Abstract
In Caenorhabditis elegans, specific Argonaute proteins are dedicated to the RNAi and microRNA pathways. To uncover how the precise Argonaute selection occurs, we designed dsRNA triggers containing both miRNA and siRNA sequences. While dsRNA carrying nucleotides mismatches can only enter the miRNA pathway, a fully complementary dsRNA successfully rescues let-7 miRNA function and initiates silencing by RNAi. We demonstrated that RDE-1 is essential for RNAi induced by the perfectly paired trigger, yet is not required for silencing by the let-7 miRNA. In contrast, ALG-1/ALG-2 are required for the miRNA function, but not for the siRNA-directed gene silencing. Finally, a dsRNA containing a bulged miRNA and a perfectly paired siRNA can enter both pathways suggesting that the sorting of small RNAs occurs after that the dsRNA trigger has been processed by Dicer. Thus, our data suggest that the selection of Argonaute proteins is affected by two molecular features: (1) the structure of the small RNA duplex; and (2) the Argonautes specific characteristics.
Keywords: Argonaute; ALG-1; ALG-2; RDE-1; RNAi, microRNA
INTRODUCTION
In animals, the RNAi and the microRNA (miRNA) pathways lead to different outcomes. The RNAi pathway degrades specifically the messenger RNA (mRNA), which is fully complementary to the siRNA embedded in the RISC complex, while miRNA, partially complementary to the untranslated region of the mRNA, will not initiate specific target cleavage but instead will abrogate protein synthesis. Among the cellular factors associated with these processes, members of the Argonaute gene family have been shown to be exclusively associated with either the miRNA or the RNAi pathway. In Drosophila melanogaster, Ago1 is associated with miRNA (Okamura et al. 2004) and Ago2 is essential for the RNAi pathway (Hammond et al. 2001). In the nematode Caenorhabditis elegans, genetic studies demonstrated that the Argonaute rde-1 gene is strictly required for RNAi (Tabara et al. 1999) and two Argonaute family members alg-1 and alg-2 are essential for the miRNA pathway but have no role into the RNAi pathway (Grishok et al. 2001). Furthermore, recent biochemical data have revealed that RDE-1 is associated with siRNAs (Yigit et al. 2006) and ALG-1 and ALG-2 are associated with miRNAs (Hutvágner et al. 2004). One reason of this disparity could be that trigger molecules are recognized by specific factors that “imprint” the RNA molecule and guide it to the appropriate Argonaute protein and consequently to the appropriate outcome.
To address the role played by the double-stranded RNA (dsRNA) triggers on the Argonaute selection, we designed different dsRNA molecules containing both miRNA and siRNA. Our analysis suggests that the molecular feature of the dsRNA triggers as well as the molecular characteristics of Argonaute proteins contribute to the proper selection of Argonaute in their respective silencing pathways in C. elegans.
RESULTS AND DISCUSSION
A small dsRNA trigger able to enter both miRNA and RNAi pathways
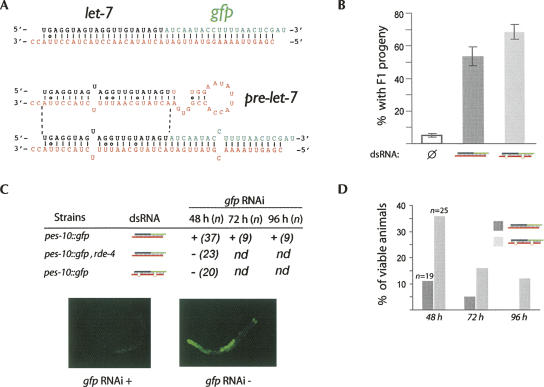
To perform this study, we have generated small dsRNA molecules that contain both the let-7 miRNA sequence and an siRNA fully complementary to the gfp mRNA (Fig. 1A). To monitor the capacity of the chimeric dsRNA molecule to generate a functional miRNA, we used a C. elegans strain that carries a thermosensitive (ts) allele of the let-7 gene (Reinhart et al. 2000). We can then provide functional copies of let-7 miRNA by injecting the chimeric dsRNA into the animals and observe if we can rescue let-7 loss-of-function phenotypes at the nonpermissive temperature. When the let-7ts animals are injected with the dsRNA chimeric trigger and then shifted to the nonpermissive temperature, more than 50% of these animals had progeny (Fig. 1B). In contrast, when noninjected L3 larvae stage animals are shifted to 25°C or when animals are injected with nonrelated dsRNA molecules (data not shown), all of the let-7ts animals display bursting vulva phenotype and only 5% of the shifted animals provide few F1 progeny (Fig. 1B). Thus, our data indicate that a fully complementary double-stranded trigger can efficiently rescue the loss of function of a specific miRNA.
FIGURE 1.
Chimeric dsRNA molecules trigger both microRNA and RNAi pathways in C. elegans. (A) Diagram of chimeric double-stranded RNA triggers. Chemically synthesized RNA oligos were annealed to generate the dsRNA molecule that contains let-7 miRNA (black strand) and an siRNA targeting gfp mRNA (green strand). The presence of a 2 nt 3′ overhang ensures precise cleavage by Dicer to generate functional 21-nt-long let-7 miRNA and GFP siRNA (Han et al. 2006). The Wobble base pairing found in the hairpin let-7 miRNA precursor molecule (Pasquinelli et al. 2000) was transferred into the chimeric dsRNA molecule structure to favor the loading of the let-7 miRNA into the miRISC complex. The asymmetry rule (Schwarz et al. 2003) has also been observed for the design of the portion of the dsRNA molecule that generates the GFP siRNA. To generate the unpaired dsRNA duplex, a portion of the stem of let-7 precursor (pre-let-7) has been reproduced into the dsRNA trigger and the mismatch at the ninth position of the sense strand has also been inserted into the GFP siRNA. (B) dsRNA molecules rescue let-7 miRNA function. The animals shifted to the nonpermissive temperature (25°C) generating more than three F1 progeny are scored as rescued. The error bars represent the 95% confidence interval from three independent experiments where between 20 and 30 animals have been scored. (C) Only the fully complementary chimeric dsRNA molecule can initiate RNAi response. The GFP signal is monitored with a GFP stereomicroscope. The (+) indicates that the RNAi is functional (absence of GFP signal) while (–) indicates that the RNAi is deficient (strong expression of GFP) in animals. Ninety-six hours after the dsRNA injection, the GFP signal is still undetectable under the microscope. The number (n) of animals observed is indicated within the parenthesis. All animals observed for each strain displayed the same GFP level. Representative pictures of injected pes-10∷gfp animals are shown to illustrate a functional (+) or deficient (–) RNAi response. (D) The let-7 miRNA rescue is rapidly attenuated. Viable animals are scored every 24 h after being transferred for 48 h at nonpermissive temperature (25°C). While nearly all animals from the let-7ts strain die after 48 h at nonpermissive temperature from a burst vulva (Reinhart et al. 2000), the phenotype is delayed in animals injected with chimeric dsRNA molecules. All injected animals with fully complementary dsRNA duplex displayed busting vulva phenotype after 96 h. Numbers (n) of animals observed are indicated.
We next examined the potential of the fully complementary dsRNA chimeric molecule to enter the RNAi pathway and initiate a specific RNA silencing response. To address this issue, we used the previously described dsRNA trigger that contains an siRNA that is fully complementary to a 21 nucleotide (nt) region of the gfp mRNA (Fig. 1A). When transgenic animals carrying the gfp gene are injected with the trigger chimera, the green fluorescence signal is rapidly silenced in the animal body (Fig. 1C). To confirm that the RNAi pathway mediates the GFP silencing, we injected the dsRNA molecule in animals carrying a null allele of the rde-4 gene, an essential component of the RNAi in C. elegans that binds dsRNA molecules (Tabara et al. 2002; Parker et al. 2006). In these animals, the injection of the dsRNA molecule did not affect the fluorescence level observed (Fig. 1C), indicating that the GFP siRNA produced from the processing of the chimeric dsRNA is entering the RNAi pathway to silence efficiently the targeted mRNA.
We therefore conclude that we have successfully generated a dsRNA trigger that can effectively rescue both miRNA function and specifically silence an mRNA through the RNAi pathway.
Mismatches in the dsRNA trigger increase the let-7 rescue but circumvent the RNAi capacity
Next, we addressed whether the nature of the dsRNA trigger may affect its capacity to be introduced into the miRNA and RNAi pathway. We then modified the fully complementary dsRNA molecule to generate a dsRNA trigger containing a single mismatch in both let-7 and GFP siRNA duplexes that mimic the one found in the stem portion of the let-7 miRNA precursor (Fig. 1A). As opposed to noninjected animals, 69% of the animal population injected with this molecule can reach the L4-adult transition and give many F1 progeny (Fig. 1B). In comparison with the fully complementary molecule, animals injected with the dsRNA trigger containing mismatches led to a better rescue of let-7 miRNA (Fig. 1B). On the other hand, the transgenic animals carrying GFP gene, and injected with the dsRNA trigger containing mismatches, did not display any RNAi response contrasting the silencing observed in animals injected with the fully complementary dsRNA molecules (Fig. 1C). The increased rescue activity of a dsRNA trigger that can only enter the miRNA pathway suggests that RNAi and miRNA initiating complexes may compete for loading their respective Argonautes with small RNAs when the dsRNA trigger carried fully base-paired nucleotides (see model in Fig. 4, below).
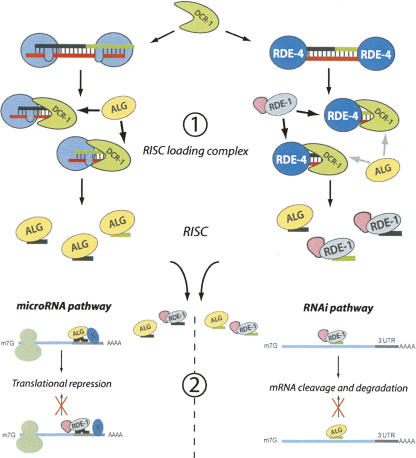
FIGURE 4.
Model for the Argonaute proteins selection in C. elegans. The sequence complementarity of the double-stranded (ds)RNA duplex represents the first step that influences the Argonaute selection (1). Fully base-paired dsRNA molecules are recognized by the dsRNA binding protein RDE-4 that recruits Dicer (DCR-1) to induce dsRNA trigger cleavage (right panel). The presence of RDE-4 on the small RNA duplex processed by DCR-1 might favor the recruitment of RDE-1 over ALGs into the RISC loading complex. Therefore, the majority of RISC complex produced by complementary siRNA duplexes are associated to RDE-1. In contrast, dsRNA duplexes containing mispaired nucleotides are not bound by RDE-4 but instead by another dsRNA binding protein that will favor the recruitment of ALGs to the RISC complex (left panel). Once Argonaute/small RNA complexes are formed, a second step of Argonaute selection occurs at the target level (2). The functionality of the Argonaute protein in either the miRNA pathway (left panel) or the RNAi pathway (right panel) may be dictated by their enzymatic activity as observed in Drosophila (Forstemann et al. 2007) and/or can be mediated by specific interactions with cellular factors associated with the targeted mRNA (X factor on the left panel) or by direct interaction with the Argonaute protein prior to the interaction with the mRNA (pink object). Thus, it is only when the small RNA/Argonaute complex is associated to the target and interacts with specific factors that the translational repression or the mRNA cleavage and degradation occur.
In addition, we also observed that the rescue of the let-7ts animals with dsRNA molecules only lasts less than 96 h after injection (72 h for the fully complementary dsRNA and 96 h with the dsRNA containing mismatches; Fig. 1D). In contrast, the fully complementary dsRNA molecule initiates an RNAi response that can be maintained throughout the life span of the injected animals (Fig. 1C; data not shown). These data suggest that the miRNA complex has an extremely rapid turnover while the siRNA complex may act during multiple rounds of mRNA cleavage. Thus, the cleavage may initiate amplification of the RNAi response by RNA-dependent RNA polymerase (Pak and Fire 2007; Sijen et al. 2007). Accordingly, we observed that the RNA-dependent RNA polymerase rrf-1 is not required for let-7 miRNA rescue (Fig. 2A) but is important for RNAi (Fig. 2B).
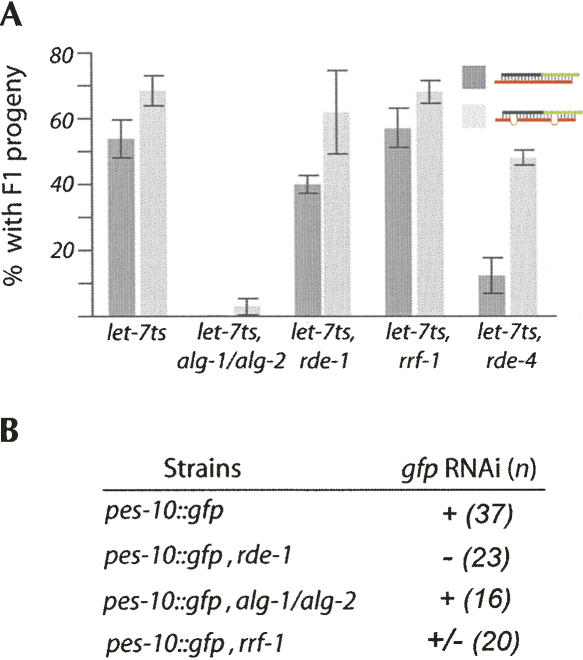
FIGURE 2.
The Argonaute proteins are sorted properly into their respective pathways. (A) The rescue of let-7 loss of function by both dsRNA triggers necessitates ALGs. Once injected by indicated dsRNA triggers, animals are scored at nonpermissive temperature for the presence of F1 progeny (greater than three). The error bars represent the 95% confidence interval (as in Fig. 1B). (B) The RNAi initiated by the fully complementary trigger requires rde-1 and necessitates partially the RNA-dependent RNA polymerase rrf-1 (GFP silencing is noted but not as efficient as in the pes-10∷gfp animals [see Fig. 1C]). Numbers (n) of animals observed are indicated. All animals observed for each strain displayed the same GFP level.
From these observations, we conclude that the presence of unpaired nucleotides in the dsRNA trigger abrogates its capacity to enter the RNAi pathway but increases its capacity to initiate the miRNA pathway.
The dsRNA trigger is not the only molecular cue that guides the specific requirement of Argonaute proteins
In RNAi, a perfect base-paired dsRNA molecule initiates the targeted mRNA degradation. On the other hand, the imperfect stem–loop structure of the miRNA leads to translational inhibition. In previous studies, we have shown that the Argonaute RDE-1 interacts with siRNAs (Yigit et al. 2006) while ALG-1 and ALG-2 Argonaute proteins are associated with the let-7 miRNA (Hutvágner et al. 2004). Therefore, the distinctive outcomes of these two RNA silencing pathways may be mediated by the molecular feature of their triggers (bulged or perfectly paired dsRNAs) or may be associated with their specific Argonaute proteins. To address this possibility, we used the fully symmetrical and perfectly paired dsRNA trigger that efficiently produced miRNA as well as RNAi responses (Fig. 1B, C). We thus generated let-7ts strains carrying either nonfunctional rde-1 gene or alg-1 and alg-2 genes and addressed whether or not rde-1 and/or alg-1/alg-2 are important for the let-7 rescue by the chimeric dsRNA molecule. Despite the fact that the dsRNA provided is perfectly base-paired, the rescue of the let-7 miRNA does not require RDE-1 but requires the presence of ALGs (Fig. 2A). Interestingly, we observed that in contrast to the chimeric molecule containing mismatches, the fully base-paired dsRNA necessitates the dsRNA binding protein RDE-4 (Fig. 2A). These data support that RDE-4 is important to promote Dicer cleavage of perfectly base-paired dsRNA trigger (see model in Fig. 4, below).
In parallel, we also tested the Argonaute proteins requirement for the RNAi-induced response by the trigger chimera. When GFP positive animals lacking functional alg-1 and alg-2 genes are injected with the dsRNA trigger, we observe a complete disappearance of the green fluorescent signal as the wild-type animals expressing GFP (Fig. 2B). However, when the animals are lacking functional alleles of the rde-1 gene, the dsRNA trigger does not alter the expression of the green fluorescence protein (Fig. 2B).
Thus, although we are using a dsRNA trigger able to enter both small RNA pathways, they still necessitate their Argonaute proteins, suggesting that RDE-1 and ALGs have molecular characteristics that confer their specific requirements for the RNAi and the miRNA pathway.
The sorting of small RNAs may occur after Dicer cleavage
So far, our data indicate that the presence of mismatched nucleotides affects the capacity of dsRNA triggers to initiate RNAi response and that a perfectly paired dsRNA molecule can effectively enter both miRNA and RNAi pathways but still requires the appropriate Argonaute proteins (ALGs or RDE-1, respectively). Thus, the sorting of small RNAs can be produced by specific dsRNA binding proteins that will commit the dsRNA trigger to either the RNAi or miRNA pathway or they can be sorted after Dicer cleavage.
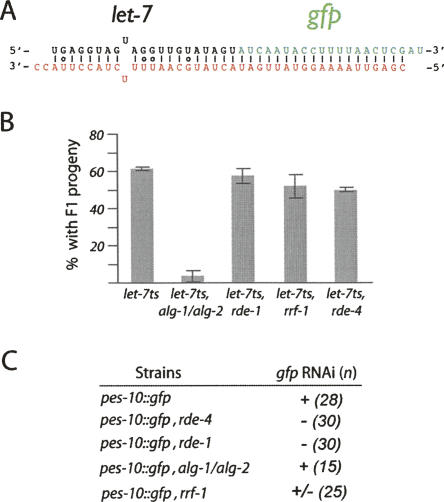
To address whether the sorting of small RNAs occurs prior to or after Dicer mediated maturation of the dsRNA trigger, we generated a dsRNA trigger that contains a bulged let-7 dsRNA and a perfectly paired GFP siRNA (Fig. 3A). As observed with other dsRNA triggers, the new dsRNA trigger can rescue the let-7 miRNA loss-of-function phenotype observed when the let-7ts allele is shifted to the nonpermissive temperature (Fig. 3B). Interestingly, even if the dsRNA trigger contained unpaired nucleotides, the presence of a perfectly paired siRNA is sufficient to initiate silencing of the targeted gfp mRNA (Fig. 3C). In addition, because the fully base-paired molecule requires RDE-4 as well as the Argonaute ALGs to rescue let-7 function (Fig. 2A), these observations strongly support that the recognition of the dsRNA trigger by dsRNA binding protein does not significantly affect the selection of Argonaute proteins.
FIGURE 3.
Sorting of small RNAs occurs after Dicer cleavage. (A) Diagram of double-stranded RNA trigger containing bulged let-7 miRNA (black strand) and perfectly paired GFP siRNA (green strand). (B) The bulged/perfectly paired dsRNA can rescue let-7 loss of function and requires ALGs. Once injected, animals are scored at nonpermissive temperature for the presence of F1 progeny (greater than three). The error bars represent the 95% confidence interval (as in Fig. 1B). (C) The dsRNA trigger containing perfectly paired siRNA can perform RNAi. The GFP silencing requires rde-1 and necessitates partially the RNA-dependent RNA polymerase rrf-1 (as observed in Fig. 2B). Numbers (n) of animals observed are indicated. All animals observed for each strain displayed the same GFP level.
Therefore, our data indicate that the sorting of small RNAs may occur after Dicer processes the dsRNA trigger in C. elegans, as very recently reported in Drosophila (Forstemann et al. 2007; Tomari et al. 2007).
The dsRNA structures and their distinct molecular features affect Argonaute protein selection
Taken together, our data indicate that the selection of Argonaute proteins occurs at two different levels. First, dsRNA molecules containing mismatches can only enter the miRNA pathway, while perfectly paired dsRNAs are the only molecules generating an Argonaute complex able to perform RNAi. Second, our experiments using a fully complementary dsRNA trigger demonstrate that, although the same molecule enters both miRNA and RNAi pathways, the specific requirement of the Argonaute genes is not altered. Third, when a dsRNA molecule contains a bulged miRNA and perfectly paired siRNA duplexes, it can effectively enter both miRNA and RNAi pathways. Thus, our data strongly suggest that the nature of the dsRNA molecule is not the only step that influences sorting of the Argonaute proteins for miRNA and RNAi pathways but their specific associations with either of these RNA silencing pathways is also influenced by their molecular features.
The Argonaute proteins are essential for all RNA silencing pathways discovered so far in eukaryotes (reviewed in Hutvagner and Simard 2008). The presence of many members of this family suggests specific roles associated with each one of them. In mammals, all four Argonaute proteins can bind miRNAs and siRNAs, but only hAgo2 has the capacity to induce endonucleolytic cleavage of the targeted mRNA (Liu et al. 2004; Meister et al. 2004). This difference may be explained by the fact that only hAgo2 possesses the catalytic residues essential for the “slicing” activity in its Piwi domain (Rivas et al. 2005). In flies, ago1 mutants are defective in the miRNA pathway but not in siRNA-directed cleavage, whereas ago2 mutants are defective in the RNAi pathway but not in miRNA silencing (Okamura et al. 2004).
As observed in flies, C. elegans has distinct requirements for Argonaute proteins in siRNA or miRNA-directed RNA silencing. Genetic studies have demonstrated that the Argonaute RDE-1 is essential for the RNAi pathway (Tabara et al. 1999) while ALG-1 and ALG-2 are important for the miRNA pathway (Grishok et al. 2001). Moreover, the overexpression of Argonautes required for the miRNA pathway cannot rescue the loss of the rde-1 gene for the RNAi response (Yigit et al. 2006). To explain this Argonaute specificity, we hypothesized that the molecular characteristics of dsRNA triggers may be crucial for the Argonaute choice. Our current findings demonstrate that a dsRNA duplex that mimics the unpaired miRNA precursor can influence its capacity to either enter the RNAi or the miRNA pathways. The capacity of a dsRNA duplex structure to partition small RNAs between Argonaute proteins has also been observed in Drosophila (Forstemann et al. 2007; Tomari et al. 2007). Using biochemical assays, they have observed that a single mismatch in the RNA duplex weakens the binding of the Ago2 loading complex (Tomari et al. 2007). Interestingly, it has been recently observed in C. elegans that the presence of mismatches in the dsRNA trigger will favor the binding of ALG-1 to the small RNA instead of the RDE-1 binding (Steiner et al. 2007). Our experiments now indicate that the presence of a single mismatch into the small RNA duplex affects the sorting of small RNAs after the cleavage of the dsRNA trigger by Dicer. We can then envision that binding of the RDE-1 loading complex to a small RNA duplex containing mismatches is less efficient, like the Ago2 loading complex, as observed in Drosophila. Thus, our data, supported by these recent independent experiments, uncover the importance of the dsRNA molecular features to sort functionally distinct Argonaute proteins in animals.
In addition, our data also demonstrate that when a fully complementary dsRNA molecule is used, small RNAs processed from that molecule can enter both miRNA and RNAi pathways. Therefore, the source of the dsRNA trigger does not affect the selective association of Argonaute proteins with either the miRNA or the siRNA pathways. Recent observations in Drosophila suggest that the difference between Ago1 (miRNA pathway) and Ago2 (RNAi pathway) may be explained by the inefficient capacity of Ago1 to induce mRNA cleavage (Forstemann et al. 2007). Therefore, it has been proposed that Ago1 requires mismatches within the seed region of the small RNA duplex to generate a functional RISC complex (Tomari et al. 2007). Despite the fact that the Piwi domains of RDE-1 and ALGs Argonautes display the critical amino acids required for endonucleolytic cleavage (Yigit et al. 2006), the contribution of their cleavage activities to the RISC loading complex has not been demonstrated yet. Contrasting with Ago1 in Drosophila, ALGs proteins can mediate translational repression with an miRNA generate from a perfectly base-paired small RNA duplex (Figs. 1B, 2A).Therefore, our data support that in C. elegans, ALGs can generate a functional RISC complex from small RNA duplexes irrespective of the presence of unpaired nucleotides within their seed regions.
The association of the Argonaute complexes with targeted mRNAs may also contribute to selecting the proper Argonaute protein. The exclusive specificity of Argonaute proteins in C. elegans in the RNAi and miRNA pathways may be attributed to unique molecular partners for RDE-1 and ALGs, as observed for human Ago1 and Ago2 (Meister et al. 2005). These specific interactions may occur prior to or during the period in which the Argonaute/small RNA complex enters in association with the mRNA target (Fig. 4).
Based on our observations and recently published experiments, we propose that the specific selection of Argonaute proteins for the miRNA and RNAi pathways in C. elegans is controlled at two different levels. Initially, the Argonaute RDE-1 favors small RNAs derived from the perfect dsRNA duplex while ALGs favor RNAs produced from the bulged dsRNA duplex (but can still accept small RNAs produced from both types of RNA duplexes). The small RNA binding specificity of these Argonaute proteins can be dictated by their respective loading complexes. For RDE-1, the specificity for the RNAi pathway may come from RDE-4, a double-stranded RNA binding protein found in complex with RDE-1 (Tabara et al. 2002; Duchaine et al. 2006); while ALGs, the Argonautes specific to the miRNA pathway, may be influenced by a yet unknown dsRNA binding protein similar to Drosophila Loquacious (Forstemann et al. 2005). In addition, the presence of a central mismatch in the small RNA duplex may destabilize the RDE-4 interaction as recently observed for R2D2, the dsRNA binding protein associated with the RNAi pathway in Drosophila (Tomari et al. 2007). Once functional Argonaute/small RNA complexes are formed, molecular interactions with specific proteins and/or with the targeted mRNA will influence their capacity to initiate either RNAi or miRNA responses (Fig. 4).
We believe that extensive biochemical studies and complex analysis of the Argonaute ALGs and RDE-1 proteins will explain how they can specifically alter protein synthesis by miRNAs and mediate specific mRNA degradation guided by siRNAs.
MATERIALS AND METHODS
Design and preparation of dsRNA molecules
All RNA oligonucleotides were obtained from Integrated DNA Technologies, Inc. Oligos sequences are the following:
5′-UGAGGUAGUAGGUUGUAUAGUAUCAAUACCUUUUAACUCGAU-3′ (let-7/GFP);
5′-CGAGUUAAAAGGUAUUGAUACUAUACAACCUACUACCUUACC-3′ (let-7/GFP reverse);
5′-CGAGUUAAAACGUAUUGAUACUAUGCAAUUUUCUACCUUACC-3′ (let-7/GFP mismatches reverse); and
5′-CGAGUUAAAAGGUAUUGAUACUAUGCAAUUUUCUACCUUACC-3′ (let-7 bulged/GFP reverse).
To generate dsRNA trigger molecules, complementary oligos were pooled and annealed (boiled at 100°C for 2 min and annealed 2h at 37°C) to obtain a final solution of double-stranded RNA at 2 mg/mL.
RNAi interference and rescued experiments
The let-7 miRNA rescue and RNAi experiments were performed by injecting dsRNA molecules (2 mg/mL) into L3 and L4 larval stage animals, respectively.
For the rescue of let-7 experiment, let-7ts (n2853) strains were injected with various dsRNA molecules at permissive temperature (15°C). After 5 h recovery, animals were isolated and placed at nonpermissive temperature and phenotypes were observed after 48 h post-injection. As control for each let-7 rescue experiment, equal numbers of uninjected animals were also shifted to the nonpermissive temperature (25°C).
For the RNAi assays, various strains expressing GFP driven by pes-10 promoter were injected. The GFP expression was monitored in injected animals after 48 h under the GFP stereomicroscope. Strong GFP silencing is observed in the pharynx and along the animal body (see Fig. 1C).
Worm strains
Animals used to generate the studied strains were: rde-1 (ne300), rde-4 (ne337), rrf-1 (pk1417), and alg-2 (ok304).
To obtain alg-1/alg-2 double mutants, alg-1 RNAi was performed into alg-2 (ok304) and let-7ts (n2853); alg-2 (ok304) as previously described in Grishok et al. (2001). L3 stage animals were injected with dsRNA (50 μg/mL) targeting alg-1 mRNA and maintained at permissive temperature, when required, for 5 h followed by injections of respective dsRNA trigger molecules. For the RNAi experiment, the dsRNA targeting alg-1 was injected in late L4 stage animals in order to bypass the let-7 phenotype observed in alg-1/alg-2 double mutants. alg-1/alg-2 double mutants as previously observed (Grishok et al. 2001) still produced dead embryos.
ACKNOWLEDGMENTS
We thank Dr. Craig Mello for discussions. We also thank Rhea Utley, Jacques Côté, Gyorgy Hutvagner, Jean-Yves Masson, and members of the laboratory for comments on the manuscript. Some nematode strains were provided by the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resources (NCRR). This work has been funded in part by the Canadian Institutes of Health Research (CIHR) and the Natural Sciences and Engineering Research Council of Canada (NSERC). M.-E.L.B. was supported by a fellowship from the NSERC and M.J.S. is a CIHR New Investigator.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.901908.
REFERENCES
- Duchaine, T.F., Wohlschlegel, J.A., Kennedy, S., Bei, Y., Conte D., Jr, Pang, K., Brownell, D.R., Harding, S., Mitani, S., Ruvkun, G., et al. Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell. 2006;124:343–354. doi: 10.1016/j.cell.2005.11.036. [DOI] [PubMed] [Google Scholar]
- Forstemann, K., Horwich, M.D., Wee, L., Tomari, Y., Zamore, P.D. Drosophila microRNAs are sorted into functionally distinct Argonaute complexes after production by Dicer-1. Cell. 2007;130:287–297. doi: 10.1016/j.cell.2007.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forstemann, K., Tomari, Y., Du, T., Vagin, V.V., Denli, A.M., Bratu, D.P., Klattenhoff, C., Theurkauf, W.E., Zamore, P.D. Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS Biol. 2005;3:e236. doi: 10.1371/journal.pbio.0030236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishok, A., Pasquinelli, A.E., Conte, D., Li, N., Parrish, S., Ha, I., Baillie, D.L., Fire, A., Ruvkun, G., Mello, C.C. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- Hammond, S.M., Boettcher, S., Caudy, A.A., Kobayashi, R., Hannon, G.J. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science. 2001;293:1146–1150. doi: 10.1126/science.1064023. [DOI] [PubMed] [Google Scholar]
- Han, J., Lee, Y., Yeom, K.H., Nam, J.W., Heo, I., Rhee, J.K., Sohn, S.Y., Cho, Y., Zhang, B.T., Kim, V.N. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- Hutvagner, G., Simard, M.J. Argonaute proteins: Key players in RNA silencing. Nat. Rev. Mol. Cell Biol. 2008;9:22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- Hutvágner, G., Simard, M.J., Mello, C.C., Zamore, P.D. Sequence-specific inhibition of small RNA function. PLoS Biol. 2004;2:e98. doi: 10.1371/journal.pbio.0020098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J., Carmell, M.A., Rivas, F.V., Marsden, C.G., Thomson, J.M., Song, J.J., Hammond, S.M., Joshua-Tor, L., Hannon, G.J. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- Meister, G., Landthaler, M., Patkaniowska, A., Dorsett, Y., Teng, G., Tuschl, T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Meister, G., Landthaler, M., Peters, L., Chen, P.Y., Urlaub, H., Luhrmann, R., Tuschl, T. Identification of novel Argonaute-associated proteins. Curr. Biol. 2005;15:2149–2155. doi: 10.1016/j.cub.2005.10.048. [DOI] [PubMed] [Google Scholar]
- Okamura, K., Ishizuka, A., Siomi, H., Siomi, M.C. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes & Dev. 2004;18:1655–1666. doi: 10.1101/gad.1210204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak, J., Fire, A. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science. 2007;315:241–244. doi: 10.1126/science.1132839. [DOI] [PubMed] [Google Scholar]
- Parker, G.S., Eckert, D.M., Bass, B.L. RDE-4 preferentially binds long dsRNA and its dimerization is necessary for cleavage of dsRNA to siRNA. RNA. 2006;12:807–818. doi: 10.1261/rna.2338706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquinelli, A.E., Reinhart, B.J., Slack, F., Martindale, M.Q., Kuroda, M.I., Maller, B., Hayward, D.C., Ball, E.E., Degnan, B., Muller, P., et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- Reinhart, B.J., Slack, F.J., Basson, M., Pasquinelli, A.E., Bettinger, J.C., Rougvie, A.E., Horvitz, H.R., Ruvkun, G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Rivas, F.V., Tolia, N.H., Song, J.J., Aragon, J.P., Liu, J., Hannon, G.J., Joshua-Tor, L. Purified Argonaute2 and an siRNA form recombinant human RISC. Nat. Struct. Mol. Biol. 2005;12:340–349. doi: 10.1038/nsmb918. [DOI] [PubMed] [Google Scholar]
- Schwarz, D.S., Hutvagner, G., Du, T., Xu, Z., Aronin, N., Zamore, P.D. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- Sijen, T., Steiner, F.A., Thijssen, K.L., Plasterk, R.H. Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science. 2007;315:244–247. doi: 10.1126/science.1136699. [DOI] [PubMed] [Google Scholar]
- Steiner, F.A., Hoogstrate, S.W., Okihara, K.L., Thijssen, K.L., Ketting, R.F., Plasterk, R.H., Sijen, T. Structural features of small RNA precursors determine Argonaute loading in Caenorhabditis elegans. Nat. Struct. Mol. Biol. 2007;14:927–933. doi: 10.1038/nsmb1308. [DOI] [PubMed] [Google Scholar]
- Tabara, H., Sarkissian, M., Kelly, W.G., Fleenor, J., Grishok, A., Timmons, L., Fire, A., Mello, C.C. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell. 1999;99:123–132. doi: 10.1016/s0092-8674(00)81644-x. [DOI] [PubMed] [Google Scholar]
- Tabara, H., Yigit, E., Siomi, H., Mello, C.C. The dsRNA binding protein RDE-4 interacts with RDE-1, DCR-1, and a DExH-box helicase to direct RNAi in C. elegans. Cell. 2002;109:861–871. doi: 10.1016/s0092-8674(02)00793-6. [DOI] [PubMed] [Google Scholar]
- Tomari, Y., Du, T., Zamore, P.D. Sorting of Drosophila small silencing RNAs. Cell. 2007;130:299–308. doi: 10.1016/j.cell.2007.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yigit, E., Batista, P.J., Bei, Y., Pang, K.M., Chen, C.C., Tolia, N.H., Joshua-Tor, L., Mitani, S., Simard, M.J., Mello, C.C. Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell. 2006;127:747–757. doi: 10.1016/j.cell.2006.09.033. [DOI] [PubMed] [Google Scholar]