Abstract
We describe a chemical coupling procedure that allows joining of two RNAs, one of which contains a site-specific base analog substitution, in the absence of divalent ions. This method allows incorporation of nucleotide analogs at specific positions even into large, cis-cleaving ribozymes. Using this method we have studied the effects of substitution of G638 in the cleavage site loop of the VS ribozyme with a variety of purine analogs having different functional groups and pKa values. Cleavage rate versus pH profiles combined with kinetic solvent isotope experiments indicate an important role for G638 in proton transfer during the rate-limiting step of the cis-cleavage reaction.
Keywords: ribozyme mechanism, RNA ligation, general acid–base catalysis
INTRODUCTION
The early observation that the small self-cleaving ribozymes (hammerhead, hairpin, hepatitis delta virus [HDV], and Neurospora Varkud satellite [VS]), typically, exhibit their maximal observed cleavage rates in the presence of divalent cations led to the proposal that these ribozymes were inherently metalloenzymes (Pyle 1993). Subsequent experiments showed that these ribozymes are capable of substantial cleavage rates even in the absence of divalent cations (Nesbitt et al. 1997; Young et al. 1997; Murray et al. 1998; Curtis and Bartel 2001; O'Rear et al. 2001; Perrotta and Been 2006; Poon et al. 2006), inspiring a broader search for alternative or additional catalytic strategies. During the past few years evidence has emerged that many ribozymes use general acid–base catalysis to cleave and/or ligate specific phosphoester linkages; however, identifying the exact roles of individual nucleotides in the chemical mechanism is still an ongoing process (Nakano et al. 2000; Pinard et al. 2001; Shih and Been 2001; Bevilacqua 2003; Bevilacqua et al. 2004; Ke et al. 2004; Das and Piccirilli 2005; Fedor and Williamson 2005; Han and Burke 2005; Perrotta et al. 2006; Wilson et al. 2006; Sigel and Pyle 2007).
Nucleotide analogs have proven very useful in investigating the roles of individual base, sugar, and phosphate functional groups in the function of an RNA. Interference approaches, in which an analog is incorporated during in vitro transcription into random positions at low frequency in a population of RNA molecules, have been used to rapidly identify positions important for folding and/or catalysis in a variety of RNAs (Eckstein 1985; Strobel 1999). Other experimental designs require a pure population of RNA molecules containing an analog introduced at one or more specific positions; this is usually accomplished by chemical synthesis (Ogilvie et al. 1988; Scaringe et al. 1990; Wincott et al. 1995). Such site-specific substitution has been used to incorporate analogs with altered functional properties for studies of RNA structure, folding and catalysis (for review, see Baum and Silverman 2007). Due to limitations of chemical synthesis, efficient site-specific incorporation is applicable only to RNAs up to about 60 nucleotides. To study larger RNAs, the short chemically synthesized RNA can sometimes be annealed with other RNAs to reconstitute a complex that is intended to mimic the important aspects of the structure of the larger, natural unimolecular cis RNA. However, the reannealed complex does not always completely recapitulate the properties of the unimolecular RNA.
Another approach to incorporating site-specific analogs into large RNAs involves enzymatically ligating the chemically synthesized RNA to the flanking region(s) of the larger RNA (which has been produced separately, either by chemical synthesis or in vitro transcription) to obtain the desired full-length RNA. Typically, the two RNAs are brought together by a complementary DNA bridging molecule and joined using either T4 DNA ligase or T4 RNA ligase (Moore and Sharp 1992; Moore and Query 2000). Silverman and coworkers recently reported a different approach in which a ribozyme catalyzes ligation of the two RNAs (Baum and Silverman 2007). All of these RNA ligation methods require long incubation times and the presence of Mg2+, conditions that are incompatible with obtaining uncleaved precursor RNA from fast-cleaving ribozymes. In special situations it has been possible to manipulate experimental conditions to allow ribozyme-mediated ligation of the short chemically synthesized oligonucleotide containing the base analog to the rest of the RNA (Hiley et al. 2002); however, a general approach for incorporating nucleotide analogs into uncleaved precursor RNAs of the larger ribozymes has been lacking.
Here we report the development of a method for joining two RNAs in the absence of Mg2+. We use this method to evaluate the effects of purine nucleotide analog substitution at position G638 in the cleavage loop of the VS ribozyme on the kinetics and mechanism of the cis-cleavage reaction.
RESULTS
We have previously found that the fast-cleaving version of the VS ribozyme, designated RG (Fig. 1A), exhibits pH- and D2O-dependent cleavage between pH 5 and 9, consistent with a catalytic mechanism that utilizes general acid–base catalysis (Zamel et al. 2004). A756 in helix VI has been implicated as one probable participant in the chemical step (Lafontaine et al. 2001b, 2002; Hiley et al. 2002; Sood and Collins 2002; Jones and Strobel 2003; Smith and Collins 2007). Previous studies have pointed to G638, in the cleavage loop of stem–loop I, as also being important for cleavage and/or ligation of the VS ribozyme: this nucleotide is strongly conserved in ligation-based in vitro selection experiments (Andersen and Collins 2000); mutation to any other base strongly decreases activity in a trans-cleaving version of the ribozyme (Wilson et al. 2007), and NMR studies of stem–loop I show G638 to be close enough to the scissile bond to potentially affect the chemical step of the reaction (Hoffmann et al. 2003).
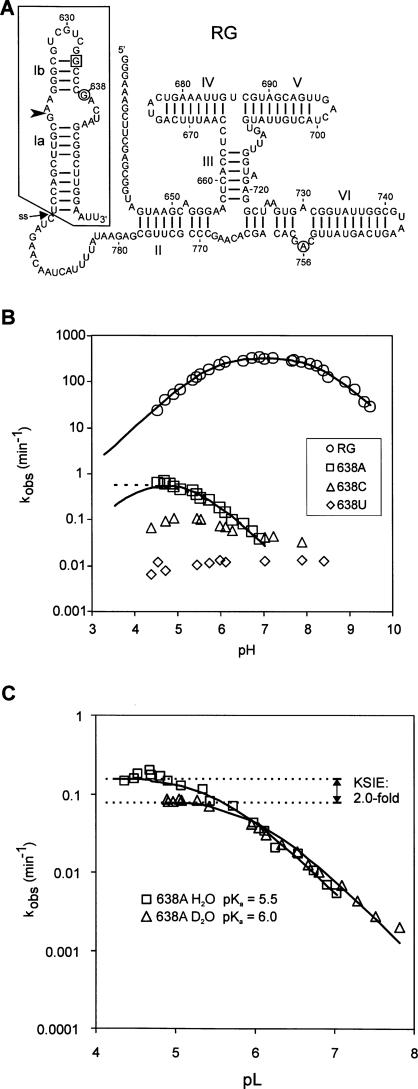
FIGURE 1.
Site-directed mutagenesis of position 638 in the RG version of the VS ribozyme. (A) Secondary structure of RG self-cleaving precursor RNA. The cleavage site is indicated by the arrowhead; the G634C substitution that enforces the active conformation of helix Ib is boxed. The putative nucleobases involved in general acid–base catalysis, G638 and A756, are circled. To incorporate nucleotide analogs at position 638 the boxed region was synthesized chemically and joined via disulfide linkage at the position indicated by the arrow labeled “ss” (see Fig. 3). (B) The effect of pH on the apparent cleavage rate constant (k obs) of wild-type and mutant RNAs with the indicated base substitutions at position 638. Where appropriate, data are fit to a model of general acid–base catalysis (solid line) or general acid-only catalysis (dashed line). Estimates of kinetic parameters are summarized in Table 1. (C) Kinetic solvent isotope effect on cleavage of the 638A mutant. k obs in H2O (squares) or D2O (circles) versus pL (pH or pD, respectively).
Site-directed mutagenesis of position 638
Figure 1B shows the effect of mutation of G638 to each of the other natural nucleobases on the pH-rate profile of the cis-cleaving RG version of the VS ribozyme. The 638C and 638U mutants cleave very slowly and are unaffected by pH; these mutants were not investigated further. In contrast, the 638A mutant exhibits a distinctive rate versus pH curve that could be interpreted in at least two ways. The simplest interpretation is that this curve describes a reaction catalyzed only by a general acid with an apparent pKa of 5.5 (Fig. 1B, dashed line), i.e., mutation of G638 to adenosine eliminates the contribution of the general base. Alternatively, the data could be fit to a general acid–base model with one apparent pKa of 5.3 and the other shifted below the pH range in which we had collected data (<4.5) (Fig. 1B, solid line). A kinetic solvent isotope effect (KSIE) of twofold on the plateau of the curve and a rightward shift of 0.5 units in the rate versus pL curve for reactions performed in D2O provide evidence that proton transfer is still the rate-limiting step in the 638A mutant (Fig. 1C). As with the wild type (Smith and Collins 2007), the magnitude of the KSIE is too small for proton inventory experiments to convincingly distinguish between transfer of one or two protons in the rate-limiting step (data not shown). Taken together, these data indicate that the identity of the nucleobase at position 638 strongly influences the rate-limiting proton transfer step of the cleavage reaction.
Cleavage activity decreases sharply below pH 4
In an attempt to distinguish between a general acid–base and general acid-only mechanism in the 638A mutant, we measured cleavage rates of wild type and 638A at even lower pH. The data in Figure 2A show that below about pH 4 the rate versus pH curves of both RNAs decrease extremely steeply, indicative of the involvement of an additional process in the overall observed cleavage reaction (see Discussion). Because the steep slope of this part of the rate versus pH curve was likely to have an impact on our ability to accurately estimate apparent pKa s in the lower pH region of the bell-shaped portion of the curve, we constructed a mathematical model to describe the rapid decrease in rate at low pH (see Materials and Methods). A steep decrease in kobs versus pH could be explained by (1) an “independent sites” model in which a ribozyme molecule is inactivated by independent protonation of any one of several nucleobases (Knitt and Herschlag 1996) or (2) a “cooperative crash” model in which protonation of one nucleobase increases either the probability of protonation of another nucleobase or the probability that such protonation will lead to loss of activity. In these models, the active and inactive states of the RNA are assumed to be in rapid equilibrium relative to the intrinsic rate of the cleavage step, k 1. Each model postulates some number, n, of relevant nucleobases, each with an average apparent pKa, pKaC, which when protonated causes loss of activity; however, the predicted shapes of the rate versus pH curves for each model are fundamentally different for the two models (Fig. 2B,C).
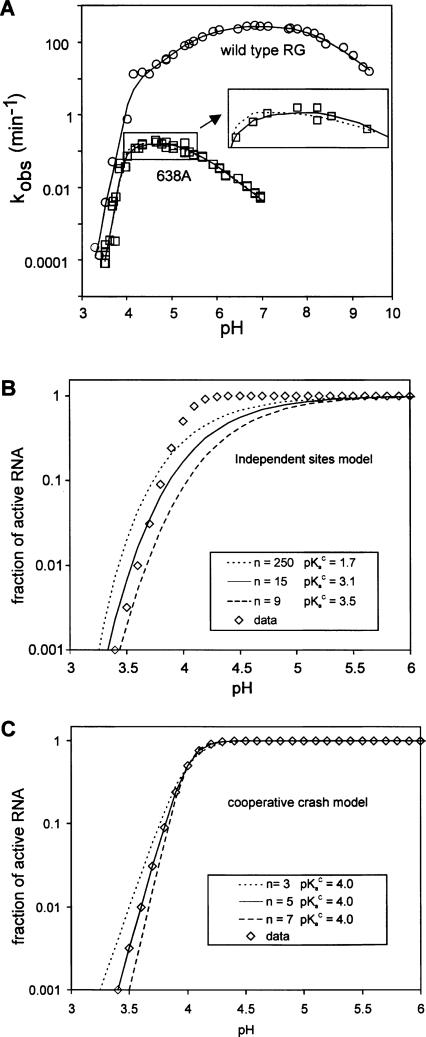
FIGURE 2.
Steep loss of self-cleavage activity at low pH. (A) The effect of low pH on k obs for wild type (circles) or G638A (squares). Data are fit to a model that combines the cooperative crash with general acid–base catalysis (solid lines) or with general acid-only catalysis (dotted line); see Materials and Methods. The inset is an enlargement with some data points removed to allow visualization of the two different fit lines. (B,C) Simulations showing the difference in shapes of the curves and the effects of parameter values for n and pKaC for the independent sites (B) or cooperative crash (C) models (Equations 4 and 5 in Materials and Methods). Data (diamonds) were simulated with the cooperative crash model (n = 5, pKaC = 4.0; where n represents the number of nucleobases, each with an apparent pKa, pKaC, that must be protonated to inactivate the ribozyme) and fit to Equation 4 or 5. Solid lines show the best fit solution using each model; additional lines are simulations with the specified values for n and pKaC.
We fit the wild-type rate versus pH data to equations describing the standard general acid–base model (Equation 4) combined with either the independent or cooperative models (Equation 6 or 7). The independent sites model did not qualitatively fit the data, even with extreme, biologically unreasonable parameter values (data not shown). In contrast, the cooperative model produced a good fit with optimal values of n = 5 and pKaC = 4.1 (Fig. 2A, solid line). Slightly higher or lower values of pKaC (±∼0.1 pH units) also fit the data almost equally well if the value of n was adjusted to compensate (±∼0.5). This analysis suggests that the wild-type ribozyme is rate-limited by general acid–base catalysis in the pH range from 4.5 to 9, and rapidly loses activity at lower pH due to a phenomenon resulting from cooperative protonation of approximately five bases with apparent pKa ≈ 4.
A similar analysis of the rate versus pH curve for 638A also showed that the steep loss in activity below pH 4 did not fit to the independent sites model (not shown) but did fit to the cooperative crash model. Combining the cooperative crash model with a general acid-only model is sufficient to explain the complete rate versus pH curve for 638A with an apparent pKaA = 5.5 for the general acid, and n = 6.7 and pKaC = 4.0 (Fig. 2A dotted line). However, the data are also consistent with a general acid–base model with one apparent pKa of 5.3 and a hypothetical second titratable group with pKa ≈ 4.1 (Fig. 2A, solid line: the solid and dotted lines are almost superimposable; kinetic parameters are summarized in Table 1). The estimate of the latter pKa is unlikely to be accurate, as this part of the rate versus pH curve is strongly affected by the steep decrease in activity caused by the cooperative inactivation involving multiple functional groups with an apparent pKaC ≈ 4.0 very similar to the predicted pKa of the second hypothetical general acid–base group (see Discussion). Even though these two models cannot be distinguished, the large change in rate versus pH curve observed with the 638A mutant further supports the hypothesis that the nucleobases at position 638 plays an important role in the rate-limiting proton transfer step of the cis-cleavage reaction.
TABLE 1.
Summary of kinetic parameters

A novel chemical ligation method to incorporate site-specific nucleotide analogs
To further investigate the role in catalysis of the nucleobases at position 638, we needed a way of selectively replacing this nucleobase with nonnatural analogs. As described in the Introduction, currently available methods require Mg2+-containing buffers that are incompatible with obtaining uncleaved precursor RNA from a fast-cleaving ribozyme such as RG. We were aware of one chemical ligation procedure that could join two modified RNAs via reductive amination in the absence of Mg2+; however, the long reaction times reported (up to 7 d) were impractical for our needs (Bellon et al. 1996). Our attempts to improve on this procedure were unsuccessful. A completely different chemistry, using a sulfhydryl-containing nucleotide to attack a nucleotide containing an activated disulfide, has been used to monitor tertiary structure within an RNA (Cohen and Cech 1997). By combining the best features of these two chemical approaches we have designed a strategy that joins two RNAs via a disulfide bond.
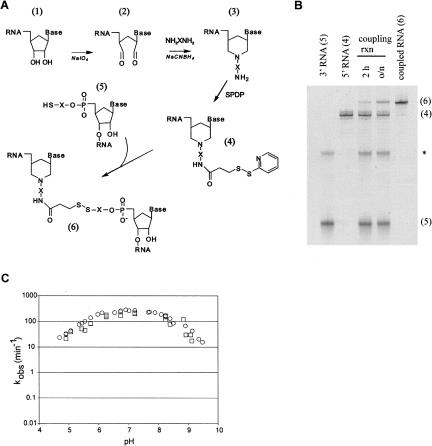
Our general approach is diagrammed in Figure 3A. Molecule 1 (the “upstream” RNA) represents any RNA whose 3′ end is to be joined to the 5′ end of molecule 5 (the “downstream” RNA). Molecule 1 is first converted to molecule 2 via oxidation with sodium periodate, then to molecule 3 via reaction with hexamethylenediamine and sodium cyanoborohydride. A sulfhydryl reactive moeity is introduced onto the 3′ amine of Molecule 3 by reaction with N-succinimidyl 3-(2-pyridyldithiol)propionate (SPDP) to make molecule 4. The downstream RNA, molecule 5, contains a 5′ SH group, prepared by chemical synthesis (although priming T7 transcription with a dinucleotide containing a 5′ sulfhydryl group may also work). When the upstream and downstream RNAs are mixed together the 5′SH group in molecule 5 attacks the sulfhydryl reactive moiety in molecule 4, joining the two RNAs via a disulfide bond to make molecule 6, the full-length RNA (Fig. 3A,B).
FIGURE 3.
Mg2+-independent RNA–RNA ligation scheme. (A) Synthesis scheme. Molecule 1, the upstream RNA, was synthesized by T7 transcription; molecule 4, the downstream RNA, was synthesized chemically and contains a nucleotide analog at position 638. X indicates a (CH2)6 linker, SPDP is N-succinimidyl 3-(2-pyridyldithiol)propionate, NH2XNH2 is hexamethylenediamine. (B) Denaturing PAGE analysis of typical input RNAs, a coupling reaction, and a gel-purified ligated precursor RNA from synthesis of RGssG (wild type); numbers in parentheses correspond to molecules numbered in the synthesis scheme; the asterisk indicates a presumed dimer of molecule 5. (C) Cleavage rate versus pH curve for RG RNA synthesized by in vitro transcription (circles; data from Fig. 1B) and RGssG prepared by the coupling scheme in A (squares).
To determine if the disulfide linkage itself had any effect on the kinetic properties of the VS ribozyme we compared cleavage time courses of the wild-type RG ribozyme that had been synthesized by in vitro transcription to those of the equivalent molecule obtained by joining a chemically synthesized stem–loop I to the rest of the ribozyme that had been synthesized by in vitro transcription. The disulfide-linked RNA is designated RGssG, where the last G indicates that guanosine is present at position 638; RNAs containing base analogs at position 638 are named in a parallel fashion (see the following section). The apparent first-order cleavage rate constant of RGssG under standard reaction conditions was the same within experimental error of that of RG synthesized by T7 transcription (3.6 versus 4.0 sec−1; data not shown). The extents of cleavage of RGssG and its nucleotide analog containing variants (described below) were somewhat lower than for in vitro transcribed RG RNA (about 60% versus 85%): chase experiments (Zamel et al. 2004) provided no evidence that this was due to an altered cleavage-ligation equilibrium; denaturing gel purification of the uncleaved RGssG RNA followed by renaturation indicated that a portion of the inactive RNA was misfolded, and suggested that the remainder may have been inactivated by side reactions during synthesis (data not shown). Comparison of the first order cleavage rate constant of the active population of the RNA versus pH for RG and RGssG showed that apparent cleavage rate constants of the two RNAs were very similar over the pH range examined, supporting the conclusion that the disulfide linkage had no detrimental effect on the kinetics of cis-cleavage (Fig. 3C).
Base analog substitution at position 638
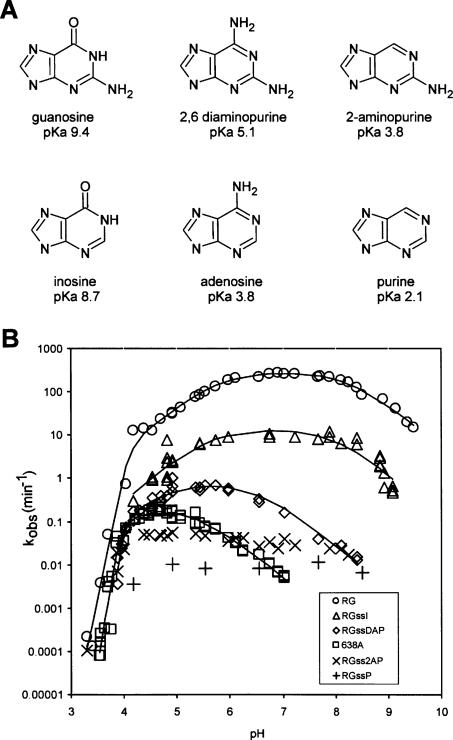
We used the above strategy to synthesize variants of RG in which G638 was replaced with purine nucleotide analogs with different functional groups and pKa values (Fig. 4A). Chemically synthesized stem–loop I variant RNAs, equivalent to molecule 5 (Fig. 1A, boxed area) containing inosine (I), 2,6-diaminopurine (DAP), 2-aminopurine (2AP), or purine (P) at position 638 were joined via disulfide linkage to the remainder of the ribozyme that had been synthesized by T7 transcription and converted to the form of molecule 4 in Figure 3. The coupled, cis-cleaving ribozymes are designated RGssI, RGssDAP, RGss2AP, and RGssP, respectively.
FIGURE 4.
Effects of pH on cleavage rate of RNAs with purine analogs at position 638. (A) Nucleoside analogs and their N1 pKa values (measured for the free nucleosides—actual pKas in RNA may be different; Moody et al. 2005; Tang et al. 2007); the state of N1 protonation at neutral pH is shown. (B) Cleavage rate versus pH curves for wild-type RG (638G), mutant 638A, and RGss analog-containing RNAs containing inosine (RGssI), 2,6-diaminopurine (RGssDAP), 2-aminopurine (RGss2AP), and purine (RGssP) synthesized as in Figure 3A.
Rate versus pH curves for the analog-containing RNAs are shown in Figure 4B. Below approximately pH 4 all RNAs examined showed the steep decrease in activity described above for wild type and 638A. In the pH range from about 4.5 to 9, where we could expect to observe indications of general acid–base catalysis, substitution with purine or 2AP caused slow, pH-independent cleavage; these analogs were not investigated further. Taken together with data described below, these observations suggest that the keto oxygen at position 6 of guanosine (and absent in purine and 2AP) and/or the protonation state of N1, whose pKa would be several pH units lower in purine or 2AP compared to the natural guanosine, plays an important role in self-cleavage.
Substitution with inosine, which has only a slightly altered N1 pKa compared to guanosine, but which lacks the 2-amino group, decreased the apparent cleavage rate by approximately an order of magnitude and shifted the rate versus pH curve slightly leftward, with apparent pKas of 5.7 and 7.9. The small decrease in cleavage rate (compared to other analogs and the G638A mutant) and the small shift in apparent pKas provide evidence that the 2-amino group of guanosine makes a measurable but small contribution to the self-cleavage reaction (see Discussion).
DAP, like adenosine, replaces the natural 6-keto group with an amino group, but the N1 pKa of DAP is 1.3 units higher than adenosine. The rate versus pH curve of RGssDAP resembles that of 638A in showing a log-linear decrease in rate as pH increases, but the curve is shifted to a higher pH by about 1.5 pH units compared to adenosine. Like the 638A mutation, the RGssDAP curve can be described by a model in which steep loss in activity at low pH is combined with either general acid catalysis only or with general acid–base catalysis, with the curve shifted substantially downward and to the left compared to wild type. The latter model fits the data somewhat better, especially in that it yields values for n and pKaC that are similar to those for the other RNAs (Table 1, n = 6.5, pKaC = 4.0). Taken together, the effects of these base analog substitutions on the pH dependence of cleavage strongly support a role for G638 in the rate-limiting proton transfer step of the self-cleavage reaction.
DISCUSSION
Site-specific base analog substitution in large ribozymes requires joining a chemically synthesized portion of RNA to the rest of the RNA to construct a self-cleaving precursor RNA with a base analog at a particular position. For many RNAs this has been accomplished using ligase enzymes that require divalent cations for activity. However, such enzymatic ligation strategies are not applicable to cis-cleaving ribozymes, especially those that cleave rapidly in divalent cations, because the precursor RNA formed by enzymatic ligation self-cleaves as fast as it is made. To overcome this problem we developed a method for joining two RNA molecules via a disulfide bond in the absence of divalent cations. The chemical ligation approach described here should be generally applicable to joining any two RNAs, or even constructing hybrid macromolecules composed of an RNA and another molecule containing a sulfhydryl group.
Using this approach we investigated the role in cis-cleavage of the nucleobase at position 638 in the cleavage loop of the VS ribozyme. Because some of the bases and base analogs used have low N1 pKa values and we observed apparent pKa s in the rate versus pH curves at low pH, a thorough analysis required measuring cleavage rates at pH values well below those typically collected when studying ribozymes. For all of the RNAs examined we observed a steep decrease in apparent cleavage rate below about pH 4.0. To obtain a mathematical description of the steep decrease in activity at low pH we fit the rate versus pH data for each RNA to a variety of models (see Materials and Methods and Results) that included a term involving some number, n, of bases with an average apparent pKa, pKaC, which when protonated inactivated the ribozyme. The best fit was obtained with models in which the cooperative protonation of five to six bases, each with an average apparent pKaC of ∼4.0, caused inactivation of the ribozyme.
Failure to include a term describing the steep decrease in activity at low pH can lead to misinterpretation of the remainder of the rate versus pH curve in certain situations. For example, a general acid-only reaction might be misinterpreted as a general acid–base reaction if data were not collected to a sufficiently low pH to correctly evaluate the decrease in activity below the apparent pKa (see Fig. 1B). Even when data are collected at enough low pH values to describe the complete curve, it is still not possible to distinguish convincingly between a general acid-only model and a general acid–base model in the remainder of the curve if the putative lower pKa of the general acid–base model has a value near pKaC; in this situation it is not even possible to accurately estimate the lower pKa in the general acid–base model because the cooperative crash dominates the curve fitting in this region. For example, the cleavage curve of the 638A mutant RNA fits almost equally well to a general acid plus crash model as it does to a general acid–base plus crash model (Fig. 1B; Table 1).
The explanation for the steep decrease in activity at low pH is likely to involve loss of RNA structure. At low pH it would be expected that some adenine and cytosine bases in any RNA would become protonated, leading to loss of tertiary structure when a sufficient number of important interactions were disrupted, and even loss of secondary structure at sufficiently low pH (Moody et al. 2005; Siegfried et al. 2007). The apparent pKa of 4.0 observed for the phenomenon causing the steep decline in activity of all of the RNA examined is consistent with protonation of adenosines and/or cytosines, which have pKa s of ∼3.8 and 4.2 as free nucleosides in solution that may be shifted slightly higher in the environment of a folded RNA (Saenger 1984; Moody et al. 2005). Similarly, a value of n = 5 or 6 seems a reasonable estimate for the number of protonatable bases that might be expected to disrupt the tertiary interactions of an RNA of the size and complexity of the VS ribozyme. Known tertiary interactions in VS RNA include three Watson–Crick base pairs between loops I and V (Rastogi et al. 1996), a U-turn motif in the III–IV–V junction (Sood and Collins 2001), interactions in the II–III–VI junction (Lafontaine et al. 2001a), and between the cleavage site loop and the 730 loop (Hiley et al. 2002).
Our observations of the effects of base analogs at position 638 in the cis-cleaving RG version of the VS ribozyme are qualitatively similar to those recently reported by Lilley and coworkers (Wilson et al. 2007) using a trans-cleaving version of the ribozyme. Some of our interpretations are tempered by data that were not reported in that earlier study, including the effects of the steep loss of activity at low pH in the data analysis and rate versus pH data for two additional base analogs (purine and 2AP). Other differences in RNA design between the two studies likely account for some of the quantitative differences in apparent rates and pKa values. For example, the substrate for the trans reaction used by Lilley and coworkers lacks helix Ia, which in cis affects the observed cleavage and ligation rates and the equilibrium between them (Rastogi and Collins 1998; Zamel et al. 2004; Smith and Collins 2007). In the current work, we used the RG version of the ribozyme that contains a stable helix Ia, additional nucleotides in the cleavage loop that strongly favor cleavage rather than ligation, and a G634C substitution that locks stem–loop Ib in the shifted conformation required for activity (Zamel et al. 2004). Despite these differences, a consistent story emerges from both studies that adds strong support to the hypothesis that G638 plays an important role in general acid–base catalysis in the VS ribozyme cleavage reaction.
In both cis and trans versions of the VS ribozyme, the removal of the exocyclic 2-amino group results in a leftward shift of the rate versus pH curve and a decrease in observed cleavage rate (compare guanosine with inosine and DAP with adenosine: the former of each pair contains the 2-amino group and has a slightly higher N1 pKa; the latter lacks the 2-amino group) (Fig. 4). The magnitude of the rate decrease is about five- to 50-fold, depending on which version of the ribozyme and what pH is compared. This rather small decrease in rate suggests that the protons on the 2-amino group are not the ones transferred during the rate-limiting step, but that this exocyclic group may play a role in positioning or affecting the pKa of the group involved in the rate-limiting step.
Removal of the keto oxygen has a severe effect on the observed cleavage rate. Comparing guanosine with 2AP, or inosine with purine, reveals a rate decrease of three to four orders of magnitude (Fig. 4). Even though these pairs of nucleotides differ structurally only by the presence or absence of the keto group, the absence of this group also leads to a decrease of about six pH units in the value of the N1 pKa. From first principles of chemistry, and from studies of other ribozymes, the N1 position of guanosine is a likely candidate for participation in the chemical step of the reaction (Pinard et al. 2001; Kuzmin et al. 2004; Han and Burke 2005; Martick and Scott 2006; Thomas and Perrin 2006; Wilson et al. 2006), leaving open the possibility that the low N1 pKa is responsible for the slow cleavage of the 2AP and purine substitutions.
Determining whether the decrease in activity of the 2AP and purine analogs is due mostly to the loss of the keto group or to the difference in N1 pKa would benefit from a guanosine analog that retains the exocylic 2-amino and 6-keto groups but lacks a protonatable atom at ring position 1. To our knowledge an analog with these characteristics, such as N1 deazaguanosine, is not available. An 8-azaguanosine analog, whose fluorescence is affected by the state of N1 protonation (Da Costa et al. 2007), may also be a useful tool if it could be incorporated by chemical synthesis. As an alternative, rather than removing the keto group, we can compare the effects of replacing the keto group with an amino group. Comparing guanosine with DAP and inosine with adenosine shows that having an amino group instead of a keto group at ring position six is less deleterious than simply removing the keto group. As above, these exocyclic substitutions also alter the N1 pKa. Indeed, observed cleavage rate follows the same order as N1 pKa: guanosine is fastest, followed by inosine, DAP, and adenosine, in keeping with the possibility that N1 pKa is an important determinant of the cleavage rate.
In both the VS and hairpin ribozymes, substitution of a putative catalytic guanosine nucleobase with an analog having a low pKa produces a rate versus pH curve that is very different from that of wild type (Pinard et al. 2001; Wilson et al. 2007; this study). For example, substitution with adenosine or DAP results in very slow cleavage at higher pH, and the log of the cleavage rate constant increases linearly as pH decreases. This is in complete contrast to what is observed with the wild-type VS and hairpin ribozymes, in which cleavage is maximal at neutral pH and decreases as pH decreases in VS, or is pH-independent in the hairpin ribozyme. These observations suggest that the corresponding guanosines may play similar roles in the catalytic mechanisms of both ribozymes.
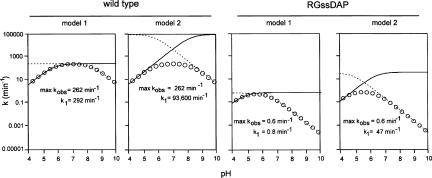
How much do the changes at position 638 affect the rate of the catalytic step of the self-cleavage reaction? This question is not as straightforward as it seems. A simple comparison of the relative maximum k obs values provides one possible answer (Table 1, column 3); however, even in a simple model of the wild-type RNA in which the group with the apparent pKa of 8.3 is assigned to G638 and the pKa at 5.8 is assigned to A756, the principle of kinetic ambiguity shows that an alternative model in which these assignments are reversed is equally consistent with the observed cleavage rate data (Jencks 1969; Bevilacqua 2003). The two models differ substantially in their estimate of k 1, the intrinsic rate of catalysis, of wild type: 292 min−1 versus 93,600 min−1 (Fig. 5A,B; Table 1). When G638 is replaced with a base analog that has a different pKa, we might have expected to see only one of the two apparent pKa s in the rate versus pH curve change; however, in all cases, both apparent pKa s were affected, indicating that analog substitutions have more widespread effects in the active site than simply altering only the pKa of nucleotide 638. For example, there are two interpretations of the changes in pKa in the DAP-containing RNA: (1) both of the wild-type pKa s have shifted to lower values (8.3 to 6.5, and 5.8 to 4.8) or (2) the pKa at 8.3 has shifted to 4.8 and the pKa at 5.8 has moved upward to 6.5 (Fig. 5C,D). These two possible models, combined with each of the two kinetically ambiguous models for wild type, produce four possible answers to the question “how much slower is catalysis in the DAP-analog RNA” (Table 1).
FIGURE 5.
Estimation of k 1, the intrinsic rate of the bond-breaking step. Simulations of each of the two kinetically ambiguous models of general acid–base catalysis for wild-type and RGssDAP using Equations 3 and 4 in Materials and Methods (Bevilacqua 2003). Circles represent simulated k obs values; dashed and dotted lines indicate the titration of the general acid or general base. Apparent pKa values are summarized in Table 1.
In some cases we can argue that not all four of the possible answers are equally likely, as two of them require reversing the assignment of which pKa is associated with the general acid and which with the general base. For example, inosine has a pKa only slightly lower than guanosine, and the apparent pKa s of the rate versus pH curve for the RNA containing inosine at position 638 (RGssI) are only slightly lower than in wild type. It seems most likely in this case that the apparent pKa s of the two titratable groups in wild type have each simply shifted slightly lower, leading to the conclusion that k 1 in wild type is either 20-fold (comparing model 1 for each RNA) or 36-fold (comparing model 2) faster than in RGssI (not 6324-fold faster, nor the counterintuitive possibility that k 1 is actually faster in RGssI, as inferred from comparison of wild-type model 2 to mutant model 1 and vice versa, respectively; Table 1). On the other hand, the more dramatic changes in the rate versus pH curve for the adenosine- or DAP-containing RNAs make it difficult to tell which pKa in the analog RNA corresponds to which in the wild type, leaving all four interpretations open, as well as the possibility (especially for the adenosine substitution) that the general base functionality has been eliminated.
A more quantitative comparison of intrinsic catalytic rates is further complicated by other factors that influence the estimation of k 1. The relationship between the observed cleavage rate and the theoretical maximal rate of a catalyzed reaction is described by k obs = F(k 1), where k obs is the experimentally estimated apparent rate constant, k 1 is the intrinsic rate constant of the bond-breaking step, and F is the fraction of RNA in the catalytically competent state. As discussed previously (Bevilacqua 2003; Smith and Collins 2007), and expanded in the current work, F itself is the product of several proportions. These include fHA + · fB −, which describes the fraction of the RNA population in which both the general acid and general base are in their functional protonated and deprotonated forms, respectively; and fC, the proportion of RNA that retains critical tertiary interactions (this proportion decreases rapidly below approximately pH 4). Also implicit is a list of potential factors, up to fn, describing the proportion of RNA in the correctly folded or docked state, the proportion in which putative electrostatic or transition state stabilizing interactions are formed, etc. So, at a given pH,
which implies that k obs will have a much smaller value than the potential intrinsic rate of catalysis (k 1) that could be achieved if all of the RNA was in the catalytically competent state.
In summary, we present evidence that the N1 and/or 6-keto groups of the guanosine nucleotide at position 638 in the cleavage loop of the VS ribozyme are important for the rate-limiting proton transfer step in the cis-cleavage reaction catalyzed by this RNA. These experiments were made possible by a novel approach for joining two pieces of RNA in the absence of divalent cations via a disulfide bond. This approach should be applicable to the study of any RNA and to hybrid molecules consisting of an RNA joined to another sulfhydryl-containing molecule.
MATERIALS AND METHODS
RNA constructs and mutants
The RG version of the VS ribozyme, derived from RS19 by including a C634G substitution that locks stem Ib in the shifted conformation required for activity (Andersen and Collins 2000), has been described previously (Smith and Collins 2007) and is shown in Figure 1A. Site-directed mutagenesis of the ribozyme-encoding plasmid was used to construct mutants containing A, C, or U at position 638; sequences were confirmed by automated DNA sequencing of the entire ribozyme region. Precursor RNAs were obtained by in vitro transcription with T7 RNA polymerase in the presence of [α-32P]GTP from plasmids linearized with EcoRI, followed by purification by denaturing gel electrophoresis and ethanol precipitation (Milligan et al. 1987; Hiley and Collins 2001).
Incorporation of base analogs was performed as outlined in Figure 3A. The 3′ RNA (molecule 5) was a chemically synthesized ribo-oligonucleotide obtained from Dharmacon, Inc. containing a base analog (Fig. 4A) at position 638 and a 5′ sulfhydryl group. The 5′ RNA was prepared by T7 RNA polymerase in vitro transcription from the RG plasmid template linearized with BglII; this yields an RNA whose 3′ end corresponds to the cytosine immediately upstream of the arrow indicated “ss” in Figure 1A (Fig. 3A, molecule 1). A representative procedure for the generation of RGss RNA is as follows. Molecule 1 (∼1 μmol RNA), was oxidized with 10 mM sodium periodate in 50 mM sodium acetate (pH 4.8) in a 50 μL volume. Reactions were incubated at room temperature for 30 min and then filtered through a micro G50 column (ProbeQuant G-50 Micro Column GE Healthcare) equilibriated in 5 mM sodium phosphate (pH 7.2) and ethanol precipitated. The 3′ amine was introduced into the oxidized RNA via reductive amination. The oxidized RNA was treated with 40 mM hexamethylenediamine dihydrochloride and 10 mM sodium cyanoborohydride in 50 mM sodium acetate (pH 4.8) in a 50 μL volume. Reactions were incubated at room temperature for 2 h and then filtered through a micro G50 column and ethanol precipitated. The activated thiol was introduced via the 3′ amine RNA using 1 mM N-succinimidyl 3-(2-pyridyldithio) propionate (SPDP; Pierce Scientific) in 10 mM sodium phosphate (pH 7.2), 150 mM NaCl, and 1 mM EDTA. Reactions were incubated at room temperature for 2 h and then filtered through a micro G50 column and ethanol precipitated. Molecule 5 was deprotected by treatment with 40 mM DTT in 10 mM sodium phosphate (pH 7.2), 150 mM NaCl, and 1 mM EDTA. After a 2-h incubation at room temperature, the DTT was removed by passing the reaction through a micro G50 column. The upstream and downstream RNAs (5 μM and 30 μM, respectively) were coupled via thiol exchange in 10 mM sodium phosphate (pH 7.2), 150 mM NaCl, and 1 mM EDTA. After overnight incubation at 4°C the reactions were quenched by addition of 2× the reaction volume of formamide RNA loading dye. RNAs were separated by electrophoresis on a 4% polyacrylamide/ 8.3 M urea gel, excised, and eluted into 2 mL of water (65°C, 1 h). Eluted RNAs were filtered through 0.8 μm/0.2 μmM Acrodiscs (Gelman Sciences). The RNAs were then ethanol precipitated and dissolved in water. The RNA concentrations for the coupling step have not been fully optimized; typical yields from the coupling step ranged from 5% to 63%. Typical reactions starting with 1 μmol of upstream RNA give rise to 50–200 pmol of purified coupled RNA.
Cis-cleavage reactions
Buffers used were lactate, acetate, MES, cacodylate, Tris, HEPES, and CAPSO. pH was adjusted by titration with NaOH or HCl; in some cases mixtures of two buffers were used to obtain the desired pH. Because the high concentration of MgCl2 affected the pH of the different buffered solutions and their concentrated stocks to different extents, pH values reported are those for 1× solutions containing all components except RNA measured at 37°C with a SympHony pH meter and calomel microelectrode (VWR Scientific Products) calibrated at 37°C. RNA was preincubated at 37°C in 1× reaction solution (40 mM buffer, 50 mM KCl, 2 mM spermidine). Reactions were initiated by mixing one volume of RNA with one volume of pre-warmed 1× reaction solution containing 400 mM MgCl2 (final concentrations of MgCl2 and RNA = 200 mM and 10 nM, respectively) either by manual pipetting or using a Kintek RQF-3 rapid quench flow instrument (Kintek Corp.), stopped, and analyzed by gel electrophoresis and Phosphorimaging as described previously (Smith and Collins 2007).
Data analysis
Data from individual self-cleavage reaction time courses were fit to a single exponential:
where fc is the fraction cleaved at time t, k is the apparent cleavage rate constant k obs, and f max is the fraction of the RNA that was cleavable. Between pH ≈ 5.5 and 4.0, cleavage curves for RG RNA became biphasic, with a decreasing fraction of RNA in the fast phase as pH decreased. In this pH region the rate of the data were fit to a double exponential:
where A 1 and A 2 are the amplitudes and k 1 and k 2 are the rate constants of the fast and slow phase, respectively. The rate constant of the fast phase, k 1, was used for subsequent analyses; k 2 was several 100-fold slower than k 1 and has not been investigated further.
Estimation of parameters from rate versus pH curves
The intrinsic rate constant of the bond-breaking step, k 1, is related to the experimentally observed apparent rate constant, k obs, by
where f active is the fraction of RNA in the functional state to perform catalysis (see below). The fraction of RNA in the protonation state capable of general acid–base or general acid-only catalysis (fab or fa, respectively) is described by Equation 4 or 5.
General acid–base catalysis (Bevilacqua 2003):
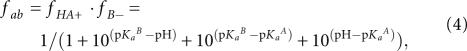 |
General acid-only catalysis:
where fHA + and fB − are the fractions of the RNA population in which the general acid and general base are protonated and deprotonated, respectively; pKaA and pKaB are the apparent pKa of the general acid and base.
To model the steep decrease in k obs at low pH, the following two equations were evaluated to describe the fraction of RNA in the active state due to pH-dependent inactivation of the ribozyme by a process independent of general acid–base catalysis.
Protonation of multiple independent sites (Knitt and Herschlag 1996):
Or cooperative protonation of multiple sites (derived from the Hill equation; Weiss 1997):
where fi or fc is the fraction of active RNA, n is the number, and pKaC is the average apparent pKa of nucleotides that must be protonated for the RNA to lose activity at low pH. Equation 7, but not Equation 6, provided an adequate fit to the data (see Results).
To calculate the fraction of active RNA, f active, at any point in the pH range, Equation 4 or 5 (which describe the fraction of RNA in the appropriate protonation state for general acid or general acid–base catalysis) was multiplied by Equation 7 (which describes the fraction of RNA in the active state affected by the low pH inactivation). For example, the wild-type data are best described by a combination of general acid–base catalysis and loss of activity by cooperative protonation at low pH:
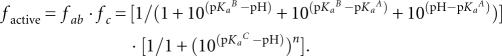 |
k obs versus pH data were fit to versions of Equation 3 where f active was described by combining Equations 4 and 7 or Equations 5 and 7. To equalize the weighting of all data points, nonlinear least squares curve fitting was performed using the natural logarithm of k obs, using the Solver add-in of Microsoft Excel 2003 (Microsoft Corp.) to simultaneously estimate optimal values of k 1, pKaA, pKaB, pKaC, and n.
ACKNOWLEDGMENTS
This research was supported by the Canadian Institutes for Health Research and the Canada Foundation for Innovation.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.936508.
REFERENCES
- Andersen, A.A., Collins, R.A. Rearrangement of a stable RNA secondary structure during VS ribozyme catalysis. Mol. Cell. 2000;5:469–478. doi: 10.1016/s1097-2765(00)80441-4. [DOI] [PubMed] [Google Scholar]
- Baum, D.A., Silverman, S.K. Deoxyribozyme-catalyzed labeling of RNA. Angew. Chem. Int. Ed. Engl. 2007;46:3502–3504. doi: 10.1002/anie.200700357. [DOI] [PubMed] [Google Scholar]
- Bellon, L., Workman, C., Scherrer, J., Usman, N., Wincott, F. Morpholino-linked ribozymes: A convergent synthetic approach. J. Am. Chem. Soc. 1996;118:3771–3772. [Google Scholar]
- Bevilacqua, P.C. Mechanistic considerations for general acid–base catalysis by RNA: Revisiting the mechanism of the hairpin ribozyme. Biochemistry. 2003;42:2259–2265. doi: 10.1021/bi027273m. [DOI] [PubMed] [Google Scholar]
- Bevilacqua, P.C., Brown, T.S., Nakano, S., Yajima, R. Catalytic roles for proton transfer and protonation in ribozymes. Biopolymers. 2004;73:90–109. doi: 10.1002/bip.10519. [DOI] [PubMed] [Google Scholar]
- Cohen, S.B., Cech, T.R. Dynamics of thermal motions within a large catalytic RNA investigated by cross-linking with thiol-disulfide interchange. J. Am. Chem. Soc. 1997;119:6259–6268. [Google Scholar]
- Curtis, E.A., Bartel, D.P. The hammerhead cleavage reaction in monovalent cations. RNA. 2001;7:546–552. doi: 10.1017/s1355838201002357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Costa, C.P., Fedor, M.J., Scott, L.G. 8-Azaguanine reporter of purine ionization states in structured RNAs. J. Am. Chem. Soc. 2007;129:3426–3432. doi: 10.1021/ja067699e. [DOI] [PubMed] [Google Scholar]
- Das, S.R., Piccirilli, J.A. General acid catalysis by the hepatitis delta virus ribozyme. Nat. Chem. Biol. 2005;1:45–52. doi: 10.1038/nchembio703. [DOI] [PubMed] [Google Scholar]
- Eckstein, F. Nucleoside phosphorothioates. Annu. Rev. Biochem. 1985;54:367–402. doi: 10.1146/annurev.bi.54.070185.002055. [DOI] [PubMed] [Google Scholar]
- Fedor, M.J., Williamson, J.R. The catalytic diversity of RNAs. Nat. Rev. Mol. Cell Biol. 2005;6:399–412. doi: 10.1038/nrm1647. [DOI] [PubMed] [Google Scholar]
- Han, J., Burke, J.M. Model for general acid–base catalysis by the hammerhead ribozyme: pH-activity relationships of G8 and G12 variants at the putative active site. Biochemistry. 2005;44:7864–7870. doi: 10.1021/bi047941z. [DOI] [PubMed] [Google Scholar]
- Hiley, S.L., Collins, R.A. Rapid formation of a solvent-inaccessible core in the Neurospora Varkud satellite ribozyme. EMBO J. 2001;20:5461–5469. doi: 10.1093/emboj/20.19.5461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiley, S.L., Sood, V.D., Fan, J., Collins, R.A. 4-thio-U cross-linking identifies the active site of the VS ribozyme. EMBO J. 2002;21:4691–4698. doi: 10.1093/emboj/cdf462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann, B., Mitchell, G.T., Gendron, P., Major, F., Andersen, A., Collins, R.A., Legault, P. NMR structure of the active conformation of the Varkud satellite ribozyme cleavage site. Proc. Natl. Acad. Sci. 2003;100:7003–7008. doi: 10.1073/pnas.0832440100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jencks, W. Catalysis and enzymology. Dover Publications Inc; New York: 1969. [Google Scholar]
- Jones, F.D., Strobel, S.A. Ionization of a critical adenosine residue in the Neurospora Varkud satellite ribozyme active site. Biochemistry. 2003;42:4265–4276. doi: 10.1021/bi020707t. [DOI] [PubMed] [Google Scholar]
- Ke, A., Zhou, K., Ding, F., Cate, J.H., Doudna, J.A. A conformational switch controls hepatitis δ virus ribozyme catalysis. Nature. 2004;429:201–205. doi: 10.1038/nature02522. [DOI] [PubMed] [Google Scholar]
- Knitt, D.S., Herschlag, D. pH dependencies of the Tetrahymena ribozyme reveal an unconventional origin of an apparent pKa . Biochemistry. 1996;35:1560–1570. doi: 10.1021/bi9521147. [DOI] [PubMed] [Google Scholar]
- Kuzmin, Y.I., Da Costa, C.P., Fedor, M.J. Role of an active site guanine in hairpin ribozyme catalysis probed by exogenous nucleobase rescue. J. Mol. Biol. 2004;340:233–251. doi: 10.1016/j.jmb.2004.04.067. [DOI] [PubMed] [Google Scholar]
- Lafontaine, D.A., Norman, D.G., Lilley, D.M. Structure, folding and activity of the VS ribozyme: Importance of the 2-3-6 helical junction. EMBO J. 2001a;20:1415–1424. doi: 10.1093/emboj/20.6.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine, D.A., Wilson, T.J., Norman, D.G., Lilley, D.M. The A730 loop is an important component of the active site of the VS ribozyme. J. Mol. Biol. 2001b;312:663–674. doi: 10.1006/jmbi.2001.4996. [DOI] [PubMed] [Google Scholar]
- Lafontaine, D.A., Wilson, T.J., Zhao, Z.Y., Lilley, D.M. Functional group requirements in the probable active site of the VS ribozyme. J. Mol. Biol. 2002;323:23–34. doi: 10.1016/s0022-2836(02)00910-5. [DOI] [PubMed] [Google Scholar]
- Martick, M., Scott, W.G. Tertiary contacts distant from the active site prime a ribozyme for catalysis. Cell. 2006;126:309–320. doi: 10.1016/j.cell.2006.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan, J.F., Groebe, D.R., Witherell, G.W., Uhlenbeck, O.C. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody, E.M., Lecomte, J.T., Bevilacqua, P.C. Linkage between proton binding and folding in RNA: A thermodynamic framework and its experimental application for investigating pKa shifting. RNA. 2005;11:157–172. doi: 10.1261/rna.7177505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, M.J., Query, C.C. Joining of RNAs by splinted ligation. Methods Enzymol. 2000;317:109–123. doi: 10.1016/s0076-6879(00)17009-0. [DOI] [PubMed] [Google Scholar]
- Moore, M.J., Sharp, P.A. Site-specific modification of pre-mRNA: The 2′-hydroxyl groups at the splice sites. Science. 1992;256:992–997. doi: 10.1126/science.1589782. [DOI] [PubMed] [Google Scholar]
- Murray, J.B., Seyhan, A.A., Walter, N.G., Burke, J.M., Scott, W.G. The hammerhead, hairpin and VS ribozymes are catalytically proficient in monovalent cations alone. Chem. Biol. 1998;5:587–595. doi: 10.1016/s1074-5521(98)90116-8. [DOI] [PubMed] [Google Scholar]
- Nakano, S., Chadalavada, D.M., Bevilacqua, P.C. General acid–base catalysis in the mechanism of a hepatitis δ virus ribozyme. Science. 2000;287:1493–1497. doi: 10.1126/science.287.5457.1493. [DOI] [PubMed] [Google Scholar]
- Nesbitt, S., Hegg, L.A., Fedor, M.J. An unusual pH-independent and metal-ion-independent mechanism for hairpin ribozyme catalysis. Chem. Biol. 1997;4:619–630. doi: 10.1016/s1074-5521(97)90247-7. [DOI] [PubMed] [Google Scholar]
- Ogilvie, K.K., Usman, N., Nicoghosian, K., Cedergren, R.J. Total chemical synthesis of a 77-nucleotide-long RNA sequence having methionine-acceptance activity. Proc. Natl. Acad. Sci. 1988;85:5764–5768. doi: 10.1073/pnas.85.16.5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Rear, J.L., Wang, S., Feig, A.L., Beigelman, L., Uhlenbeck, O.C., Herschlag, D. Comparison of the hammerhead cleavage reactions stimulated by monovalent and divalent cations. RNA. 2001;7:537–545. doi: 10.1017/s1355838201002461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrotta, A.T., Been, M.D. HDV ribozyme activity in monovalent cations. Biochemistry. 2006;45:11357–11365. doi: 10.1021/bi061215+. [DOI] [PubMed] [Google Scholar]
- Perrotta, A.T., Wadkins, T.S., Been, M.D. Chemical rescue, multiple ionizable groups, and general acid–base catalysis in the HDV genomic ribozyme. RNA. 2006;12:1282–1291. doi: 10.1261/rna.14106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinard, R., Hampel, K.J., Heckman, J.E., Lambert, D., Chan, P.A., Major, F., Burke, J.M. Functional involvement of G8 in the hairpin ribozyme cleavage mechanism. EMBO J. 2001;20:6434–6442. doi: 10.1093/emboj/20.22.6434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon, A.H., Olive, J.E., McLaren, M., Collins, R.A. Identification of separate structural features that affect rate and cation concentration dependence of self-cleavage by the Neurospora VS ribozyme. Biochemistry. 2006;45:13394–13400. doi: 10.1021/bi060769+. [DOI] [PubMed] [Google Scholar]
- Pyle, A.M. Ribozymes: A distinct class of metalloenzymes. Science. 1993;261:709–714. doi: 10.1126/science.7688142. [DOI] [PubMed] [Google Scholar]
- Rastogi, T., Collins, R.A. Smaller, faster ribozymes reveal the catalytic core of Neurospora VS RNA. J. Mol. Biol. 1998;277:215–224. doi: 10.1006/jmbi.1997.1623. [DOI] [PubMed] [Google Scholar]
- Rastogi, T., Beattie, T.L., Olive, J.E., Collins, R.A. A long-range pseudoknot is required for activity of the Neurospora VS ribozyme. EMBO J. 1996;15:2820–2825. [PMC free article] [PubMed] [Google Scholar]
- Saenger, W. Principles of nucleic acid structure. Springer-Verlag; New York: 1984. [Google Scholar]
- Scaringe, S.A., Francklyn, C., Usman, N. Chemical synthesis of biologically active oligoribonucleotides using β-cyanoethyl protected ribonucleoside phosphoramidites. Nucleic Acids Res. 1990;18:5433–5441. doi: 10.1093/nar/18.18.5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih, I.H., Been, M.D. Involvement of a cytosine side chain in proton transfer in the rate-determining step of ribozyme self-cleavage. Proc. Natl. Acad. Sci. 2001;98:1489–1494. doi: 10.1073/pnas.98.4.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegfried, N.A., Metzger, S.L., Bevilacqua, P.C. Folding cooperativity in RNA and DNA is dependent on position in the helix. Biochemistry. 2007;46:172–181. doi: 10.1021/bi061375l. [DOI] [PubMed] [Google Scholar]
- Sigel, R.K., Pyle, A.M. Alternative roles for metal ions in enzyme catalysis and the implications for ribozyme chemistry. Chem. Rev. 2007;107:97–113. doi: 10.1021/cr0502605. [DOI] [PubMed] [Google Scholar]
- Smith, M.D., Collins, R.A. Evidence for proton transfer in the rate-limiting step of a fast-cleaving Varkud satellite ribozyme. Proc. Natl. Acad. Sci. 2007;104:5818–5823. doi: 10.1073/pnas.0608864104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sood, V.D., Collins, R.A. Functional equivalence of the uridine turn and the hairpin as building blocks of tertiary structure in the Neurospora VS ribozyme. J. Mol. Biol. 2001;313:1013–1019. doi: 10.1006/jmbi.2001.5119. [DOI] [PubMed] [Google Scholar]
- Sood, V.D., Collins, R.A. Identification of the catalytic subdomain of the VS ribozyme and evidence for remarkable sequence tolerance in the active site loop. J. Mol. Biol. 2002;320:443–454. doi: 10.1016/s0022-2836(02)00521-1. [DOI] [PubMed] [Google Scholar]
- Strobel, S.A. A chemogenetic approach to RNA function/structure analysis. Curr. Opin. Struct. Biol. 1999;9:346–352. doi: 10.1016/S0959-440X(99)80046-3. [DOI] [PubMed] [Google Scholar]
- Tang, C.L., Alexov, E., Pyle, A.M., Honig, B. Calculation of pKas in RNA: On the structural origins and functional roles of protonated nucleotides. J. Mol. Biol. 2007;366:1475–1496. doi: 10.1016/j.jmb.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Thomas, J.M., Perrin, D.M. Active site labeling of G8 in the hairpin ribozyme: Implications for structure and mechanism. J. Am. Chem. Soc. 2006;128:16540–16545. doi: 10.1021/ja063942y. [DOI] [PubMed] [Google Scholar]
- Weiss, J.N. The Hill equation revisited: Uses and misuses. FASEB J. 1997;11:835–841. [PubMed] [Google Scholar]
- Wilson, T.J., Ouellet, J., Zhao, Z.Y., Harusawa, S., Araki, L., Kurihara, T., Lilley, D.M. Nucleobase catalysis in the hairpin ribozyme. RNA. 2006;12:980–987. doi: 10.1261/rna.11706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, T.J., McLeod, A.C., Lilley, D.M. A guanine nucleobase important for catalysis by the VS ribozyme. EMBO J. 2007;26:2489–2500. doi: 10.1038/sj.emboj.7601698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wincott, F., DiRenzo, A., Shaffer, C., Grimm, S., Tracz, D., Workman, C., Sweedler, D., Gonzalez, C., Scaringe, S., Usman, N. Synthesis, deprotection, analysis and purification of RNA and ribozymes. Nucleic Acids Res. 1995;23:2677–2684. doi: 10.1093/nar/23.14.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young, K.J., Gill, F., Grasby, J.A. Metal ions play a passive role in the hairpin ribozyme catalysed reaction. Nucleic Acids Res. 1997;25:3760–3766. doi: 10.1093/nar/25.19.3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamel, R., Poon, A., Jaikaran, D., Andersen, A., Olive, J., De Abreu, D., Collins, R.A. Exceptionally fast self-cleavage by a Neurospora Varkud satellite ribozyme. Proc. Natl. Acad. Sci. 2004;101:1467–1472. doi: 10.1073/pnas.0305753101. [DOI] [PMC free article] [PubMed] [Google Scholar]