Abstract
Dendritic cells provide a critical link between innate and adaptive immunity and are essential to prime a naive T-cell response. The transition from immature dendritic cells to mature dendritic cells involves numerous changes in gene expression; however, the role of post-transcriptional changes in this process has been largely ignored. Tristetraprolin is an AU-rich element mRNA-binding protein that has been shown to regulate the stability of a number of cytokines and chemokines of mRNAs. Using TTP immunoprecipitations and Affymetrix GeneChips, we identified 393 messages as putative TTP mRNA targets in human dendritic cells. Gene ontology analysis revealed that ∼25% of the identified mRNAs are associated with protein synthesis. We also identified six MHC Class I alleles, five MHC Class II alleles, seven chemokine and chemokine receptor genes, indoleamine 2,3 dioxygenase, and CD86 as putative TTP ligands. Real-time PCR was used to validate the GeneChip data for 15 putative target genes and functional studies performed for six target genes. These data establish that TTP regulates the expression of DUSP1, IDO, SOD2, CD86, and MHC Class I-B and F via the 3′-untranslated region of each gene. A novel finding is the demonstration that TTP can interact with and regulate the expression of non-AU-rich element-containing messages. The data implicate TTP as having a broader role in regulating and limiting the immune response than previously suspected.
Keywords: human, dendritic cells, cell differentiation, molecular biology, gene regulation
INTRODUCTION
Dendritic cells (DCs) are professional antigen-presenting cells (APCs) that prime naive T-cell responses and help tailor selective immune responses to individual classes of pathogens (Banchereau and Steinman 1998; Proietto et al. 2004; de Jong et al. 2005). Immature DCs (iDCs) exhibit a phenotype characterized by a high phagocytic capacity and low expression of co-stimulatory molecules such as CD40, CD80, and CD86 (Mahnke et al. 2002). Capture of microbial pathogens or necrotic tissue by iDCs results in activation, initiating the process of DC maturation. Functionally, mature DCs cease to capture new antigen and up-regulate expression of co-stimulatory molecules and MHC Class I and II in order to stimulate an antigen-specific T-cell response (Banchereau and Steinman 1998). At the molecular level, DC maturation results in dramatic changes in the gene expression profile in the cell. Most studies to date have focused on changes in transcription (Tureci et al. 2003; McIlroy et al. 2005). However, modulation of gene expression occurs not only at the level of transcription, but also at post-transcriptional levels such as changes in mRNA stability and translation. To date, the role of post-transcriptional processes in DC maturation have not been studied in detail.
Recent work suggests the existence of post-transcriptional operons that coordinate gene expression (Keene and Tenenbaum 2002; Keene and Lager 2005; Moore 2007). The post-transcriptional operon model proposes that mRNAs are coordinately controlled through trans-acting RNA-binding proteins (RBPs) that interact with specific cis-acting regulatory elements encoded in mRNA, usually the 3′-untranslated region (3′-UTR). The best characterized mRNA cis-acting regulatory element is the AU-rich element (ARE) (Chen et al. 1994; Zubiaga et al. 1995), located in the 3′-UTR of many cytokine and proto-oncogene mRNAs. It is estimated that 5% – 8% of the genes in the human genome have AREs in their 3′-UTRs suggesting the post-transcriptional regulatory network controlled by ARE binding proteins may be extremely complex (Bakheet et al. 2006).
The importance of ARE-mediated regulation of gene expression is illustrated by the inflammatory cytokine tumor necrosis factor α (TNF-α). Mice with a germline deletion of the TNF-α ARE spontaneously develop pathologies indistinguishable from rheumatoid arthritis and Crohn's disease (Kontoyiannis et al. 1999). Macrophages and T-cells from these mice produced threefold to 10-fold more TNF-α protein than their wild-type counterparts, and these effects were mediated solely through increased TNF-α mRNA stability and translation.
One of the best-characterized trans-acting ARE-binding proteins is the zinc finger protein tristetraprolin (TTP), also known as Nup475, TIS11, G0S24, and ZFP36. TTP has been shown to regulate the mRNA stability of a number of inflammatory mediators including TNF-α, GM-CSF, IL-2, IL-3, IL-6, CCL2, CCL3, iNOS, COX2, as well as its own mRNA (Carballo et al. 2000; Stoecklin et al. 2000; Boutaud et al. 2003; Brooks et al. 2004; Phillips et al. 2004; Linker et al. 2005; Ogilvie et al. 2005). In addition to innate immune activating stimuli such as LPS, recent work has demonstrated that TTP expression is induced by IFNγ (Sauer et al. 2006). TTP then limited IFNγ induction of the pro-inflammatory genes TNF-α, IL-6, CCL2, and CCL3, by destabilizing the mRNAs of these genes (Sauer et al. 2006). Thus, TTP operates as part of a negative feedback loop to limit the inflammatory response.
In order to systematically identify TTP mRNA targets as well as changes in the post-transcriptional program(s) that occur upon DC maturation, we generated iDCs and mDCs from normal human donors, immunoprecipitated TTP using a modification of the RIP-chip method (Tenenbaum et al. 2000; Gerber et al. 2004; Penalva et al. 2004; André et al. 2006; Townley-Tilson et al. 2006), and analyzed the bound targets on Affymetrix U133A GeneChips. This screen identified 100 iDC specific, 109 mDC specific, and 185 common TTP targets. We validated the GeneChip data by performing new TTP immunoprecipitations followed by real-time PCR for 15 mRNA targets. Finally, we demonstrate that TTP can functionally regulate the expression of six genes in a 3′-UTR-specific manner. Three of the genes, dual specific phosphatase 1 (DUSP1), indolamine 2,3 dioxygenase (IDO), and superoxide dismutase 2 (SOD2), contain ARE elements in their 3′-UTRs. CD86, a critical co-stimulatory molecule that is up-regulated upon DC maturation, is regulated by TTP, although it does not have a classic ARE in its 3′-UTR. Finally, we identified a conserved 36-nucleotide (nt) region that does not contain an ARE but is present in all six MHC Class I alleles. This element is sufficient to confer TTP regulation, demonstrating that TTP can alter mRNA stability through a non-ARE-containing target sequence. Together, these data establish TTP as a critical regulator of DC maturation with a broad role in immune regulation.
RESULTS
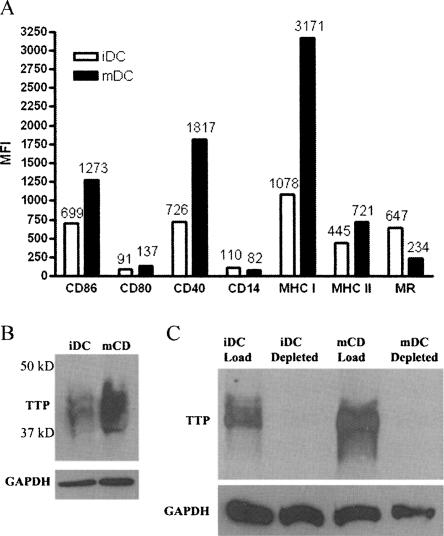
Human DCs were generated using a standard protocol of leukapheresis, countercurrent elutriation, and differentiation of the monocyte fraction with GM-CSF and IL-4 for 7 d (Guyre et al. 2002). The resulting immature DCs were split with half receiving LPS (2.5 μg/mL) and CD40L (10 μg/mL) for 24 h, and half left untreated. Consistent with prior data, maturation with LPS and CD40L resulted in increased expression of CD86, CD80, CD40, and MHC Class I, and a decrease in mannose receptor and CD14 expression (Fig. 1A; Banchereau and Steinman 1998). Human iDCs express lower levels of TTP than mDCs (Fig. 1B). Figure 1C demonstrates that TTP protein was effectively depleted from the DC lysate by immunoprecipitation with the CARP3 anti-TTP antibody (Brooks et al. 2002).
FIGURE 1.
Phenotype of iDCs and mDCs and TTP expression. Monocyte-derived dendritic cells were generated using IL-4 and GM-CSF. (A) Flow cytometric analysis of the surface marker expression on a representative set of human iDCs and mDCs used for immunoprecipitations. Mean fluorescent intensity (MFI) for CD86, CD80, CD40, MHC Class I, and Class II increase with maturation. MFI for CD14, and mannose receptor (MR) decreased with maturation. (B) Cytoplasmic lysates generated from iDCs and mDCs were immunoblotted for TTP protein expression. TTP expression is higher in iDCs than mDCs. (C) Total cytoplasmic and total cytoplasmic lysate depleted of TTP by IP were immunoblotted for TTP protein. TTP was cleared from the iDC and mDC lysates (iDC depleted, mDC depleted). Immunoblot for GAPDH served as a loading control.
Following immunoprecipitation, RNA was isolated from the TTP immunoprecipitate with Trizol and hybridized to Affymetrix U133A human GeneChips. Previous RIP-Chip experiments in human cells have demonstrated that the minimal amount of RNA isolated with protein A beads, pre-immune serum, or peptide blocked antibody does not result in enrichment of highly abundant mRNAs such as β-actin, histones, or ribosomal protein mRNAs. Microarray analysis of these control IPs has not found consistent enrichment of specific mRNAs when analyzed by multiple different computational methods. In light of these data, we did not analyze the RNA isolated from the pre-immune serum/protein A bead complex.
RIP-Chip analysis of TTP
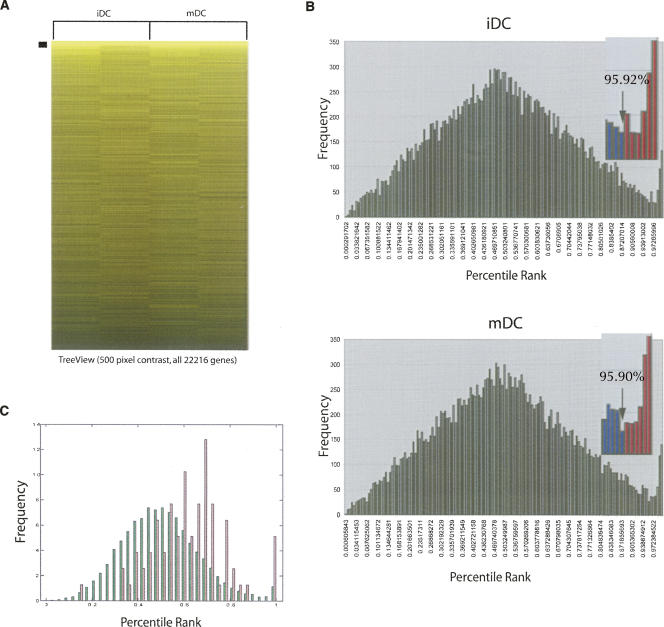
Affymetrix data from the GeneChips (two iDC, two mDC) were analyzed using the protocols developed for the analysis of targets of the histone stem–loop binding protein (SLBP) (Townley-Tilson et al. 2006) with modifications for application to Affymetrix GeneChips. The color-coded heat map of the net signal intensities from the four Affymetrix U133A human GeneChips is shown in Figure 2A. The signal intensity for each gene on each GeneChip was sorted by net signal intensity and each element on the array assigned a percentile rank. The mean percentile rank was then calculated from the duplicate GeneChip analyses and graphed in a distribution histogram showing the gene frequency for each percentile rank bin. The distribution of percentile ranks shows a bimodal distribution, and the cutoff for enriched genes was established at the trough between the bimodal distributions (Townley-Tilson et al. 2006). Genes with percentile rank above 95.92% in the iDCs and above 95.90% in the mDCs were considered significantly enriched. This resulted in 285 genes identified as putative TTP targets in iDCs and 294 genes identified in the mDCs (Fig. 2B).
FIGURE 2.
Identification of TTP targets by RIP-Chip. Messenger RNAs enriched in each RIP-Chip experiment were identified by assigning each element on the microarray a percentile rank on each of the GeneChips analyzed. These were subsequently used to calculate the mean percentile rank for both experiments. (A) TreeView image representing all 22,216 genes found on the array, sorted by percentile rank. The image's pixel settings are at 500 contrast; however, enriched genes are detected at values of greater than 10,000. The distinct band at the top represents the significantly enriched genes, as the color intensity correlates to the net intensity of the gene on the array. (B) Graphs of the distribution histograms of the mean percentile ranks for the two iDC RIP-Chip experiments (top panel) and mDC RIP-Chip experiments (bottom panel). The distribution in each is bimodal, showing a tail at the high percentile ranks (bin size = 0.006706). Putative iDC targets were defined as those that fall to the right of this trough with a percentile rank above 95.92% (the distribution of these genes is red in the histogram inset). Putative mDC targets were defined as those that fall to the right of this trough with a percentile rank above 95.90% (the distribution of these genes is red in the histogram inset). (C) Histogram showing the distribution of net intensity for all genes on the microarray (green) and the genes that have a GO biological process annotation of Protein Synthesis (pink). The Y-axis is the percentage of genes plotted at a given intensity value The percentile rank distributions are plotted on the X-axis across all four experiments (bin size = 0.33) as a percentage of all genes (green; 22,216 genes) or as a percentage of only the protein synthesis genes (pink; 78 genes).
Combining the two sets of genes resulted in a total of 393 unique genes. These include 100 TTP targets specifically enriched in the iDC IPs, 109 TTP targets specifically enriched in the mDC IPs, and 185 TTP targets enriched in both the iDC and mDC IPs. A striking feature of the identified genes is the large fraction encoding ribosomal proteins (86 genes) or translation factors (14 genes). Together, genes associated with protein translation account for >25% of the identified TTP targets. GO∷TermFinder analysis of the putative TTP targets identified the GO biological process term protein synthesis as significantly enriched (Bonferonni corrected p value = 0.00580). All genes on the microarray were plotted along with those genes with the GO term protein synthesis, demonstrating their clear enrichment at higher intensities (Fig. 2C). In addition to genes involved in protein synthesis, the six expressed MHC Class I genes (HLA-A, HLA-B, HLA-C, HLA-E, HLA-F, and HLA-G) were identified as were seven chemokine-related genes (CCL13, CCL17, CCL18, CCL22, CCR7, CKLFSF6, and MIF). The complete list of targets is presented in Supplemental Figure 1.
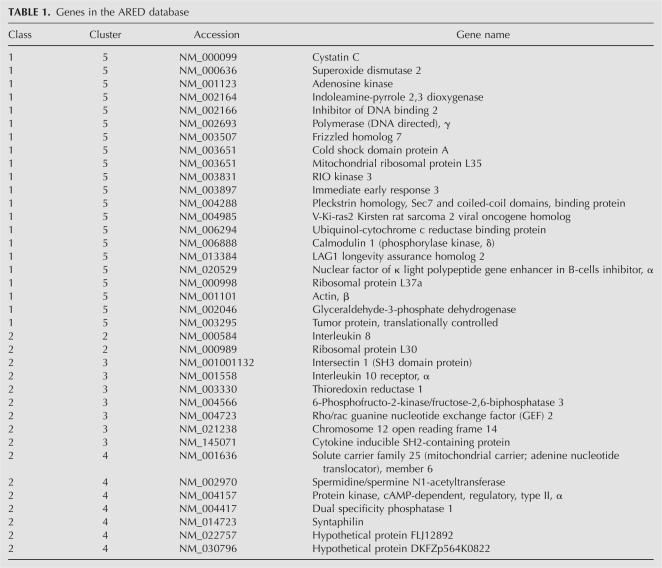
TTP has been shown to bind AU-rich elements (ARE) located in the 3′-UTR of unstable mRNAs. We searched the ARE Database version 2 (ARED2) (Bakheet et al. 2003) to determine which of the 393 identified genes contain an ARE. Surprisingly, only 37 of the 393 genes identified as putative TTP ligands were present in the ARED2 database (Table 1). Of the ARE containing genes, the majority (57%) were Class I, Group 5 AREs, which contain several individual AUUUA motifs dispersed through the 3′-UTR. The remaining genes were Class II ARE messages, split between Group 2 (5%), Group 3 (19%), and Group 4 (19%). These messages contain repeats of the AUUUA sequence (AUUUAUUUAUUUA), with Group 2 containing four repeats, Group 3 containing three repeats, and Group 4 containing two repeats. Recent work examining changes in global mRNA stability in TTP+/+ and TTP−/− fibroblasts identified 306 transcripts with altered mRNA stability, 33 of which contained AREs (Lai et al. 2006). The low percentage of ARE-containing genes is consistent with TTP binding to additional non-ARE mRNA sequence elements, as predicted using in vitro binding studies (Worthington et al. 2002), as well as with TTP interacting with mRNA ligands via a protein bridge, as reported for the iNOS mRNA (Linker et al. 2005).
TABLE 1.
Genes in the ARED database
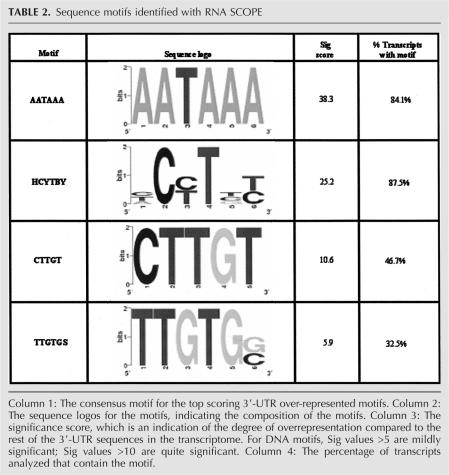
In light of the low percentage of ARE-containing genes identified, we employed a modified version of the SCOPE algorithm (SCOPE) (Carlson et al. 2007; Chakravarty et al. 2007) to search for 3′-UTR motifs present in the identified TTP ligands. This RNA version of SCOPE is under development and operates in a manner similar to SCOPE, but for RNA. Due to a number of expressed sequence tags and chromosome open reading frames contained on the U133A chip, the RNA SCOPE analysis included 290 genes (Supplemental Fig. 1). The motif with the highest significance identified was the polyadenylation signal AATAAA, present in 84.1% of the messages (Table 2). The motif present in the highest percentage of messages was HCYTBY, found in 87.5% of mRNAs, but because of its extreme degeneracy its relevance is unclear. The other motifs identified were two overlapping elements: CTTGT and TTGTGS, present in 46.7% and 32.5% of the messages, respectively. Combined, at least one of these elements was present in 53.8% of the messages.
TABLE 2.
Sequence motifs identified with RNA SCOPE
Confirmation of TTP mRNA ligands
The RIP-Chip analysis reduced the number of possible TTP ligands in DCs from 22,216 to 393. This analysis is a target discovery tool and it is important to note that the use of bimodal curves results in the inclusion of some false positives in the data set and the exclusion of some true positives. In this particular analysis, we did not analyze standard deviations of the RIP-chip targets for the separate mDC and iDC experiments.
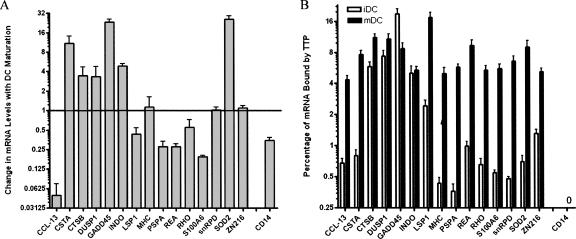
In order to validate the RIP-Chip findings, we performed a separate set of immunoprecipitations and assayed for the presence of 15 putative TTP ligands and one negative control (CD14) by real-time PCR (Fig. 3). One milligram of iDC and mDC cytoplasmic lysate was pre-cleared with pre-immune serum. The lysate was then split in half, with total cellular RNA isolated from half and TTP-RNA complexes immunoprecipitated from the other half. Figure 3A presents the relative mRNA level for each gene from the total analyte mRNA. The data are graphed as the change in message level with DC maturation for each gene, using the CT level in the iDC samples as the reference. Thus, expression levels greater than 1 indicate higher expression levels in mDC, while expression levels less than 1 indicate higher expression levels in iDCs. The changes in gene expression seen here are consistent with previous reports for MHC Class I (Kuchtey et al. 2005), IDO (von Bubnoff et al. 2004; Braun et al. 2005), and CD14 (Rieser et al. 1998).
FIGURE 3.
Confirmation of TTP targets by Real-Time PCR. iDC and mDC cytoplasmic lysate was pre-cleared with pre-immune serum. Total RNA was then isolated from half of the supernatant, and the other half was used for new TTP IPs and the captured RNA isolated. Total and TTP IP RNA samples were then screened for the presence of 15 putative TTP targets identified by RIP-Chip and one negative control (CD14) using Real-Time PCR. (A) Change in total mRNA level with DC maturation. The relative mRNA level of each gene is calculated using ΔΔCt. (B) The percentage of each mRNA bound by TTP in iDCs and mDCs. Values range from a low of 0.36% to a high of 18.8%. No CD14 signal was detected by real-time PCR in the TTP immunoprecipitate.
Next, we determined the percentage of mRNA bound by TTP in iDCs and mDCs for 16 genes (Fig. 3B). These data compare the amount of RNA in the TTP IP with total RNA for that cell. The percent of mRNA bound by TTP in iDCs and mDCs ranged from as little as 0.36% for PSPA in iDCs, to a high of 18.8% for GADD45, also in iDCs. No CD14 mRNA was captured with the TTP immunoprecipitation. Examining the highest percent interaction for each gene gives a range of 4.4% for CCL-13 and the previously stated 18.8% for GADD45. This range is consistent with our previous observation that TTP binds to ∼6%–8% of the TNF-α mRNA in macrophages (S.A. Brooks, unpubl.). It should be noted that even with the histone stem–loop-binding protein, which has a dissociation constant (Kd) of 1.5 nm (Battle and Doudna 2001), the percentage of histone mRNA recovered from purified polyribosomes by IP, typically, ranges from 25% to 40%. In contrast, TTP has a dissociation constant of ∼20 nm for ARE, 10 times higher than SLBP (Brewer et al. 2004). Ten genes (CCL-13, CSTA, MHC Class I, PSPA, REA, RHO, S100A6, snRPD, SOD2, ZN2216) show significantly more mRNA bound in mDCs. Five genes (CTSB, DUSP1, GADD45, INDO, LSP1) are bound by TTP in iDCs and mDCs. Together, these data support our findings from the RIP-Chip analysis.
Functional effect of TTP on 3′-UTR mediated reporter expression
We have previously established a model system to study TTP function using transient transfections of gene-specific 3′-UTRs cloned downstream from a luciferase reporter and a TTP expression construct. This method establishes TTP-mediated post-transcriptional gene regulation, but does not discriminate the level of post-transcriptional regulation. However, to date, all data indicate that TTP modulates message stability and not message translation, and we assume that TTP is operating in the same manner here.
The 3′-UTRs of three of the validated ARE-containing messages, DUSP1, IDO, and SOD2, were cloned into the 3′-UTR of the luciferase reporter pGL3-Control. These constructs were then used to assess the ability of TTP to reduce luciferase expression in a transfection assay using HEK293 cells and RAW264.7 cells (Fig. 4). HEK293 cells are a human cell line that expresses very little if any endogenous TTP (Brooks et al. 2002, 2004; Rigby et al. 2005). RAW264.7 cells are a mouse macrophage cell line and provide a more functionally relevant cell for these studies. In addition to wild-type human TTP, we utilized a TTP zinc finger mutant (TTP-Zn-FM) that lacks the ability to bind to mRNA (Lai et al. 2002). In the RAW cells, we also employed a mouse TTP expression construct, which behaved identically to human TTP in all cases (data not shown).
FIGURE 4.
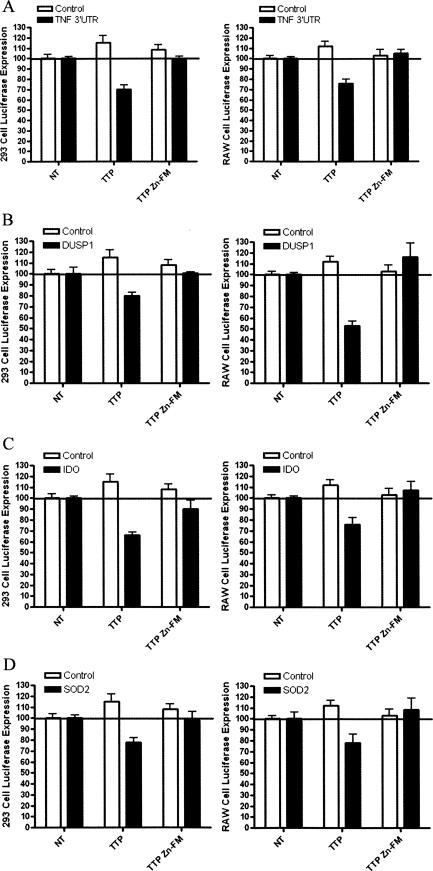
Functional characterization of TTP mRNA ligands. The functional effect of TTP transfection on 3′-UTR-mediated luciferase expression was examined in HEK 293 and RAW264.7 cells, using wild-type (TTP) and Zinc-Finger Mutant (TTP Zn-FM), which cannot bind mRNA (n = 4). (A) Consistent with previous work, TTP transfection had little effect on pGL3 control luciferase expression. Wild-type TTP transfection resulted in a significant decrease in full-length TNF-α 3′-UTR luciferase reporter expression in both 293 (p < 0.001) and RAW (p < 0.001) cells, while the TTP Zn-FM did not alter TNFα 3′-UTR-mediated expression. (B) Wild-type TTP significantly inhibited DUSP1 3′-UTR luciferase expression in both 293 (p < 0.001) and RAW (p < 0.01) cells. The TTP Zn-FM did not alter DUSP1 3′-UTR luciferase expression. (C) Wild-type TTP significantly inhibited IDO 3′-UTR luciferase expression in both 293 (p < 0.001) and RAW (p < 0.05) cells. The TTP Zn-FM did not alter IDO 3′-UTR luciferase expression. (D) Wild-type TTP significantly inhibited SOD2 3′-UTR luciferase expression in both 293 (p < 0.01) and RAW (p < 0.01) cells. The TTP Zn-FM did not alter SOD2 3′-UTR luciferase expression.
Luciferase expression from the pGL3 control luciferase reporter was not altered by TTP co-transfection (Fig. 4A). Consistent with previous work, luciferase expression from a human TNFα 3′-UTR (nucleotides 855–1643) containing reporter was significantly inhibited with wild-type TTP co-transfection (p < 0.001 two-tailed t-test) (Brooks et al. 2002, 2004; Rigby et al. 2005). Mutation of the TTP zinc fingers (TTP Zn-FM), which are required for RNA binding, resulted in a loss of TTP function, consistent with previous work (Lai et al. 2002; Brooks et al. 2004; Rigby et al. 2005).
DUSP1 is a MAPK phosphatase that binds to activated MAPKs and dephosphorylates them at threonine and tyrosine residues. In macrophages, DUSP1 expression is increased by activating factors such as LPS and peptidoglycan (Chen et al. 2002; Shepherd et al. 2004), as well as by immunosuppressive factors such as dexamethasone and IL-10 (Hammer et al. 2005a,b; Zhao et al. 2005). The DUSP1 mRNA contains a Class 2 ARE (Table 1). DUSP1 3′-UTR luciferase expression was significantly inhibited by co-transfection of wild-type TTP in both 293 (p < 0.001) and RAW (p < 0.01) cells. Transfection of the TTP zinc finger mutant was no different from no co-transfected TTP in 293 cells and resulted in a slight increase in luciferase-DUSP 3′-UTR reporter in RAW264.7 cells (Fig. 4B).
Indoleamine 2,3-dioxygenase (IDO) is responsible for converting tryptophan to kynurenines. It is expressed in a wide variety of tissues and is up-regulated by IFNγ. IDO enzymatic activity correlates with reduced T-cell-mediated responses, and mDCs that have functional IDO enzyme activity can be potent suppressors of T-cell responses in vivo and in vitro (Mellor and Munn 2004). The IDO 3′-UTR contains a Class 1 ARE. IDO 3′-UTR-mediated luciferase expression (Fig. 4C) was significantly inhibited with wild-type TTP transfection in both 293 (p < 0.001) and RAW cells (p < 0.05). There was no effect of the TTP Zn-FM mutant transfection in either cell type.
Superoxide dismutase 2 (SOD2) is an antioxidant enzyme involved in cellular protection from reactive oxygen species. SOD catalyzes the dismutation reaction of superoxide radical anion (O−2) to hydrogen peroxide. SOD2 mRNA contains a Class 1 ARE. TTP transfection significantly reduced SOD2 3′-UTR luciferase expression in both 293 (p < 0.01) and RAW (p < 0.01) cells with wild-type TTP, while the TTP Zn-FM mutant had no effect (Fig. 4D).
TTP regulates CD86 expression
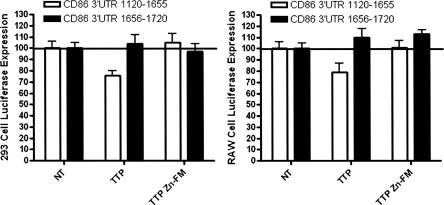
CD86 is not in the ARE database and does not contain a canonical ARE nonamer (A/UUAUUUAUA/U) sequence. However, CD86 does contain two ARE-like sequences—(nucleotides 1583–1589) UAUUUAU and (nucleotides 2552–2560) UUAUUUUAU. We cloned the human CD86 3′-UTR into pGL3 control in two parts: CD86 3′-UTR 1120–1655, which contains the first ARE-like element, and CD86 3′-UTR 1656–2720, which contains the second of the ARE-like sequences. In 293 cells, TTP transfection resulted in a 24% decrease in luciferase expression with the CD86 3′-UTR 1120–1655 (p < 0.05) (Fig. 5). TTP transfection did not result in a significant change in the luciferase-CD86–3′-UTR 1656–2720 construct. Comparable results were obtained with RAW cells. TTP transfection reduced CD86 3′-UTR 1120–1655 luciferase expression by 21% (p < 0.05) but had no effect on CD86 3′-UTR 1656–2720 (Fig. 5). Transfection of TTP-Zn-FM did not result in significant changes to either of the report constructs. We conclude that TTP regulation of CD86 expression requires a cis-element located between nucleotides 1120 and 1655, which is likely to be the UAUUUAU sequence at nucleotides 1583–1589. However, we cannot exclude the presence of other cis-elements within this 3′-UTR fragment.
FIGURE 5.
Functional characterization of CD86. The functional effect of TTP expression on CD86 3′-UTR-mediated luciferase expression was examined in 293 and RAW cells, using wild-type (TTP), Zinc-Finger Mutant (TTP Zn-FM) (n = 4). Transfection of wild-type TTP resulted in a 24% decrease in CD86 3′-UTR nucleotides 1120–1655 in 293 cells (p < 0.05) and a 21% decrease in RAW264.7 cells (p < 0.05). The TTP Zn-FM binding mutant had no effect on CD86 3′-UTR nucleotides 1120–1655. There was no effect of TTP transfection on CD86 3′-UTR nucleotides 1656–2720.
TTP regulates MHC Class I expression via a non-ARE-dependent mechanism
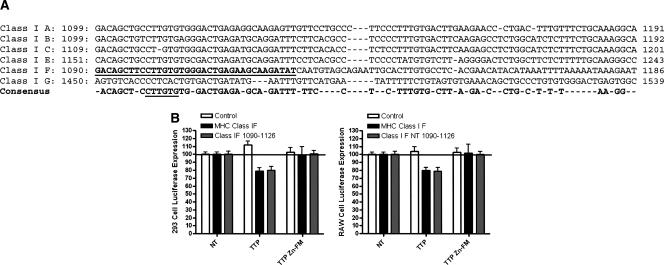
Finally, we examined the effect of TTP on MHC Class I 3′-UTR-mediated expression. MHC Class I is not in the ARE database, and there are no “ARE-like” sequences in the 3′-UTR. The GeneChip data indicated that TTP interacted with all six MHC Class I mRNAs (A, B, C, E, F, G). We first performed an alignment of the 3′-UTRs of the six MHC Class I genes (Fig. 6A). The 3′-UTRs range in length from 720 nt (allele G) to 99 nt (allele F). Alignment of the MHC Class I Allele 3′-UTRs identified a single relatively short region of overlap that coincides with the 99-nt Class I-F 3′-UTR. Interestingly, this element occurs early in the 3′-UTR of five of the six alleles. Only HLA Class I-G, which contains the longest 3′-UTR (720 nt), is the sequence located more than 150 nt downstream from the stop codon.
FIGURE 6.
Functional effect of TTP on MHC Class IF expression. (A) Alignment of the 3′-UTRs of the six expressed MHC Class I molecules. The 36-nt region from MHC Class IF is indicated in bold underline. The cis-element identified by RNA SCOPE is underlined in the consensus sequence. (B) Wild-type TTP significantly inhibited (p < 0.05) luciferase expression from the 99-nt MHC Class IF 3′-UTR as well as the 36-nt region from 1090 to 1126. The TTP Zn-FM mutant had no effect on luciferase expression (n = 4).
We cloned the MHC Class I-F allele into the pGL3 reporter and performed transfection experiments as above (Fig. 6B). TTP transfection resulted in a 21% reduction of luciferase expression in 293 cells (p < 0.05) and a 20% reduction in RAW cells (p < 0.05). Next, we cloned the 36 nt from 1090 to 1125 that contained the region of highest homology in Figure 6A. An examination of the sequence of this element (GACAGCTTCCTTGTGTGGGACTGAGAAGCAAGATAT) reveals that it contains almost the exact sequence motifs (CTTGT, TTGTGS) identified in the RNA SCOPE analysis (Table 2). The 36-nt element was sufficient to confer the same degree of TTP sensitivity as the full-length Class IF 3′-UTR. Once again, transfection of TTP-Zn-FM had no effect on the reporter.
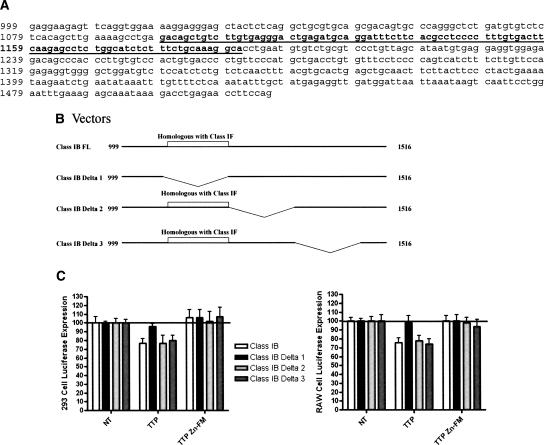
In order to establish that the region identified in the MHC Class I-F allele was necessary as well as sufficient, we cloned the MHC Class I-B 3′-UTR into the pGL3-Control luciferase vector and generated a series of deletions (Fig. 7A). The MHC Class I-B 3′-UTR is 517 nt long. The region of homology with Class I-F is indicated in bold underline in Figure 7A. We made three deletion mutants from the full-length MHC Class I-B 3′-UTR (Fig. 7B). MHC B Delta 1 deletes nucleotides 1104–1251 and eliminates all but the first four nucleotides of the sequence that aligns with MHC Class I-F. MHC B Delta 2 deletes nucleotides 1251–1324. MHC B Delta 3 deletes nucleotides 1324–1461 (Fig. 7B). Figure 7C demonstrates that wild-type TTP is able to inhibit wild-type MHC Class I-B 3′-UTR luciferase expression in both 293 and RAW 264.7 cells. Deletion of the Class I-B region that aligns with Class I-F (MHC Class I-B Delta 1) eliminates the effect of TTP transfection. MHC B Delta 2 and Delta 3 both retain TTP responsiveness. These data in combination with Figure 6 demonstrate that TTP regulates MHC Class I expression via a non-ARE cis-element.
FIGURE 7.
Identification of the TTP cis-element in MHC Class IB. (A) MHC Class IB 3′-UTR. Underline bold sequences are nucleotides that align with MHC Class IF. (B) MHC Class IB vectors constructs used to identify the TTP-binding site. The region of homology with MHC Class IF is indicated. (Class IB FL) Full-length Class IB 3′-UTR nucleotides 999–1517; (Class IB Delta 1) Class IB 3′-UTR with nucleotides 1104–1251 deleted. This region corresponds to the region of homology with MHC Class IF. (Class IB Delta 2) Class IB 3′-UTR with nucleotides 1251–1324 deleted; (Class IB Delta 3) Class IB 3′-UTR with nucleotides 1324–1461 deleted. (C) Wild-type TTP significantly inhibited (p < 0.05) luciferase expression from wild-type, Class IB Delta 2, and Class IB Delta 3 constructs. TTP inhibition was lost with the Delta 1 construct (n = 4). The TTP Zn-FM mutant had no effect on any of the luciferase constructs.
DISCUSSION
Dendritic cells are crucial regulators of immune responses serving as a link between the innate and adaptive immune systems. The transition from immature DC to mature DC represents a critical switch from anti-inflammatory to pro-inflammatory status in the DC. We sought to identify the in vivo TTP mRNA targets in human monocyte-derived iDCs and mDCs using TTP IP followed by Affymetrix GeneChips analysis (RIP-Chip).
Several previous efforts have been undertaken to identify TTP targets, using SELEX to identify TTP mRNA-binding sites, and examining changes in mRNA stability in TTP−/− fibroblasts (Worthington et al. 2002; Lai et al. 2006). We view these studies as complementary to the work presented here, each having its own strengths and weaknesses. In vitro binding assays such as SELEX identify sequences bound by TTP with high affinity but may miss lower-affinity binding. These assays also exclude the involvement of other proteins in TTP–mRNA interaction, as is the case for the protein KSRP (Linker et al. 2005). Microarray-based mRNA decay assays establish a functional effect, but not the direct involvement of TTP, even in TTP−/− cells, as the loss of TTP could alter the expression of proteins that themselves may regulate mRNA stability. For example, DUSP1, which we identified as a TTP ligand, operates to limit p38 phosphorylation (Hammer et al. 2005a,b). Since modulation of p38 activity is involved in regulating mRNA stability, the downstream effects of altered DUSP1 mRNA stability in TTP−/− animals is likely to have an impact on both TTP and non-TTP mRNA targets. The RIP-Chip assay employed here establishes that TTP likely interacts with a given mRNA, but does not by itself establish a functional consequence of this interaction. Additionally, the messages identified may represent the subset of messages that have survived the action of TTP and not the complete set of messages with which TTP interacts. Indeed, this may account for the difference in messages we identify as TTP ligands in iDCs and mDCs. Regardless, the inability of the RIP-Chip assay to identify every TTP ligand in a cell does not invalidate the identified, validated, and functionally characterized ligands, just as the shortcomings associated with SELEX and Chip-based decay assays do not invalidate those assays.
Our analysis of the TTP mRNA ligands using RNA SCOPE did not identify the ARE, which is present in 12.8% of the messages examined, as occurring significantly above the background level of 8% (Bakheet et al. 2006). However, it is also likely that the small number of ARE-containing mRNA targets identified is the consequence of their higher rate of turnover, resulting in a lower percentile rank. Supporting this, we did not identify TNF-α above the threshold cut for either mDCs or iDC but did confirm that the TNF-α mRNA was present in the TTP IP as determined by real-time PCR in Figure 3 (data not shown). We have previously demonstrated that proteasome inhibition inhibits TTP function, at least with regard to TNF-α mRNA stability. While beyond the scope of this manuscript, studies are planned to perform the RIP-Chip analysis comparing cells treated with and without proteasome inhibitors.
The large number of non-ARE-containing messages we identified supports the hypothesis that TTP interacts with non-ARE-containing messages. The number and percent of ARE-containing messages we identified in iDCs (27, 9.5%) and mDCs (29, 9.9%) is comparable to the number and percentage of ARE-containing messages identified with altered mRNA stability in TTP−/− fibroblasts (33, 10.78%). Several non-ARE-containing binding motifs were identified in SELEX experiments, and we demonstrate that TTP both interacts with and functionally regulates MHC Class I expression. Additionally, while KSRP, which serves as a protein bridge between iNOS and TTP (Linker et al. 2005), interacts with the ARE, it is plausible that TTP interacts with RNA-binding proteins that have specificity for other RNA cis-elements, expanding the pool of messages with which TTP could interact.
The RNA SCOPE analysis identified a non-ARE sequence element, CTTGTG, present in 53.8% of the messages. This element is also present in the 36-nt region we functionally identified as conferring TTP responsiveness in the MHC Class I 3′-UTR. Analysis of only the MHC Class I genes with RNA SCOPE reveals a more precisely defined sequence element that we show is sufficient to confer TTP regulation of a luciferase reporter construct. Our findings are consistent with a previous report demonstrating that MHC Class I gene expression is regulated at the level of mRNA stability (Kuchtey et al. 2005). The magnitude of the effect we see in the functional studies is consistent with our previous work and that of others (Brooks et al. 2004; Stoecklin et al. 2004; Rigby et al. 2005; Deleault et al. 2008). While these changes may appear modest, small changes in the message stability can have dramatic consequences. For example, inhibiting TTP function in monocytes with LPS results in a change in TNF-α mRNA stability from 37 min to 56 min, a 51% change in stability (Deleault et al. 2008). Unlike most cells in the body, MHC Class I expression in DCs is a highly regulated process (Banchereau and Steinman 1998). Proper immune function is critically dependent on antigen-specific DC activation of T-cells. It seems likely that TTP regulation of MHC Class I stability represents an important control point for the increase in MHC Class I expression that occurs with DC maturation. Whether TTP regulation of MHC Class I is specific to DCs or other specialized antigen-presenting cells is unclear, however, TTP regulation of TNF-α is limited to myeloid cells even though lymphoid cells express both TNF-α and TTP (Carballo et al. 1997).
The large number of ribosomal and translation factors identified was unexpected, although L37a contained an ARE. It is possible that modulating ribosomal protein expression represents an underappreciated site of regulatory control. Additionally, there are numerous reports in the literature of extra-ribosomal functions for ribosomal proteins. For example, S19 forms a homodimer that serves as a monocyte chemotactic factor that is thought to participate in the pathology of chronic inflammatory diseases such as rheumatoid arthritis (Yamamoto 2000). During development, mutation of ribosomal proteins can result in very selective effects that cannot be explained as simply disregulated protein synthesis. In Drosophila, mutation of S2 causes arrest of oogenesis (Cramton and Laski 1994), mutation of S6 results in melatonic tumors and hypertrophied hematopoetic organs (Watson et al. 1992), and L19 mutants fail to develop normal wing blades (Hart et al. 1993). Basal expression of ribosomal protein expression varies across tissues, in a way that does not necessarily correlate with cell division (Thomas et al. 2000). There are seven different S27 message isoforms all with identical coding regions but different tissue distributions, consistent with post-transcriptional regulation (Thomas et al. 2000). Ribosomal proteins L7 and S11 are overexpressed in colorectal carcinomas (Kasai et al. 2003), while the L13 protein, which has an identical protein-coding region to the breast basic conserved 1 protein (BBC1), is overexpressed in gastrointestinal cancer cells (Kobayashi et al. 2006). RNAi knockdown of L13 significantly enhanced the sensitivity of these cells to DNA damage, while exogenous addition of L13 expression into cells inhibited chemosensitivity (Kobayashi et al. 2006). Finally, a recent report identified several ribosomal proteins as specific markers of juvenile idiopathic arthritis (Allantaz et al. 2007). While beyond the focus of this study, further characterization of the role of TTP in the regulation of ribosomal protein expression should provide critical insights.
In this way, TTP may operate as part of a post-transcriptional operon, functioning with other RBPs to regulate DC maturation at the post-transcriptional level. By coordinately regulating the stability and translation of mRNAs from multiple different genes, post-transcriptional operons allow for the coordinated expression of functionally related groups of genes (Keene and Tenenbaum 2002; Keene and Lager 2005; Moore 2007). Recent work examining the Puf family of RBPs in both Saccharomyces cerevisiae and Drosophila melanogaster has demonstrated common themes for the messages bound (Gerber et al. 2004; André et al. 2006). In S. cerevisiae, each of the five Puf proteins interacted with mRNAs whose proteins had common functions and subcellular localization (Gerber et al. 2004). In D. melanogaster, the Puf gene PUMILIO interacted with mRNAs encoding functionally related proteins, but the set of messages bound was distinct at different developmental stages (André et al. 2006). Similar data have been demonstrated in mammalian cell lines for the ELAV family of RBPs (Tenenbaum et al. 2000; Penalva et al. 2004). The composition of mRNAs detected in HuB-mRNP complexes changed in P19 embryonal carcinoma stem cells after induction of neuronal differentiation with retinoic acid (Tenenbaum et al. 2000). DC maturation likely involves similar mechanisms with multiple RBPs interacting with distinct subsets of messages, to coordinate the series of changes that occur, from changes in phagocytosis to increasing expression of MHC and co-stimulatory molecules. Indeed, post-transcriptional operons may fine-tune DC maturation to specify a specific immune response to individual classes of pathogens.
Conclusion
Our data indicate that TTP interacts with about 300 messages in iDCs and in mDCs. These data establish that TTP can interact with and regulate the expression of non-ARE-containing mRNAs, specifically MHC Class I mRNAs. A number of the TTP mRNA ligands identified have been previously established as critically involved in DC maturation and function, including MHC Class I, CD86, several chemokine and chemokine receptors, and IDO. This study establishes TTP as a having a broader role in regulating the immune response than previously suspected.
MATERIALS AND METHODS
Materials
Reagents
Lipopolysaccaride (LPS) (E. coli 026:B6) was purchased from Sigma. IL-4, GM-CSF, and CD40L were purchased from Peprotech. Cell culture bags were purchased from American Fluoroseal. The antibodies for CD11c, CD86, CD80, CD40, CD14, MHC I, MHC II, and mannose receptor (MR) and the corresponding isotype controls were purchased from Beckman Coulter.
Leukocyte harvest and isolation
All procedures were approved by the Institution Review Board for Human Subjects. Normal human leukocytes were collected on a Cobe Spectra Apheresis cell separator, according to the Dartmouth-Hitchcock Medical Center, SOP # DR III D, using the white blood cell (WBC) procedure for preparation of monocytes (MNC), for 85 min (Wallace et al. 2000). Highly purified populations of human lymphocytes and monocytes were obtained from the leukapheresis product by countercurrent elutriation of WBCs. Lymphocytes and monocytes were isolated by counterflow centrifugation cell elutriation (J6M/E Beckman centrifuge with JE 5.0 rotor) using a 6.5 mL Sanderson chamber (Wallace et al. 2000). Monocytes were used immediately as outlined below.
Generation and maturation of dendritic cells
Differentiation: Monocytes were incubated in VueLife bags in RPMI containing 10% FCS and GM-CSF (10 ng/mL) and IL-4 (20 ng/mL) for 7 d, receiving additional GM-CSF and IL-4 as above on day 4 (Guyre et al. 2002). Maturation: iDCs were harvested, washed, and cultured in the presence of LPS (2.5 mg/mL) and CD40L (10 mg/mL), for 24 h at 37°C in VueLife bags. Flow cytometry: Analysis by flow cytometry was completed using a FACSCalibur (Becton Dickinson). iDCs and mDCs were examined by flow cytometry for surface staining with directly conjugated monoclonal antibodies to CD14, CD40, CD45, CD80, CD86, Mannose Receptor, and MHC Class I and II molecules using a mAb against CD11c as a lineage-specific gate.
Cell lysis and immunoprecipitation
DCs were washed three times with ice-cold 1× phosphate-buffered saline and resuspended in 1 mL of ice-cold buffer A (10 mM Tris-HCl at pH 7.6, 1 mM KAc, 1.5 mM MgAc, 2 mM DTT, 10 μL/mL HALT protease inhibitor). Cells were lysed with a Teflon pestle homogenizer at 1500 rpm and centrifuged at 12,000g for 10 min to pellet the nuclei; the supernatant was collected, and protein was quantified using a BCA assay. For immunoblotting, cytoplasmic lysates were boiled in 2× loading buffer for 3 min, resolved by SDS-PAGE, and electrotransferred to nitrocellulose. Immunoblotting was performed for TTP using the CARP-3 antibody (a generous gift from William Rigby) and GAPDH (6C5; American Research Products).
For all immunoprecipitations, equal quantities of protein from iDCs and mDCs were used. TTP RNA complexes were recovered by immunoprecipitation using an Anti-TTP (CARP-3) antibody. Affinity CARP-3 or pre-immune serum was bound to 1 mg of protein A–Sepharose beads by incubating overnight at 4°C with continuous rotation in IP buffer (10 mM Tris-HCl at pH 7.6, 1.5 mM MgCl2, 100 mM NaCl, 0.5% Triton X-100, 10 μL/mL HALT protease inhibitor); unbound material was removed with three washes with 500 μL of IP buffer. In order to remove proteins and RNAs that might bind nonspecifically to the protein A–Sepharose or antibody, each lysate was pre-cleared by incubating with the pre-immune serum bound beads for 2 h at 4°C. The pre-immune beads were pelleted, and the supernatant was removed and added to the CARP-3 antibody beads, which were incubated for 2 h at 4°C with continuous rotation. Anti-CARP3 beads and their bound complexes were recovered by centrifugation and washed six times with IP buffer. RNA was isolated from the immunoprecipitate with Trizol (Invitrogen) and used either for GeneChip analysis or real-time PCR.
Affymetrix GeneChip screen and analysis
U133A Affymetrix GeneChips, representing 22,284 genes, were hybridized according to the manufacturer's protocol in the Dartmouth Microarray Core facility. Affymetrix data from the four GeneChips (two iDC, two mDC) were analyzed using methods previously described for analysis of histone stem–loop-binding protein (SLBP) targets, with modifications for application to Affymetrix GeneChips (Townley-Tilson et al. 2006). Affymetrix control spots (n = 68) were removed from the set of genes and not considered further. Each array was normalized by linear scaling such that the median value on each array was the same. The genes on each array were first sorted by their net signal intensity. Each gene on the array was then assigned a percentile rank based on its net signal intensity relative to all other genes on the array. Sorting by percentile rank resulted in genes with the greatest net intensity receiving the highest percentile rank. The percentile ranks were averaged between the two iDC and mDC experiments to produce a mean percentile rank for every gene in the two experiments; the distributions are presented graphically as a histogram with genes segregated into bin classes based on their percentile ranks (see Fig. 2). The graphical representation shows a bimodal distribution. A cutoff was established that divided the two distributions. Significantly enriched genes were defined as genes to the right of the bimodal distribution (percentile rank > 95.92%) (Lieb et al. 2001; Townley-Tilson et al. 2006). Gene Ontology (GO) TermFinder was used to determine significantly over-represented biological processes (Boyle et al. 2004), and these results are displayed graphically with MatLab software using the net signal intensity for all genes receiving a GO Biological Process annotation versus the net signal intensity of all genes on the microarray (Awad et al. 2004). Java TreeView software was used to display a color-coded heat map image of the net signal intensity values of all genes on the microarray (Saldanha 2004).
Real-Time PCR
Following Trizol extraction, mRNA was quantified by spectrophotometry and treated with amplification grade RNase free DNase (Invitrogen). Reverse transcription was performed using Superscript II (Invitrogen), 100 ng of mRNA, and oligo d(T) according to the manufacturer's protocol. For real-time PCR, human primer pairs were purchased from Superarray, along with the 2× RT2 Real-Time SYBR Green (Superarray). Reactions were performed in triplicate according to the manufacturer's protocol.
Transient transfections and luciferase assays
Orientation of the cDNA inserts and the integrity of the DNA sequences were confirmed by sequencing using the ABI Prism Dye Terminator Cycle Sequencing kit (Perkins-Elmer Corp.), and searched using the BLAST search program against the published sequence on the NCBI Database (Altschul et al. 1990). The pcDNA 3.1 His-C-TTP (TTP), ZnFM, and murine TTP expression constructs were generated as previously described (Brooks et al. 2002, 2004; Rigby et al. 2005). The pGL3 luciferase constructs contain the full-length DUSP1 3′-UTR nucleotides (Accession NM_004417), IDO 3′-UTR nucleotides 1326–1655 (Accession NM_002164), and SOD2 3′-UTR nucleotides 673–1026 (Accession NM_000636). The CD86 3′-UTR nucleotides 1119–2797 (Accession NM_006889) were cloned in two parts as indicated in the text. Two pGL3 MHC Class I-F 3′-UTR (Accession NM_018950) constricts were made: full-length (1090–1188) and MHC Class I-F Binding Site (1090–1126). Five pGl3 MHC Class I-B 3′-UTR (NM_005514) constructs were made: full-length (999–1518), Delta 1 (999–1103 and 1252–1518), Delta 2 (999–1251 and 1323–1518), Delta 3 (999–1323 and 1456–1518), and Delta 4 (999–1103, 1252–1323, 1456–1518).
Human embryonic kidney (HEK) 293 cell and RAW 264.7 cell transfections were performed as previously described (Brooks et al. 2002, 2004; Rigby et al. 2005), using 1 μg of luciferase construct and 25 ng of TTP expression construct or pcDNA 3.1. Transfections were performed in triplicate with at least four experiments. Cells were lysed 24 h after serum addition in 100 μL of 1× luciferase lysis buffer (Promega), and 20 μL of each sample was read in a luminometer according to the manufacturer's protocol. For each luciferase vector, a two-tailed t-test was performed comparing the effect of TTP of TTP Zn-FM co-transfection with pcDNA3.1 transfection. Unless otherwise indicated, error bars represent the standard error of the mean.
SUPPLEMENTAL DATA
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
This work was supported by a Merit Review and a Hitchcock Foundation Award to S.A.B. M.L.W. was supported by HHMI Biomedical Research Support Award #76200-560801 to Dartmouth College. We thank Khalid S.A. Khabar for advice on the ARE database and Michael Fanger for support in the generation of the human dendritic cells.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.748408.
REFERENCES
- Allantaz, F., Chaussabel, D., Stichweh, D., Bennett, L., Allman, W., Mejias, A., Ardura, M., Chung, W., Wise, C., Palucka, K., et al. Blood leukocyte microarrays to diagnose systemic onset juvenile idiopathic arthritis and follow the response to IL-1 blockade. J. Exp. Med. 2007;204:2131–2144. doi: 10.1084/jem.20070070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- André, P., Gerber, A.P., Luschnig, S., Krasnow, M.A., Brown, P.O., Herschlag, D. Genome-wide identification of mRNAs associated with the translational regulator PUMILIO in Drosophila melanogaster . Proc. Natl. Acad. Sci. 2006;103:4487–4492. doi: 10.1073/pnas.0509260103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awad, I.A., Rees, C.A., Hernandez-Boussard, T., Ball, C.A., Sherlock, G. Caryoscope: An Open Source Java application for viewing microarray data in a genomic context. BMC Bioinformatics. 2004;15:151. doi: 10.1186/1471-2105-5-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakheet, T., Williams, B.R., Khabar, K.S. ARED 2.0: An update of AU-rich element mRNA database. Nucleic Acids Res. 2003;31:421–423. doi: 10.1093/nar/gkg023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakheet, T., Williams, B.R.G., Khabar, K.S. ARED 3.0: The large and diverse AU-rich transcriptome. Nucleic Acids Res. 2006;34:D111–D114. doi: 10.1093/nar/gkj052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banchereau, J., Steinman, R.M. Dendritic cells and the control of immunity. Nature. 1998;392:245–252. doi: 10.1038/32588. [DOI] [PubMed] [Google Scholar]
- Battle, D.J., Doudna, J.A. The stem–loop binding protein forms a highly stable and specific complex with the 3′ stem–loop of histone mRNAs. RNA. 2001;7:123–132. doi: 10.1017/s1355838201001820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutaud, O., Dixon, D.A., Oates, J.A., Sawaoka, H. Tristetraprolin binds to the COX-2 mRNA 3′-untranslated region in cancer cells. Adv. Exp. Med. Biol. 2003;525:157–160. doi: 10.1007/978-1-4419-9194-2_32. [DOI] [PubMed] [Google Scholar]
- Boyle, E.I., Weng, S., Gollub, J., Jin, H., Botstein, D., Cherry, J.M., Sherlock, G. GO∷TermFinder—Open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics. 2004;20:3710–3715. doi: 10.1093/bioinformatics/bth456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun, D., Longman, R.S., Albert, M.L. A two-step induction of indoleamine 2,3 dioxygenase (IDO) activity during dendritic-cell maturation. Blood. 2005;106:2375–2381. doi: 10.1182/blood-2005-03-0979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer, B.Y., Malicka, J., Blackshear, P.J., Wilson, G.M. RNA sequence elements required for high affinity binding by the zinc finger domain of tristetraprolin: conformational changes coupled to the bipartite nature of Au-rich MRNA-destabilizing motifs. J. Biol. Chem. 2004;279:27870–27877. doi: 10.1074/jbc.M402551200. [DOI] [PubMed] [Google Scholar]
- Brooks, S.A., Connolly, J.E., Diegel, R.J., Fava, R.A., Rigby, W.F. Analysis of the function, expression, and subcellular distribution of human tristetraprolin. Arthritis Rheum. 2002;46:1362–1370. doi: 10.1002/art.10235. [DOI] [PubMed] [Google Scholar]
- Brooks, S.A., Connolly, J.E., Rigby, W.F. The role of mRNA turnover in the regulation of tristetraprolin expression: Evidence for an extracellular signal-regulated kinase-specific, AU-rich element-dependent, autoregulatory pathway. J. Immunol. 2004;172:7263–7271. doi: 10.4049/jimmunol.172.12.7263. [DOI] [PubMed] [Google Scholar]
- Carballo, E., Gilkeson, G.S., Blackshear, P.J. Bone marrow transplantation reproduces the tristetraprolin-deficiency syndrome in recombination activating gene-2 −/−mice. Evidence that monocyte/macrophage progenitors may be responsible for TNFα overproduction. J. Clin. Invest. 1997;100:986–995. doi: 10.1172/JCI119649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carballo, E., Lai, W.S., Blackshear, P.J. Evidence that tristetraprolin is a physiological regulator of granulocyte-macrophage colony-stimulating factor messenger RNA deadenylation and stability. Blood. 2000;95:1891–1899. [PubMed] [Google Scholar]
- Carlson, J.M., Chakravarty, A., DeZiel, C.E., Gross, R.H. SCOPE: A web server for practical de novo motif discovery. Nucleic Acids Res. 2007;35:W259–W264. doi: 10.1093/nar/gkm310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarty, A., Carlson, J.M., Khetani, R.S., Gross, R.H. A novel ensemble learning method for de novo computational identification of DNA binding sites. BMC Bioinformatics. 2007;8:249. doi: 10.1186/1471-2105-8-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C.Y., Chen, T.M., Shyu, A.B. Interplay of two functionally and structurally distinct domains of the c-fos AU-rich element specifies its mRNA-destabilizing function. Mol. Cell. Biol. 1994;14:416–426. doi: 10.1128/mcb.14.1.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, P., Li, J., Barnes, J., Kokkonen, G.C., Lee, J.C., Liu, Y. Restraint of proinflammatory cytokine biosynthesis by mitogen-activated protein kinase phosphatase-1 in lipopolysaccharide-stimulated macrophages. J. Immunol. 2002;169:6408–6416. doi: 10.4049/jimmunol.169.11.6408. [DOI] [PubMed] [Google Scholar]
- Cramton, S.E., Laski, F.A. string of pearls encodes Drosophila ribosomal protein S2, has Minute-like characteristics, and is required during oogenesis. Genetics. 1994;137:1039–1048. doi: 10.1093/genetics/137.4.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong, E.C., Smits, H.H., Kapsenberg, M.L. Dendritic cell-mediated T cell polarization. Springer Semin. Immunopathol. 2005;26:289–307. doi: 10.1007/s00281-004-0167-1. [DOI] [PubMed] [Google Scholar]
- Deleault, K.M., Skinner, S.J., Brooks, S.A. Tristetraprolin regulates TNF TNF-α mRNA stability via a proteasome dependent mechanism involving the combined action of the ERK and p38 pathways. Mol. Immunol. 2008;45:13–24. doi: 10.1016/j.molimm.2007.05.017. [DOI] [PubMed] [Google Scholar]
- Gerber, A.P., Herschlag, D., Brown, P.O. Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLoS Biol. 2004;2:e79. doi: 10.1371/journal.pbio.0020079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyre, C.A., Fisher, J.L., Waugh, M.G., Wallace, P.K., Tretter, C.G., Ernstoff, M.S., Barth R.J., Jr Advantages of hydrophobic culture bags over flasks for the generation of monocyte-derived dendritic cells for clinical applications. J. Immunol. Methods. 2002;262:85–94. doi: 10.1016/s0022-1759(02)00015-7. [DOI] [PubMed] [Google Scholar]
- Hammer, M., Mages, J., Dietrich, H., Servatius, A., Howells, N., Cato, A.C.B., Lang, R. Dual specificity phosphatase 1 (DUSP1) regulates a subset of LPS-induced genes and protects mice from lethal endotoxin shock. J. Environ. Monit. 2005a;203:15–20. doi: 10.1084/jem.20051753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer, M., Mages, J., Dietrich, H., Schmitz, F., Striebel, F., Murray, P.J., Wagner, H., Lang, R. Control of dual specificity phosphatase-1 (DUSP1) expression in activated macrophages by interleukin-10 (IL-10) Eur. J. Immunol. 2005b;35:2991–3001. doi: 10.1002/eji.200526192. [DOI] [PubMed] [Google Scholar]
- Hart, K., Klein, T., Wilcox, M. A Minute encoding a ribosomal protein enhances wing morphogenesis mutants. Mech. Dev. 1993;43:101–110. doi: 10.1016/0925-4773(93)90028-v. [DOI] [PubMed] [Google Scholar]
- Kasai, H., Nadano, D., Hidaka, E., Higuchi, K., Kawakubo, M., Sato, T.A., Nakayama, J. Differential expression of ribosomal proteins in human normal and neoplastic colorectum. J. Histochem. Cytochem. 2003;51:567–574. doi: 10.1177/002215540305100502. [DOI] [PubMed] [Google Scholar]
- Keene, J.D., Lager, P.J. Post-transcriptional operons and regulons co-ordinating gene expression. Chromosome Res. 2005;13:327–337. doi: 10.1007/s10577-005-0848-1. [DOI] [PubMed] [Google Scholar]
- Keene, J.D., Tenenbaum, S.A. Eukaryotic mRNPs may represent post-transcriptional operons. Mol. Cell. 2002;9:1161–1167. doi: 10.1016/s1097-2765(02)00559-2. [DOI] [PubMed] [Google Scholar]
- Kobayashi, T., Sasaki, Y., Oshima, Y., Yamamoto, H., Mita, H., Suzuki, H., Toyota, M., Tokino, T., Itoh, F., Imai, K., et al. Activation of the ribosomal protein L13 gene in human gastrointestinal cancer. Int. J. Mol. Med. 2006;18:161–170. [PubMed] [Google Scholar]
- Kontoyiannis, D., Pasparakis, M., Pizarro, T.T., Cominelli, F., Kollias, G. Impaired on/off regulation of TNF biosynthesis in mice lacking TNF AU-rich elements: Implications for joint and gut-associated immunopathologies. Immunity. 1999;10:387–398. doi: 10.1016/s1074-7613(00)80038-2. [DOI] [PubMed] [Google Scholar]
- Kuchtey, J., Chefalo, P.J., Gray, R.C., Ramachandra, L., Harding, C.V. Enhancement of dendritic cell antigen cross-presentation by CpG DNA involves type I IFN and stabilization of class I MHC mRNA. J. Immunol. 2005;175:2251. doi: 10.4049/jimmunol.175.4.2244. [DOI] [PubMed] [Google Scholar]
- Lai, W.S., Kennington, E.A., Blackshear, P.J. Interactions of CCCH zinc finger proteins with mRNA: Nonbinding tristetraprolin mutants exert an inhibitory effect on degradation of AU-rich element-containing mRNAs. J. Biol. Chem. 2002;277:9606–9613. doi: 10.1074/jbc.M110395200. [DOI] [PubMed] [Google Scholar]
- Lai, W.S., Parker, J.S., Grissom, S.F., Stumpo, D.J., Blackshear, P.J. Novel mRNA targets for tristetraprolin (TTP) identified by global analysis of stabilized transcripts in TTP-deficient fibroblasts. Mol. Cell. Biol. 2006;26:9196–9208. doi: 10.1128/MCB.00945-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieb, J.D., Liu, X., Botstein, D., Brown, P.O. Promoter-specific binding of Rap1 revealed by genome-wide maps of protein–DNA association. Nat. Genet. 2001;28:327–334. doi: 10.1038/ng569. [DOI] [PubMed] [Google Scholar]
- Linker, K., Pautz, A., Fechir, M., Hubrich, T., Greeve, J., Kleinert, H. Involvement of KSRP in the post-transcriptional regulation of human iNOS expression-complex interplay of KSRP with TTP and HuR. Nucleic Acids Res. 2005;33:4813–4827. doi: 10.1093/nar/gki797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahnke, K., Schmitt, E., Bonifaz, L., Enk, A.H., Jonuleit, H. Immature, but not inactive: The tolerogenic function of immature dendritic cells. Immunol. Cell Biol. 2002;80:477–483. doi: 10.1046/j.1440-1711.2002.01115.x. [DOI] [PubMed] [Google Scholar]
- McIlroy, D., Tanguy-Royer, S., Le Meur, N., Guisle, I., Royer, P.J., Leger, J., Meflah, K., Gregoire, M. Profiling dendritic cell maturation with dedicated microarrays. J. Leukoc. Biol. 2005;78:794–803. doi: 10.1189/jlb.0105029. [DOI] [PubMed] [Google Scholar]
- Mellor, A.L., Munn, D.H. IDO expression by dendritic cells: Tolerance and tryptophan catabolism. Nat. Rev. Immunol. 2004;4:762–774. doi: 10.1038/nri1457. [DOI] [PubMed] [Google Scholar]
- Moore, M.J. From birth to death: The complex lives of eukaryotic mRNAs. Science. 2007;309:1514–1518. doi: 10.1126/science.1111443. [DOI] [PubMed] [Google Scholar]
- Ogilvie, R.L., Abelson, M., Hau, H.H., Vlasova, I., Blackshear, P.J., Bohjanen, P.R. Tristetraprolin down-regulates IL-2 gene expression through AU-rich element-mediated mRNA decay. J. Immunol. 2005;174:953–961. doi: 10.4049/jimmunol.174.2.953. [DOI] [PubMed] [Google Scholar]
- Penalva, L.O., Tenenbaum, S.A., Keene, J.D. Gene expression analysis of messenger RNP complexes. Methods Mol. Biol. 2004;257:125–134. doi: 10.1385/1-59259-750-5:125. [DOI] [PubMed] [Google Scholar]
- Phillips, K., Kedersha, N., Shen, L., Blackshear, P.J., Anderson, P. Arthritis suppressor genes TIA-1 and TTP dampen the expression of tumor necrosis factor alpha, cyclooxygenase 2, and inflammatory arthritis. Proc. Natl. Acad. Sci. 2004;101:2011–2016. doi: 10.1073/pnas.0400148101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proietto, A.I., O'Keeffe, M., Gartlan, K., Wright, M.D., Shortman, K., Wu, L., Lahoud, M.H. Differential production of inflammatory chemokines by murine dendritic cell subsets. Immunobiology. 2004;209:163–172. doi: 10.1016/j.imbio.2004.03.002. [DOI] [PubMed] [Google Scholar]
- Rieser, C., Ramoner, R., Böck, G., Deo, Y.M., Höltl, L., Bartsch, G., Thurnher, M. Human monocyte-derived dendritic cells produce macrophage colony-stimulating factor: Enhancement of c-fms expression by interleukin-10. Eur. J. Immunol. 1998;28:2283–2288. doi: 10.1002/(SICI)1521-4141(199808)28:08<2283::AID-IMMU2283>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Rigby, W., Roy, K., Collins, J., Rigby, S., Connolly, J., Bloch, D., Brooks, S. Structure/function analysis of tristetraprolin (TTP): p38 stress-activated protein kinase and lipopolysaccharide stimulation do not alter TTP function. J. Immunol. 2005;174:7883–7893. doi: 10.4049/jimmunol.174.12.7883. [DOI] [PubMed] [Google Scholar]
- Saldanha, A.J. Java Treeview—Extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- Sauer, I., Schaljo, B., Vogl, C., Gattermeier, I., Kolbe, T., Müller, M., Blackshear, P.J., Kovarik, P. Interferons limit inflammatory responses by induction of tristetraprolin. Blood. 2006;107:4790–4797. doi: 10.1182/blood-2005-07-3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd, E.G., Zhao, Q., Welty, S.E., Hansen, T.N., Smith, C.V., Liu, T. The function of mitogen-activated protein kinase phosphatase-1 in peptidoglycan-stimulated macrophages. J. Biol. Chem. 2004;279:54023–54031. doi: 10.1074/jbc.M408444200. [DOI] [PubMed] [Google Scholar]
- Stoecklin, G., Ming, X.F., Looser, R., Moroni, C. Somatic mRNA turnover mutants implicate tristetraprolin in the interleukin-3 mRNA degradation pathway. Mol. Cell. Biol. 2000;20:3753–3763. doi: 10.1128/mcb.20.11.3753-3763.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoecklin, G., Stubbs, T., Kedersha, N., Wax, S., Rigby, W.F., Blackwell, T.K., Anderson, P. MK2-induced tristetraprolin:14–3–3 complexes prevent stress granule association and ARE-mRNA decay. EMBO J. 2004;23:1313–1324. doi: 10.1038/sj.emboj.7600163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenenbaum, S.A., Carson, C.C., Lager, P.J., Keene, J.D. Identifying mRNA subsets in messenger ribonucleoprotein complexes by using cDNA arrays. Proc. Natl. Acad. Sci. 2000;97:14085–14090. doi: 10.1073/pnas.97.26.14085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, E.A., Alvarez, C.E., Sutcliffe, J.G. Evolutionarily distinct classes of S27 ribosomal proteins with differential mRNA expression in rat hypothalamus. J. Neurochem. 2000;74:2259–2267. doi: 10.1046/j.1471-4159.2000.0742259.x. [DOI] [PubMed] [Google Scholar]
- Townley-Tilson, W.H.D., Pendergrass, S.A., Marzluff, W.F., Whitfield, M.L. Genome-wide analysis of mRNAs bound to the histone stem–loop binding protein. RNA. 2006;12:1853–1867. doi: 10.1261/rna.76006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tureci, O., Bian, H., Nestle, F.O., Raddrizzani, L., Rosinski, J.A., Tassis, A., Hilton, H., Walstead, M., Sahin, U., Hammer, J. Cascades of transcriptional induction during dendritic cell maturation revealed by genome-wide expression analysis. FASEB J. 2003;17:836–847. doi: 10.1096/fj.02-0724com. [DOI] [PubMed] [Google Scholar]
- von Bubnoff, D., Bausinger, H., Matz, H., Koch, S., Häcker, G., Takikawa, O., Bieber, T., Hanau, D., de la Salle, H. Human epidermal Langerhans cells express the immunoregulatory enzyme indoleamine 2,3-dioxygenase. J. Invest. Dermatol. 2004;123:298–304. doi: 10.1111/j.0022-202X.2004.23217.x. [DOI] [PubMed] [Google Scholar]
- Wallace, P.K., Romet-Lemonne, J.L., Chokri, M., Kasper, L.H., Fanger, M.W., Fadul, C.E. Production of macrophage-activated killer cells for targeting of glioblastoma cells with bispecific antibody to FcγRI and the epidermal growth factor receptor. Cancer Immunol. Immunother. 2000;49:493–503. doi: 10.1007/s002620000142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson, K.L., Konrad, K.D., Woods, D.F., Bryant, P.J. Drosophila homolog of the human S6 ribosomal protein is required for tumor suppression in the hematopoietic system. Proc. Natl. Acad. Sci. 1992;89:11302–11306. doi: 10.1073/pnas.89.23.11302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worthington, M.T., Pelo, J.W., Sachedina, M.A., Applegate, J.L., Arseneau, K.O., Pizarro, T.T. RNA binding properties of the AU-rich element-binding recombinant Nup475/TIS11/tristetraprolin protein. J. Biol. Chem. 2002;277:48558–48564. doi: 10.1074/jbc.M206505200. [DOI] [PubMed] [Google Scholar]
- Yamamoto, T. Molecular mechanism of monocyte predominant infiltration in chronic inflammation: Mediation by a novel monocyte chemotactic factor, S19 ribosomal protein dimer. Pathol. Int. 2000;50:863–871. doi: 10.1046/j.1440-1827.2000.01132.x. [DOI] [PubMed] [Google Scholar]
- Zhao, Q., Shepherd, E.G., Manson, M.E., Nelin, L.D., Sorkin, A., Liu, Y. The role of mitogen-activated protein kinase phosphatase-1 in the response of alveolar macrophages to lipopolysaccharide: Attenuation of proinflammatory cytokine biosynthesis via feedback control of p38. J. Biol. Chem. 2005;280:8108. doi: 10.1074/jbc.M411760200. [DOI] [PubMed] [Google Scholar]
- Zubiaga, A.M., Belasco, J.G., Greenberg, M.E. The nonamer UUAUUUAUU is the key AU-rich sequence motif that mediates mRNA degradation. Mol. Cell. Biol. 1995;15:2219–2230. doi: 10.1128/mcb.15.4.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]