022. [DOI] [PubMed] [Google Scholar]
- Duncan JE, Warrior R. The cytoplasmic dynein and kinesin motors have interdependent roles in patterning the Drosophila oocyte. Curr Biol. 2002;12:1982–91. doi: 10.1016/s0960-9822(02)01303-9. [DOI] [PubMed] [Google Scholar]
- Ephrussi A, Dickinson LK, Lehmann R. Oskar organizes the germ plasm and directs localization of the posterior determinant nanos. Cell. 1991;66:37–50. doi: 10.1016/0092-8674(91)90137-n. [DOI] [PubMed] [Google Scholar]
- Forrest KM, Gavis ER. Live imaging of endogenous RNA reveals a diffusion and entrapment mechanism for nanos mRNA localization in Drosophila. Curr Biol. 2003;13:1159–68. doi: 10.1016/s0960-9822(03)00451-2. [DOI] [PubMed] [Google Scholar]
- Goodrich JS, Clouse KN, Schupbach T. Hrb27C, Sqd and Otu cooperatively regulate gurken RNA localization and mediate nurse cell chromosome dispersion in Drosophila oogenesis. Development. 2004;131:1949–58. doi: 10.1242/dev.01078. [DOI] [PubMed] [Google Scholar]
- Guichet A, Peri F, Roth S. Stable anterior anchoring of the oocyte nucleus is required to establish dorsoventral polarity of the Drosophila egg. Dev Biol. 2001;237:93–106. doi: 10.1006/dbio.2001.0354. [DOI] [PubMed] [Google Scholar]
- Guillaud L, Bosc C, Fourest-Lieuvin A, Denarier E, Pirollet F, Lafanechere L, Job D. STOP proteins are responsible for the high degree of microtubule stabilization observed in neuronal cells. J Cell Biol. 1998;142:167–79. doi: 10.1083/jcb.142.1.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutzeit HO. Time-lapse film analysis of cytoplasmic streaming during late oogenesis of Drosophila. J Embryol Exp Morphol. 1982:101–111. [Google Scholar]
- Hudson AM, Cooley L. Understanding the function of actin-binding proteins through genetic analysis of Drosophila oogenesis. Annu Rev Genet. 2002;36:455–88. doi: 10.1146/annurev.genet.36.052802.114101. [DOI] [PubMed] [Google Scholar]
- Jankovics F, Sinka R, Lukacsovich T, Erdelyi M. MOESIN crosslinks actin and cell membrane in Drosophila oocytes and is required for OSKAR anchoring. Curr Biol. 2002;12:2060–5. doi: 10.1016/s0960-9822(02)01256-3. [DOI] [PubMed] [Google Scholar]
- Januschke J, Gervais L, Dass S, Kaltschmidt JA, Lopez-Schier H, St Johnston D, Brand AH, Roth S, Guichet A. Polar transport in the Drosophila oocyte requires Dynein and Kinesin I cooperation. Curr Biol. 2002;12:1971–81. doi: 10.1016/s0960-9822(02)01302-7. [DOI] [PubMed] [Google Scholar]
- Johansson HE, Dertinger D, LeCuyer KA, Behlen LS, Greef CH, Uhlenbeck OC. A thermodynamic analysis of the sequence-specific binding of RNA by bacteriophage MS2 coat protein. Proc Natl Acad Sci U S A. 1998;95:9244–9. doi: 10.1073/pnas.95.16.9244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamal A, Goldstein LS. Principles of cargo attachment to cytoplasmic motor proteins. Curr Opin Cell Biol. 2002;14:63–8. doi: 10.1016/s0955-0674(01)00295-2. [DOI] [PubMed] [Google Scholar]
- Kelley RL. Initial organization of the Drosophila dorsoventral axis depends on an RNA-binding protein encoded by the squid gene. Genes Dev. 1993;7:948–60. doi: 10.1101/gad.7.6.948. [DOI] [PubMed] [Google Scholar]
- Kloc M, Etkin LD. RNA localization mechanisms in oocytes. J Cell Sci. 2005;118:269–82. doi: 10.1242/jcs.01637. [DOI] [PubMed] [Google Scholar]
- Lehmann R, Nusslein-Volhard C. Abdominal segmentation, pole cell formation, and embryonic polarity require the localized activity of oskar, a maternal gene in Drosophila. Cell. 1986;47:141–52. doi: 10.1016/0092-8674(86)90375-2. [DOI] [PubMed] [Google Scholar]
- Li M, McGrail M, Serr M, Hays TS. Drosophila cytoplasmic dynein, a microtubule motor that is asymmetrically localized in the oocyte. J Cell Biol. 1994;126:1475–94. doi: 10.1083/jcb.126.6.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDougall N, Clark A, MacDougall E, Davis I. Drosophila gurken (TGFalpha) mRNA localizes as particles that move within the oocyte in two dynein-dependent steps. Dev Cell. 2003;4:307–19. doi: 10.1016/s1534-5807(03)00058-3. [DOI] [PubMed] [Google Scholar]
- Manseau LJ, Schupbach T. cappuccino and spire: two unique maternal-effect loci required for both the anteroposterior and dorsoventral patterns of the Drosophila embryo. Genes Dev. 1989;3:1437–52. doi: 10.1101/gad.3.9.1437. [DOI] [PubMed] [Google Scholar]
- Neuman-Silberberg FS, Schupbach T. The Drosophila dorsoventral patterning gene gurken produces a dorsally localized RNA and encodes a TGF alpha-like protein. Cell. 1993;75:165–74. [PubMed] [Google Scholar]
- Neuman-Silberberg FS, Schupbach T. Dorsoventral axis formation in Drosophila depends on the correct dosage of the gene gurken. Development. 1994;120:2457–63. doi: 10.1242/dev.120.9.2457. [DOI] [PubMed] [Google Scholar]
- Neuman-Silberberg FS, Schupbach T. The Drosophila TGF-alpha-like protein Gurken: expression and cellular localization during Drosophila oogenesis. Mech Dev. 1996;59:105–13. doi: 10.1016/0925-4773(96)00567-9. [DOI] [PubMed] [Google Scholar]
- Norvell A, Kelley RL, Wehr K, Schupbach T. Specific isoforms of squid, a Drosophila hnRNP, perform distinct roles in Gurken localization during oogenesis. Genes Dev. 1999;13:864–76. doi: 10.1101/gad.13.7.864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palazzo A, Ackerman B, Gundersen GG. Cell biology: Tubulin acetylation and cell motility. Nature. 2003;421:230. doi: 10.1038/421230a. [DOI] [PubMed] [Google Scholar]
- Pokrywka NJ, Stephenson EC. Microtubules are a general component of mRNA localization systems in Drosophila oocytes. Dev Biol. 1995;167:363–70. doi: 10.1006/dbio.1995.1030. [DOI] [PubMed] [Google Scholar]
- Polesello C, Delon I, Valenti P, Ferrer P, Payre F. Dmoesin controls actin-based cell shape and polarity during Drosophila melanogaster oogenesis. Nat Cell Biol. 2002;4:782–9. doi: 10.1038/ncb856. [DOI] [PubMed] [Google Scholar]
- Queenan AM, Barcelo G, Van Buskirk C, Schupbach T. The transmembrane region of Gurken is not required for biological activity, but is necessary for transport to the oocyte membrane in Drosophila. Mech Dev. 1999;89:35–42. doi: 10.1016/s0925-4773(99)00196-3. [DOI] [PubMed] [Google Scholar]
- Rom I, Faicevici A, Almog O, Neuman-Silberberg FS. Drosophila Dynein light chain (DDLC1) binds to gurken mRNA and is required for its localization. Biochim Biophys Acta. 2007;1773:1526–33. doi: 10.1016/j.bbamcr.2007.05.005. [DOI] [PubMed] [Google Scholar]
- Saunders C, Cohen RS. The role of oocyte transcription, the 5′UTR, and translation repression and derepression in Drosophila gurken mRNA and protein localization. Mol Cell. 1999;3:43–54. doi: 10.1016/s1097-2765(00)80173-2. [DOI] [PubMed] [Google Scholar]
- Spradling AC, Rubin GM. Transposition of cloned P elements into Drosophila germ line chromosomes. Science. 1982;218:341–7. doi: 10.1126/science.6289435. [DOI] [PubMed] [Google Scholar]
- St Johnston D. Moving messages: the intracellular localization of mRNAs. Nat Rev Mol Cell Biol. 2005;6:363–75. doi: 10.1038/nrm1643. [DOI] [PubMed] [Google Scholar]
- Valegard K, Murray JB, Stonehouse NJ, van den Worm S, Stockley PG, Liljas L. The three-dimensional structures of two complexes between recombinant MS2 capsids and RNA operator fragments reveal sequence-specific protein-RNA interactions. J Mol Biol. 1997;270:724–38. doi: 10.1006/jmbi.1997.1144. [DOI] [PubMed] [Google Scholar]
- Van Buskirk C, Schupbach T. Versatility in signalling: multiple responses to EGF receptor activation during Drosophila oogenesis. Trends Cell Biol. 1999;9:1–4. doi: 10.1016/s0962-8924(98)01413-5. [DOI] [PubMed] [Google Scholar]
- Weil TT, Forrest KM, Gavis ER. Localization of bicoid mRNA in Late Oocytes Is Maintained by Continual Active Transport. Dev Cell. 2006;11:251–62. doi: 10.1016/j.devcel.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Wieschaus E, M JL, Gehring W. fs(1)K10, a germline-dependent female sterile mutation causing abnormal chorion morphology in Drosophila melanogaster. Roux's Arch Dev Biol. 1978;184:75–82. doi: 10.1007/BF00848670. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Time-lapse movie of microtubules surrounding the oocyte nucleus of a stage 10 egg chamber from tau-GFP flies. The egg chamber was treated with colchicine and colcemid at time 0′ and subsequently filmed for 40 minutes. There appears to be no obvious depolymerization or breakages of the microtubules.
(A) Adult female grk*RFP flies were fed for 24 hours ethanol mixed in with yeast paste. Stage 9 egg chamber shows properly localized grk*RFP at the dorsal anterior corner. (B-C) grk*RFP flies were fed for 24 hours colchicine and colcemid mixed in with yeast paste. (B) Stage 8 egg chamber shows mislocalized grk*RFP in the oocyte and an accumulation of fluorescence in the nurse cells. (C) Stage 9 egg chamber shows a misplaced oocyte nucleus with no surrounding grk*RFP.
(A-C) Images of egg chambers stage 6 (A), early 8 (B) and mid-late 9 (C) from grkHGFP expressing flies that correspond to the FRAP movies found in supplemental figures 4-6. The black boxes cover the region that is bleached during the FRAP experiments. Panels (D-G) show images of grk*GFP at the dorsal anterior corner (D) pre bleached, (E) post bleached, (F) 2 μm up the plane of focus and (G) 2μm down the plane of focus. A white box outlines the area bleached (OON – oocyte nucleus).
Time-lapse FRAP movie of a stage 6 from grk*GFP flies under the conditions used for quantification found in the Methods section. Movie corresponds to Fig. S3A.
Time-lapse FRAP movie of a early stage 8 egg chamber from grk*GFP expressing flies. Movie corresponds to Fig. S3B.
Time-lapse FRAP movie of a mid-late stage 9 egg chamber from grk*GFP flies. Movie corresponds to Fig. S3C. As oogenesis progresses the amount of fluorescence recovery decreases.
Image of grk*GFP;sqd1 egg chamber with a black box covering the ventral-anterior area photobleached during FRAP experiments.
Time-lapse FRAP movie from the ventral-anterior corner of a grk*GFP;sqd1 egg chamber showing significant fluorescence recovery of the mislocalized mRNA.
FRAP experiments were performed on stage 6 and 7 egg chambers from K10; grkHGFP and grk*GFP;sqd1. The XY scatter plot shows the data point spread of recovery from the different fly lines. Posterior localized grk*GFP in K10 mutants show a high degree of variability, particularly during stage 6, whereas, the sqd1 mutants show a general increase in recovery during both stages (p=0.004 for sqd1 stage 7). A minimum of five FRAP experiments were taken for each data point. All error bars represent standard deviations