Abstract
The HIV-1 ribonucleoprotein (RNP) contains the major structural protein, pr55Gag, viral genomic RNA, as well as the host protein, Staufen1. In this report, we show that the nonsense-mediated decay (NMD) factor UPF1 is also a component of the HIV-1 RNP. We investigated the role of UPF1 in HIV-1-expressing cells. Depletion of UPF1 by siRNA resulted in a dramatic reduction in steady-state HIV-1 RNA and pr55Gag. Pr55Gag synthesis, but not the cognate genomic RNA, was efficiently rescued by expression of an siRNA-insensitive UPF1, demonstrating that UPF1 positively influences HIV-1 RNA translatability. Conversely, overexpression of UPF1 led to a dramatic up-regulation of HIV-1 expression at the RNA and protein synthesis levels. The effects of UPF1 on HIV-1 RNA stability were observed in the nucleus and cytoplasm and required ongoing translation. We also demonstrate that the effects exerted by UPF1 on HIV-1 expression were dependent on its ATPase activity, but were separable from its role in NMD and did not require interaction with UPF2.
Keywords: AIDS, HIV-1, UPF1, RNA stability
INTRODUCTION
A principal player in nonsense-mediated mRNA decay (NMD), a process by which RNAs that harbor premature termination codons are cleared from cells, is upframeshift protein 1, or UPF1. UPF1, an ATP-dependent RNA helicase of the SFI superfamily, is required for NMD and for efficient translation termination at nonsense codons in eukaryotes. UPF1 is also involved at various levels of RNA fate such as RNA splicing, transport, translation, and mRNA turnover (Atkin et al. 1995; Lykke-Andersen et al. 2000; Mendell et al. 2002; Nazarenus et al. 2005; Wilkinson, 2005). UPF1 is a component of several functional ribonucleoprotein (RNP) complexes in cells. In NMD, for example, UPF1 is recruited by UPF2, which is, in turn, recruited by UPF3 to make up an NMD RNP complex on RNA. UPF1 is also a component of the SMG-1-UPF1-eRF1-eRF3 (SURF) RNP complex (Kashima et al. 2006) that is functionally important during the activation of NMD by linking the terminating ribosome to downstream exon junction complexes. This RNP is also important for RNA degradation via NMD and translational activity. Recent results have uncovered a role for an additional RNP complex that is formed when UPF1 and the double-stranded (ds) RNA-binding protein Staufen1 interact on an RNA. Staufen1 can bind a structured RNA sequence in the 3′-UTR of an mRNA and can recruit UPF1 to shunt an mRNA into a decay pathway called Staufen1-mediated decay (SMD) (Kim et al. 2005).
Staufen1 has roles in RNA trafficking, transport, and translation. It is found in neuronal RNA trafficking granules that contain RNA and many types of proteins including motor proteins, translation factors, and several proteins involved in the fate of RNA in the cell (Cochrane et al. 2006). Barentsz (Btz) is one such co-resident protein in RNA trafficking granules or RNPs, and it was demonstrated that Staufen1 and Btz interact (Macchi et al. 2003). Btz was later found to be a component of the exon junction complex (EJC), a complex that is deposited upstream of exon–exon junctions prior to splicing of primary transcripts. Furthermore, in experiments in which Btz was knocked down by siRNA, Btz, like UPF1, was found to be an important factor for NMD (Palacios et al. 2004). These results point toward tight functional links between EJC proteins in splicing, RNA trafficking, and RNA metabolism (Hachet and Ephrussi 2001; Mohr et al. 2001).
HIV-1 RNA metabolism is controlled by a variety of cis-acting sequences in its 9-kb HIV-1 genomic RNA (Mouland et al. 2003). It was first noted that the genomic HIV-1 RNA has an AU-rich codon bias and a rare codon usage (Kotsopoulou et al. 2000). Multiple purine-rich sequences named cis-repressive sequences or instability sequences (CRS/INS) were also identified in the genomic RNA (Schwartz et al. 1992). These sequences are bound by hnRNP A1, polypyrimidine tract binding protein (PTB), poly(A)-binding protein (PABP), and polypyrimidine tract-binding protein-associated splicing factor (PSF) to regulate HIV-1 gene expression at the post-transcriptional level (Black et al. 1996; Zolotukhin et al. 2003). Tethering of other proteins such as the K homology splicing regulatory protein to HIV-1 RNA can also elicit RNA destabilization (Chou et al. 2006).
Rev is the key mediator of nucleocytoplasmic transport of the unspliced, genomic, and singly spliced HIV-1 RNAs via a specific interaction with the Rev-responsive element (RRE) in these RNAs (Malim et al. 1988). This is achieved via the CRM1-dependent pathway (Askjaer et al. 1998). The Rev–RRE interaction counteracts the activities of the CRS/INS by conferring stability to viral transcripts and also promotes polysomal loading in the cytoplasm (D'Agostino et al. 1992; Schwartz et al. 1992). Rev can also inhibit splicing of genomic RNA (Pomerantz et al. 1992), but in its absence, HIV-1 genomic RNA is retained in the nucleus and is completely spliced to generate 2-kb RNAs. The post-transcriptional regulation of HIV-1 gene expression implicates cis-acting RNA sequences and the activities of viral and host cell RNA-binding proteins. HIV-1 may coopt the functions of these proteins, and this may offer the virus a replicative advantage and/or the ability to enter a state of low or latent expression in the host (Cullen 2003).
The association of Staufen1 to proteins belonging to the EJC and implicated in NMD was intriguing to our group. Our observation that the abundance of UPF1 was enhanced in the HIV-1 RNP led us to examine the function of UPF1 during HIV-1 gene expression. In this study, we depleted cells of UPF1 by siRNA and show that while UPF1 knockdown resulted in suppression of NMD as expected, there was also a catastrophic decrease in HIV-1 RNA and pr55Gag expression. We rescued pr55Gag expression using an siRNA-insensitive UPF1 expression construct, but HIV-1 mRNA levels could not be rescued, indicating that UPF1 enhances HIV-1 mRNA translatability. Conversely, overexpression of UPF1 led to enhanced levels of pr55Gag and steady-state HIV-1 RNA. These effects were found in both nucleus and cytoplasm and were found to be dependent on ongoing translation of the genomic (or gag) mRNA. Finally, we demonstrate that UPF1's role in HIV-1 expression is separable from its role in NMD. This study identifies novel functions for UPF1 in the maintenance of HIV-1 RNA stability and protein synthesis.
RESULTS
Identification of UPF1 in the HIV-1 RNP
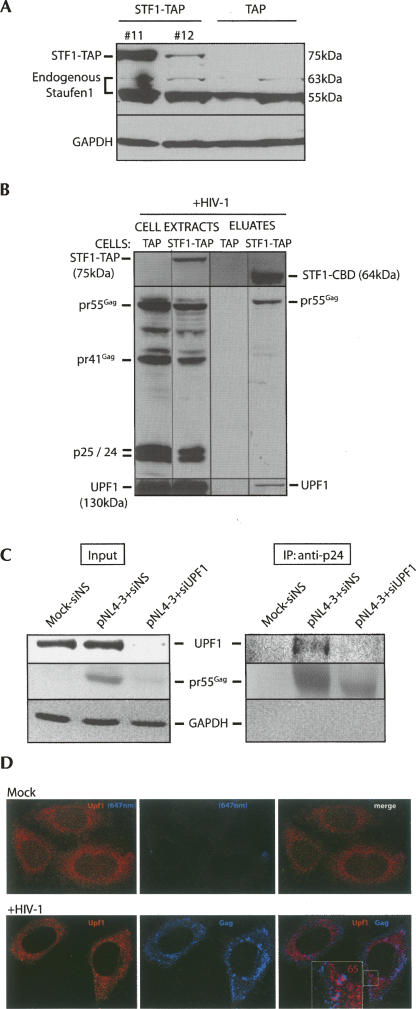
In order to characterize cellular binding partners of Staufen1, we generated a cell line that stably expresses a 75-kDa TAP-tagged Staufen1 protein following transfection of a CMV/Staufen1-TAP construct that is described in detail elsewhere (Villace et al. 2004). Of 12 independent clones, two expressed Staufen1-TAP at high levels (Fig. 1A, #11,#12). A control stable cell line was also clonally expanded that expressed the TAP tag alone (Fig. 1A, TAP control cell line). The expression of TAP in these cell lines was confirmed using mouse anti-Protein A antisera (M. Milev and A.J. Mouland, unpubl.). Staufen1-TAP clone #11 was selected because Staufen1 was expressed at levels similar to those observed for endogenous Staufen155kDa. We investigated the binding partners of Staufen1 using the TAP purification method as described previously (Villace et al. 2004). TAP- and Staufen1-TAP-expressing cell lines were transfected with HIV-1 proviral DNA and processed for TAP purification at 40 h post-transfection. Western blot analysis was performed to identify Staufen1-TAP before affinity purification (Staufen1-TAP) and post-elution to identify the fusion protein between Staufen1 and the remaining calmodulin binding domain (Staufen1-CBD) to the TEV protease cleavage site. The 75-kDa Staufen1-TAP was well expressed in cells (Fig. 1B). Staufen1-CBD was identified in the Staufen1-TAP extracts to demonstrate efficient affinity purification. Previously, we showed that Staufen1 principally binds the precursor, 55-kDa Gag, pr55Gag (Chatel-Chaix et al. 2004); therefore, we validated this assay by probing the Staufen1-TAP affinity eluates for Gag proteins. An anti-p24 antiserum was used to detect pr55Gag as well as the mature Gag proteins, pr41 and p25/p24. Pr55Gag and mature Gag products were detected in cell lysates, but only pr55Gag was detected in the eluates. This result validated the assay and demonstrates the selectivity of this virus–host interaction (Chatel-Chaix et al. 2004). The results are representative of five experiments.
FIGURE 1.
Long-term expression of Staufen1-TAP and identification of UPF1 in the HIV-1 RNP. (A) Stable Staufen1-TAP (STF1-TAP)- (#11 and #12) and TAP-expressing 293T cell lines were created. Expression was verified by Western blot analysis for Staufen1 identifying Staufen1-TAP fusion (MW = 75 kDa) and endogenous Staufen1 (55 kDa and 63 kDa) proteins. Staufen1-TAP expression for clones #11 and #12 is shown. The identity of TAP-expressing cell lines was verified by PCR of harvested genomic DNA (data not shown). (B) Cells were mock transfected or transfected with HIV-1 provirus, and TAP affinity purification was performed. Full-length fusion protein (Staufen1-TAP), truncated, TEV protease cleaved Staufen1 containing the calmodulin binding domain (Staufen1-CBD), and viral proteins were verified by Western blotting of cell extracts before (left) and after (right) tandem affinity purification. The identification of pr55Gag as a Staufen1-binding partner and not mature Gag proteins in the eluates was used for validation of the assay. UPF1 was identified in Staufen1-TAP, but not in TAP eluates (four experiments, bottom). (C) pr55Gag was immunoprecipitated from mock (+siNS), pNL4-3+siNS-, or pNL4-3+siUPF1-transfected cells using a monoclonal anti-p24 antisera as described in the Materials and Methods. UPF1, pr55Gag, and GAPDH [as loading control in cell lysates and negative control in immunoprecipitation (IP) lanes] were identified by Western blotting analyses in input cell lysates and Gag immunoprecipitates (IP). (D) To determine the cellular localization of Gag and UPF1, cells were mock transfected (top panels) or transfected with pNL4-3 (bottom panels), followed by LSCM analyses for UPF1 and Gag. The number in the inset in HIV-1-expressing cells represents the colocalization coefficient (%) of the UPF1 signal (red fluorescence) found to colocalize with Gag (blue fluorescence signal) at the cell periphery indicated by magenta-colored regions and calculated as described in the Materials and Methods.
We then determined if we could identify UPF1 in the HIV-1 RNP complex. UPF1 was abundantly expressed in cell lysates, and a strong band corresponding to UPF1 (130 kDa) was detected in the eluates derived from Staufen1-TAP-expressing cells, but not from TAP control cell lines. This was confirmed by mass spectrometry of the eluates (M. Milev and A.J. Mouland, in prep.). We also probed for a binding partner of UPF1, UPF2, but we could not detect it even though UPF3 was detectable in the Staufen1 eluates by Western blot analyses (data not shown).
The presence of UPF1 in the pr55Gag complex was then confirmed by immunoprecipitation, and effects of UPF1 depletion on the interaction were examined. Cells were mock transfected, transfected with HIV-1 proviral DNA, pNL4-3 both with a nonsilencing, control siRNA (siNS) or transfected with pNL4-3 and siRNA to UPF1 (siUPF1) to deplete UPF1 from cells (Fig. 1C). In cell lysates, a UPF1 signal was found in siNS-transfected cells (in both mock and pNL4–3 transfected), but it was dramatically reduced in UPF1-depleted cells. Pr55Gag expression was abundant in siNS-treated cells, but decreased by 75% in UPF1-depleted cells (see additional data in Fig. 2). Glyceraldehyde-3′-phosphate dehydrogenase (GAPDH) served as a loading control, and the amount of GAPDH in the input samples did not differ. Pr55Gag was immunoprecipitated from cell lysates, and the immunocomplexes were fractionated on SDS-PAGE gels followed by Western blotting for UPF1, pr55Gag, and GAPDH. A prominent UPF1 signal was observed in the pr55Gag immunoprecipitate from pNL4-3/siNS-treated cells, but not from mock-transfected or UPF1-depleted cells (Fig. 1C). Pr55Gag levels were decreased in siUPF1 conditions, and this was also reflected by the negligible signal for UPF1 in the immunoprecipitate. The absence of GAPDH in the Gag immunoprecipitate demonstrated that the interaction was selective. Based on the loading and signal intensities, we calculated that ∼0.1% and 5% of cellular UPF1 and Gag, respectively, was found in the Staufen1-TAP eluates. The abundance of pr55Gag in Staufen1-TAP eluates is consistent with our live-cell interaction analyses in which a small proportion of Gag was shown to interact with Staufen1 (Chatel-Chaix et al. 2004).
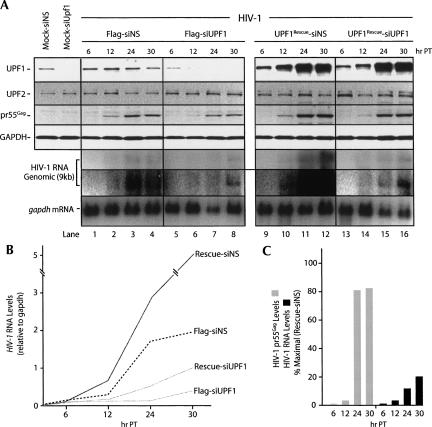
FIGURE 2.
UPF1 expression is essential for NMD and HIV-1 RNA stability. HeLa cells expressing a TCR-β transgene harboring either a wild-type codon (PTC−, 3C1 cells) or an introduced nonsense codon (PTC+, 7C3 cells) as depicted in A (adapted from Mühlemann et al. 2001) were tested for their NMD responsiveness by inhibiting translation by a brief treatment with cycloheximide (CHX). Both cell lines were mock transfected, treated with CHX, or transfected with siNS as described in the Materials and Methods. Expression of UPF1, GAPDH (as loading control), TCR-β, and gapdh mRNA levels were examined at 30 h post-transfection by Western and Northern analyses. Two exposures for TCR-β mRNA are shown. (B,C) 7C3 cells were transfected with siNS (odd-numbered lanes) or siUPF1 (even-numbered lanes) for 24 h. They were then mock transfected (lanes 1,2), or transfected with pNL4-3 (wild-type HIV-1, lanes 3–8), with Flag (lanes 3,4), UPF1WT (lanes 5,6), or a UPF1Rescue (1 μg, lanes 7,8). UPF1, UPF2 (as loading control), pr55Gag, and GAPDH (as loading control) levels were monitored by Western blot analysis. (C) TCR-β, gapdh (as loading control), and HIV-1 RNAs were assessed by Northern blot analysis. All three HIV-1 RNA species (unspliced genomic, 9 kb; singly spliced, 4 kb; and multiply spliced, 2 kb) are shown here.
Laser scanning confocal microscopy (LSCM) was performed to identify the cellular locations of UPF1 and Gag in HIV-1-expressing cells. In mock-transfected cells, UPF1 exhibited diffuse, cytoplasmic staining (Fig. 1D, top panels) and there was no detectable signal for Gag. When HIV-1 was expressed by transfection of a proviral DNA construct, UPF1 staining was again mostly diffuse in the cytoplasm, and Gag was found in the cytoplasm as well as at the cell periphery, presumably representing assembly sites for HIV-1. We calculated that ∼10%–20% of the UPF1 signal colocalized with Gag when analyzing entire cells. However, in almost 70% of cells with detectable Gag expression, UPF1 was also found to colocalize with Gag at the cell periphery in the form of small punctae (Fig. 1D, bottom panels). Sixty-five percent of the UPF1 in this region of the cell strictly overlapped with the Gag. These results put a proportion of UPF1 and Gag in the same cellular locale and would explain why UPF1 coimmunoprecipitates with Gag. The result also suggests that UPF1 may have a continued role during virus assembly.
UPF1 depletion results in decreased pr55Gag synthesis and a catastrophic loss of HIV-1 RNA
To assess the function of UPF1 in HIV-1-expressing cells, siRNA was used to deplete cells of UPF1. Stable HeLa cell lines were used that contain a T-cell receptor-β (TCR-β) minigene with either a natural termination codon (PTC−, 3C1) or a premature termination codon (PTC+, 7C3) in the second VDJ exon (Fig. 2A; Carter et al. 1995). In order to confirm the sensitivity to ongoing translation of the TCR-β NMD substrate (Li et al. 1997; Shibuya et al. 2004), both cell lines were mock transfected or treated with cycloheximide (CHX). The TCR-β and gapdh mRNAs were analyzed by Northern blotting. As shown in Figure 2A, the TCR-β mRNA lacking a PTC was abundantly expressed in 3C1 cells. However, in PTC+ cells, levels of the TCR-β mRNA were increased only when cells were treated with CHX to block translation, demonstrating the dependence of NMD on ongoing translation. UPF1 expression levels were constant in these cell lines, showing that the NMD effect of CHX treatment was not caused by impaired UPF1 expression.
These cell lines were employed to explore the effects of UPF1 on HIV-1 expression and to have a homologous control for UPF1 function in NMD. 7C3 cells were mock transfected with siNS or siRNA to UPF1 (siUPF1) without proviral DNA (Mock-siNS, Mock-siUPF1), or with proviral pNL4-3 DNA (Fig. 2B, lanes 3–8) and Flag DNA (empty vector) and siNS or siUPF1 (Flag-siNS or Flag-siUPF1), UPF1WT, and siNS or siUPF1 (UPF1WT -siNS or UPF1WT -siUPF1) or UPF1Rescue and siNS or siUPF1 (UPF1Rescue-siNS or UPF1Rescue-siUPF1). UPF1, UPF2, pr55Gag, and GAPDH expression levels were assessed by Western blot analysis. UPF1 knockdown was efficient and routinely ranged from 85% to 98% in most experiments (Fig. 2B, lanes 2,4,6) even when UPF1 was supplied in trans (Fig. 2B, lane 6). When cells were transfected with siUPF1, a significant reduction in pr55Gag was observed (Fig. 2B, cf. lanes 3,4). Cells either expressing UPF1WT or the siRNA-insensitive, UPF1Rescue in trans led to a threefold to fourfold up-regulation of pr55Gag expression levels (Fig. 2B, lanes 5,7). In cells depleted of UPF1, however, UPF1Rescue expression modestly rescued pr55Gag synthesis by 60% (±10%, SD; seven experiments) relative to UPF1Rescue-siNS-transfected cells, indicating that UPF1 expression was important for the observed effects on pr55Gag expression (Fig. 2B, lane 8). The dramatic effects of UPF1 depletion on pr55Gag synthesis were reflected by a corresponding 20-fold decrease in virus production (data not shown). These effects on HIV-1 gene expression were absolutely identical in 3C1, parental HeLa, and other human epithelial cell types (data not shown). Similar results were obtained using another set of siRNAs providing further evidence that the observed results were not due to off-target effects.
When TCR-β and HIV-1 RNA levels were quantitated by Northern analyses, PTC-containing TCR-β mRNA levels responded accordingly such that they were up-regulated when cells were depleted of UPF1 (Fig. 2C, lanes 2,4,6,8). In contrast, HIV-1 mRNA levels were dramatically down-regulated to near undetectable levels (Fig. 2C, lanes 4,6; Supplemental Fig. S1).
Overexpression of UPF1 by expression of UPF1WT or UPF1Rescue, up-regulated HIV-1 RNA severalfold, corresponding to UPF1's effects on pr55Gag synthesis (Fig. 2B, lanes 5,7). UPF1Rescue was not able to efficiently rescue the HIV-1 RNA levels, despite the modest 60%–70% rescue of TCR-β NMD-mediated degradation (Fig. 2C, lane 8). In contrast, a modest rescue was obtained for pr55Gag synthesis (Fig. 2B, lane 8), suggesting that UPF1 influenced HIV-1 mRNA translation efficiency. These results are representative of at least 10 experiments. Increasing doses of UPF1Rescue in transfections partially rescued pr55Gag levels (Supplemental Fig. S2).
A more detailed analysis was performed to evaluate the importance of UPF1 expression at several time points following transfection. Cells were transfected with either the empty vector Flag or UPF1Rescue in the presence of siNS or siUPF1. Protein and RNA analyses were performed at 6, 12, 24, and 30 h post-transfection. UPF1 expression was stable, and pr55Gag and HIV-1 RNA levels increased with time in siNS- and Flag-transfected cells (Fig. 3A, lanes 1–4). There was a time-dependent knockdown of UPF1 mediated by siUPF1, and this resulted in dramatically reduced levels of steady-state HIV-1 mRNA and pr55Gag expression (Fig. 3A, lanes 5–8). Overexpression of UPF1Rescue (in the presence of siNS) greatly enhanced UPF1 levels and enhanced pr55Gag and steady-state HIV-1 genomic RNA levels 3.4-fold and fivefold, respectively (±10%, SD in both cases) (Fig. 3A, lanes 9–12). However, while the rescue of UPF1 expression in siUPF1-depleted cells was efficient and resulted in enhanced pr55Gag synthesis, there was not a corresponding change in the abundance of the levels of steady-state HIV-1 RNA (Fig. 3A, lanes 13–16). The line graph shown in Figure 3B summarizes the relative changes in HIV-1 RNA at each time point. The relative rescue ratios presented in Figure 3C show that despite an almost 80% rescue of pr55Gag levels (relative to maximal expression levels obtained with UPF1Rescue-siNS), the levels of HIV-1 RNA only reached 10%–20% at the latest time points studied. These results demonstrate that UPF1 expression almost completely rescues pr55Gag synthesis, but it is not sufficient to efficiently rescue defects at the RNA level.
FIGURE 3.
Time-course effects of siUPF1 and UPF1 overexpression on HIV-1 pr55Gag and RNA levels. HeLa cells were transfected with siNS or siUPF1 for 24 h. They were then transfected with pNL4-3 (HIV-1) and Flag or UPF1Rescue (2 μg) and again with siNS or siUPF1 (Time 0). At 6, 12, 24, and 30 h post-transfection (hr PT), cells were harvested for Western and Northern analyses. (A) A set of mock-transfected cells (no pNL4-3) with siNS or siUPF1 is shown on the left at 30 h post-transfection. Expression levels of UPF1, UPF2, pr55Gag, and GAPDH (as loading control) were monitored by Western analyses. Expression levels of HIV-1 genomic RNA and gapdh mRNA (as loading control) were assayed by Northern blotting. Two exposures of the results for HIV-1 genomic RNA are shown. Flag and (lanes 1–4) siNS- or (lanes 5–8) siUPF1-transfected cells; UPF1Rescue and (lanes 9–12) siNS- or (lanes 13–16) siUPF1-transfected cells. (B) Genomic RNA levels based on densitometric scanning of the lower exposure shown in A were expressed relative to those of gapdh mRNA at each time point post-transfection. (C) The levels of (gray bars) HIV-1 pr55Gag and (black bars) RNA were expressed relative to the corresponding expression levels in the UPF1Rescue-siNS treatment group that were considered maximal.
Effects on HIV-1 expression require ongoing translation of gag mRNA
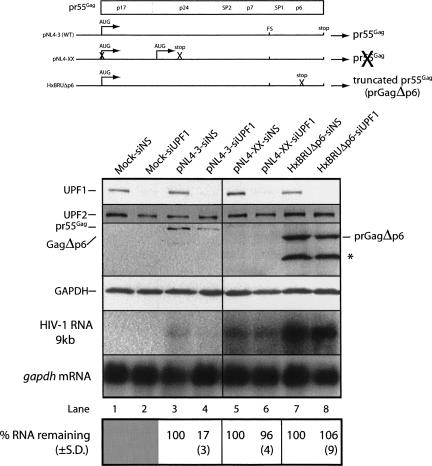
We then determined if the observed effects of UPF1 depletion required ongoing HIV-1 RNA translation. We approached this question by expressing proviral HIV-1 DNAs that express RNAs that are programmed to either terminate translation immediately following gag RNA translation initiation or at a downstream site. We mock transfected cells (no proviral DNA), or transfected cells with wild-type pNL4-3, pNL4-XX (a proviral DNA harboring two PTCs in the gag ORF: the first preventing gag RNA translation initiation at the bona fide initiator AUG and the other programming translation termination following reinitiation at a downstream AUG) or with HxBRUΔp6 (harboring a PTC in the C-terminal p6 region following the ribosomal frameshifting site) (Fig. 4). In each condition, cells were treated with either siNS or siUPF1. UPF1 knockdown was extremely efficient, reaching >90% in all cases (Fig. 4, even-numbered lanes). siUPF1 had the expected negative effects on pr55Gag synthesis and steady-state RNA levels in cells expressing wild-type pNL4-3 (Fig. 4, lane 4). Pr55Gag synthesis in cells expressing pNL4-XX was aborted due to the in-frame stop codons. In cells expressing HxBRUΔp6, UPF1 depletion had little effect on the synthesis of the Gag gene product expressed from this DNA construct, prGagΔp6. In pNL4-XX- and HxBRUΔp6-expressing cells, steady-state mRNA levels were not modulated when UPF1 was depleted from cells (Fig. 4, lanes 6,8). These results indicate that the translation of the gag ORF or the completion of the translation of this gag ORF is necessary for the UPF1-mediated stabilization of HIV-1 RNAs. This result was corroborated by the observation that Gag expression was also negatively influenced by UPF1 depletion with the use of the pNL4-X proviral DNA in which translation of the wild-type AUG is blocked, but in which gag translation reinitiates at a downstream AUG (Poon et al. 2002; L. Ajamian and A.J. Mouland, unpubl.). Thus, PTCs within the gag ORF are not recognized by the NMD machinery and are required for the destabilization of the RNA when UPF1 is depleted. These results also demonstrate that UPF1 depletion has no effect on LTR-mediated transactivation.
FIGURE 4.
UPF1 depletion affects actively translating HIV-1 genomic (gag) RNA. HeLa cells were transfected with siNS or siUPF1 for 24 h. HeLa cells were then mock transfected or transfected with pNL4-3, pNL4-XX, or HxBRUΔp6 proviral constructs depicted in the top panel and again with siNS or siUPF1 for each condition. In pNL4-3, the ATG initiation codon (ATG) is indicated, and translation of the RNA generates pr55Gag protein. Translation from this codon in pNL4-XX is mutated, preventing translation initiation at this site, but translation can initiate at a downstream initiation codon. A stop codon was introduced downstream from this internal site to prevent Gag synthesis (Poon et al. 2002). HxBRUΔp6 possesses a stop codon preventing completion of p6 synthesis and results in the synthesis of a truncated Gag protein (prGagΔp6). Virus produced from HxBRUΔp6-expressing cells show demonstrable budding and maturation defects (Gottlinger et al. 1991). Cells were harvested at 30 h. UPF1, UPF2, pr55Gag, truncated prGagΔp6, and GAPDH were assessed by Western blot analyses. Genomic RNA and gapdh mRNAs were quantitated by Northern blotting as described in the Materials and Methods. The ribosomal frameshift site (FS) is indicated on the RNAs. The abundance of the genomic RNA was related to gapdh mRNA level (loading control), and levels were related to signal intensities found in siNS-treated cells expressed as a percentage of the amount remaining (±SD) calculated from 10 experiments. The asterisk (*) identifies a processed form of Gag that is found only in the HxBRUΔp6-expressing cells.
RNA destabilizing effects of UPF1 depletion in the nucleus
We next explored the cellular compartment in which the effects of UPF1 were being exerted. We took advantage of the nuclear retention of the HIV-1 genomic RNA when the regulatory protein Rev is not expressed. siNS- or siUPF1-treated cells were mock transfected, transfected with a wild-type HIV-1 proviral DNA (HxBRU), or transfected with a Rev− proviral DNA, pcMRev(−). Cells were harvested, and protein and RNA expression levels were monitored by Western and Northern analyses. Efficient knockdown of UPF1 resulted in decreased pr55Gag expression as shown earlier [the expression of pcMRev(−) does not lead to any appreciable pr55Gag synthesis] (Fig. 5A). Analysis of steady-state mRNA levels using a pol-specific cDNA probe revealed that, even during the expression of the pcMRev(−) HIV-1, steady-state levels of genomic RNA were efficiently knocked down by 85% as a result of UPF1 depletion (Fig. 5B). When the completely spliced 2-kb RNAs were assessed by Northern blot analysis, they were found to be relatively unaffected by siUPF1 conditions in the Rev− context.
FIGURE 5.
Unspliced HIV-1 RNA is sensitive to UPF1 depletion in the nucleus. HeLa cells were transfected with siNS or siUPF1 for 24 h. HeLa cells were then mock transfected or transfected with HxBRU or the Rev-defective provirus, pcMRev(−), with either siNS or siUPF1 for each condition. (A) Western blot analysis of UPF1, pr55Gag, and GAPDH. (B) Northern blot analysis of HIV-1 unspliced 9-kb and 2-kb RNAs identified using a pol-specific cDNA probe or a 5′ TAR-specific probe, respectively. (C) FISH analyses of HIV-1 genomic RNA (green fluorescence in all panels) in cells transfected with HxBRU or pcMRev(−) in siNS or siUPF1 knockdown conditions as indicated above each panel. Cells were counterstained with DAPI to identify nuclei (blue). Merged images are shown including transmitted light/phase contrast to highlight cell periphery. Red arrows indicate nontransfected cells. These results are representative of three independently performed experiments.
In order to ensure that the genomic RNA was confined to the nucleus in these experiments, FISH analysis was performed. An identical probe sequence to that used for Northern analysis of the 9-kb RNA was used to identify genomic RNA by FISH. In nontransfected cells, there was no detectable signal for HIV-1 genomic RNA. In cells treated with siNS exhibiting expression of HIV-1, genomic RNA had a punctate cellular distribution as shown in our earlier work (Levesque et al. 2006). In pcMRev(−)-expressing cells, genomic RNA was confined to the nucleus and was abundantly expressed in siNS-treated cells, while in UPF1-depleted, pcMRev(−)-expressing cells, the RNA was confined to the nucleus, and the apparent RNA staining intensity was dramatically reduced (Fig. 5C). Since genomic RNA levels were significantly reduced in the absence or presence of Rev expression, we can conclude that both cytoplasmic and nuclear HIV-1 RNA are stabilized by UPF1 expression. Consistently, overexpression of UPF1 enhances steady-state levels of genomic RNA in the Rev− background (data not shown). It remains to be shown if the effects on nuclear RNA stability are due to common mechanisms involving UPF1.
UPF1 enhances HIV-1 RNA expression independently of its NMD function
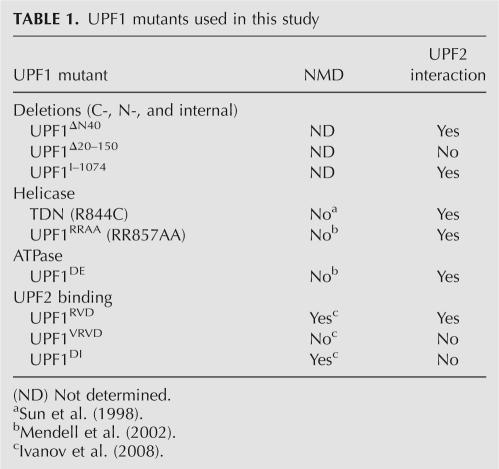
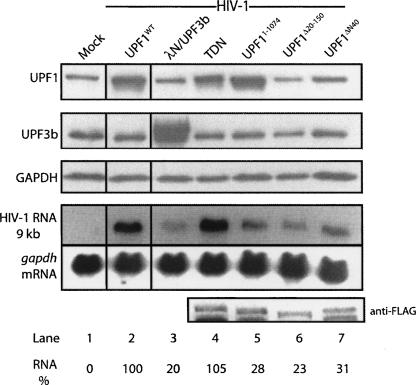
UPF1 is involved in the regulation of a number of cellular processes besides its central function in NMD. To identify the regions of UPF1 that are required for the stimulation of HIV-1 RNA expression, we overexpressed a number of UPF1 mutants and analyzed their effects on HIV-1 pr55Gag and RNA levels (Table 1). As shown earlier, overexpression of UPF1WT led to a dramatic increase of HIV-1 RNA (Fig. 6, lane 2). In contrast, a severalfold overexpression of UPF3b had little effect on steady-state HIV-1 RNA levels (Fig. 6, lane 3), indicating that the effects on HIV-1 genomic RNA are specific to UPF1. To our surprise, the RNA helicase trans-dominant negative (TDN) mutant UPF1(R844C) that is well established to be ineffective in NMD in mammalian cells (Sun et al. 1998), stimulated HIV-1 RNA expression to similar levels achieved with UPF1WT (Fig. 6, lane 4), indicating that UPF1 enhances HIV-1 RNA expression independently of its NMD function. In contrast, N-terminal (UPF1Δ20–150, UPF1ΔN40) and C-terminal (UPF11–1074) truncations and deletions of UPF1 were unable to enhance levels of HIV-1 RNA expression (Fig. 6, lanes 5–7).
TABLE 1.
UPF1 mutants used in this study
FIGURE 6.
Selective up-regulation of HIV-1 RNA levels by an intact UPF1 protein. Cells were (lane 1) mock transfected or co-transfected with (lane 2) HIV-1 proviral DNA (pNL4-3, HIV-1) and UPF1WT, (lane 3) an expression construct for UPF3b with a λN-N-terminal tag (λN/UPF3b), (lane 4) TDN (R844C), or Flag-tagged expression constructs coding for deletion mutants of UPF1 including (lane 5) the C-terminal deletion mutant, UPF1I–1074, (lane 6) the internally deleted UPF1Δ20–150, and (lane 7) the N-terminal deleted UPF1ΔN40. At 30 h post-transfection, cells were harvested for Western blot analysis for UPF1, UPF3b, and GAPDH (loading control). RNA was also isolated from cell extracts, and Northern blotting was performed to measure steady-state levels of HIV-1 and gapdh mRNA. Because the primary anti-UPF1 antibody did not efficiently recognize the truncation and deletion mutants, Western blotting using an anti-Flag antibody was employed to identify the Flag-tagged UPF1 proteins expressed in trans (bottom panel). The percentages represent genomic RNA levels related to the signal obtained in UPF1WT and HIV-1-expressing cells. The results shown are representative of three independently performed experiments.
We attempted to further map the determinants for UPF1 up-regulation using UPF2 binding mutants (UPF1RVD, UPF1DI) and with a mutant with a similar mutation in the UPF2-binding region (UPF1RVD) that retains its ability to bind UPF2 (Table 1). All of these mutants up-regulated pr55Gag expression levels (and RNA levels) (data not shown) similar to that obtained using intact UPF1 proteins (Fig. 7). Therefore, the association of UPF1 with UPF2, an event that is important for NMD though not strictly required (Table 1; Ivanov et al. 2008), was not required for the enhancement in HIV-1 RNA and pr55Gag expression levels.
FIGURE 7.
UPF2 binding is not necessary for UPF1-mediated up-regulation of HIV-1. Cells were (lane 1) mock transfected or co-transfected with HIV-1 proviral DNA (pNL4-3, HIV-1) with Flag, TDN (R844C), UPF1RVD, or UPF2-binding-deficient mutants UPF1VRVD or UPF1DI. At 30 h post-transfection, cells were harvested for Western blot analysis for UPF1, UPF3b (as loading control), pr55Gag, and GAPDH (as loading control). Cell-associated pr25, a product of pr55Gag maturation, is shown to demonstrate up-regulation more clearly than the signals obtained for pr55Gag. The percentages represent the amount of pr55Gag related to the signal obtained in Flag and HIV-1-expressing cells. The values from three independently performed experiments did not vary by more than 17%.
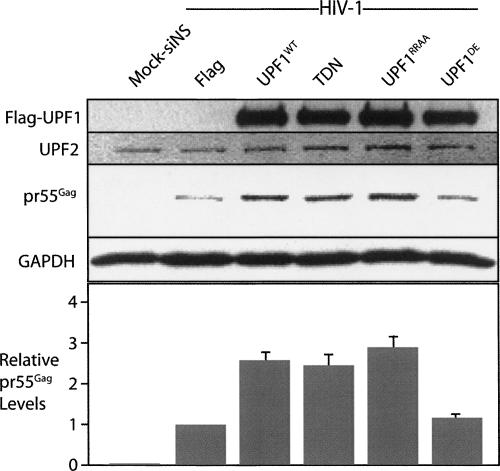
We then performed experiments to establish if the effect of UPF1 on HIV-1 expression was due to the ATPase activity of UPF1 using a well-characterized ATPase mutant whose expression also blocks NMD. We overexpressed TDN, an additional helicase trans-dominant mutant, UPF1RRAA, and the ATPase mutant, UPF1DE. UPF1WT, TDN, and UPF1RRAA up-regulated pr55Gag severalfold (Fig. 8) to levels that quantitatively correlated with genomic RNA levels (data not shown). The overexpression of UPF1DE, however, only led to a 10% increase in pr55Gag levels on average from three experiments (Fig. 8), which indicated the importance of UPF1 ATPase activity in HIV-1 expression.
FIGURE 8.
ATPase activity of UPF1 is required for HIV-1 up-regulation. Cells were (lane 1) mock transfected or co-transfected with HIV-1 proviral DNA (pNL4-3, HIV-1) with Flag, UPF1WT, TDN (R844C), UPF1RRAA, and UPF1DE. At 30 h post-transfection, cells were harvested for Western blot analysis for UPF1 expressed in trans using an anti-Flag epitope tag, UPF2 (as loading control), Gag, and GAPDH (as loading control). The relative levels of pr55Gag are related to the signal obtained in Flag and HIV-1-expressing cells, set to 1. The histogram shows the averages (±SD).
DISCUSSION
Using complementary biochemical approaches including tandem affinity chromatography, immunoprecipitation, and confocal microscopy, we demonstrated that UPF1 is a component of the HIV-1 RNP, which also includes pr55Gag, genomic RNA, and the host protein Staufen1. UPF1 coeluted with HIV-1 pr55Gag from a Staufen1-TAP column. Further analysis revealed that UPF1 coimmunoprecipitated with pr55Gag, and also a proportion of cellular UPF1 colocalized with pr55Gag at the plasma membrane, assembly sites for HIV-1. A strong correlation between HIV-1 expression and this localization pattern was found in almost 70% of HIV-1-expressing cells, while in cells that did not have detectable HIV-1 expression, UPF1 was dispersed in the cytoplasm. These results suggest that UPF1 acts in the context of the HIV-1 RNP and that HIV-1 coopts UPF1 function in cells.
The most important finding of this study lies in the identification of a novel effect of UPF1 on HIV-1 RNA metabolism. siRNA-mediated depletion of UPF1 led to a dramatic negative effect on steady-state HIV-1 RNA levels (Figs. 2–5; Supplemental Fig. S1), indicating that UPF1 is a principal player in maintaining the stability of HIV-1 RNA. The striking regulation of UPF1 on HIV-1 RNA is novel. Nevertheless, a role for UPF1 in the control of RNA stability of cellular mRNAs is not without precedence. For example, UPF1 was shown to affect the stability of both PTC- and non-PTC-containing mRNAs in earlier work (Bhattacharya et al. 2000; He and Jacobson 2001). Recent studies identified a role for UPF1 in the stability of the intronless histone H2a mRNA in a cell-cycle-dependent manner in mammalian cells (Kaygun and Marzluff 2005) and an effect on Xist RNA, but in this latter case, this RNA is entirely restricted to the nucleus (Ciaudo et al. 2006). While there is at least one similarity between NMD and the effects of UPF1 on H2A and HIV-1 RNA, including the dependence on UPF1 expression and ongoing translation (Fig. 4), HIV-1 RNA degradation does not appear to be due to UPF1-mediated NMD. First, there was no increase in HIV-1 RNA when cells were treated with siUPF1. Second, current data support the notion that NMD is dependent on splicing and the deposition of an EJC (Zhang et al. 1998; Maquat and Li 2001), suggesting that genomic RNA would not be a substrate, which is consistent with our findings. Third, despite evidence that extended 3′-UTRs may be recognized for EJC-dependent NMD (Muhlrad and Parker 1999; Buhler et al. 2006), our data demonstrate that UPF1 expression protects unspliced RNA from degradation, even with its almost 7-kb 3′-UTR. Fourth, recent work shows that the order of cotranscriptional intron removal from HIV-1 genomic RNA does not generate substrates for NMD (Bohne et al. 2005). And finally, introducing a PTC in the HIV-1 gag ORF does not lead to its degradation (Fig. 4). We cannot completely rule out the possibility, however, that the effects on HIV-1 RNA and Gag expression are an indirect consequence of inhibiting NMD, for example, especially since UPF1 depletion can alter the expression levels of many genes (Mendell et al. 2004).
Our data reveal that the effects of UPF1 on HIV-1 Gag expression are not dependent on its association to UPF2, an interaction that is appreciated to be important, but not critical, for NMD (Fig. 7; Table 1; Ivanov et al. 2008). While NMD is still active in HIV-1-expressing cells (Fig. 2), the regulation of HIV-1 expression by UPF1, which is independent of UPF2, may be achieved in the context of a distinct RNP. The association of UPF1 with the HIV-1 RNP, although potentially mediated via Staufen1 (Kim et al. 2005), does not require UPF2 to enhance expression. In fact, our Western and mass spectrometry analyses of the composition of Staufen1 complexes also confirmed that UPF2 is not detectable in Staufen1-TAP eluates (M. Milev and A.J. Mouland, unpubl.). The absence of UPF2 may favor the ability of UPF1 to bind RNA and perhaps that of HIV-1 (Chamieh et al. 2008), but this will require further analysis.
Most of the UPF1 NMD mutants, including the helicase transdominants and the UPF2-binding mutants (Table 1), up-regulated HIV-1 RNA (Figs. 7, 8). While pr55Gag expression levels increased when TDN and UPF1RRAA were overexpressed, the ATPase-/NMD-deficient mutant UPF1DE did not (Fig. 8). Expression of this mutant increases the size and number of P-bodies, increases the level of normal transcripts in P-bodies, promotes the recycling of P-body components and compromises RNA-binding capacity of UPF1 (Bhattacharya et al. 2000; Wilkinson 2005; Cheng et al. 2007). Because P-bodies are translationally silent and are sites of mRNA storage and degradation, we speculate that HIV-1 genomic RNA could be directed to PBs for storage explaining why pr55Gag levels do not change when we express this ATPase mutant. This idea is echoed by a recent study showing that PB-sequestered ARE-mRNAs prevent translation on polysomes, thereby silencing ARE-mRNA function even when mRNA decay is delayed (Franks and Lykke-Andersen 2007).
The lack of correspondence between HIV-1 RNA and pr55Gag expression levels in our rescue experiments support an enhancing role for UPF1 on HIV-1 gag mRNA translation (Figs. 2, 3; Supplemental Fig. S2). The incomplete rescue of RNA levels is indicative that UPF1 could function at a specific time in the late expression stages, and the RNA defect, due to UPF1 depletion, cannot be rescued. Nevertheless, UPF1 still maintains quite a strong effect on gag mRNA translation. The role of UPF1 in HIV-1 RNA translation is supported by its association to polysomes (Atkin et al. 1995; Pal et al. 2001), its regulation of ribosomal frameshifting (Harger and Dinman 2004), and its possible roles in translation termination (Muhlrad and Parker 1999). Nevertheless, the mechanism by which UPF1 enhances pr55Gag expression levels is not well understood, but could be due to its association to sequences within the gag ORF since the tethering of many NMD factors including UPF1 was shown to mediate translational enhancement (Nott et al. 2004). Translational enhancement could also be caused by a UPF1-mediated recruitment of residual HIV-1 RNA (from a pool that might not be translated) into polyribosomes or even by enhanced ribosome recycling following translation termination, a proposed role for UPF1 in earlier work (Nott et al. 2004). The near complete rescue of pr55Gag expression indicates that UPF1 is a principal regulator of viral structural protein synthesis. Moreover, these results support the notion that separable roles for UPF1 in HIV-1 RNA degradation and translation exist.
Ongoing translation of the gag ORF was necessary for HIV-1 RNA degradation that ensues when UPF1 is depleted from cells (Fig. 4). The translational enhancing effects mediated by UPF1 may, in fact, have two functions: (1) to enhance pr55Gag synthesis; and (2) to prevent RNA degradation due to stalling ribosomes. Furthermore, at first sight, the results showing that the gag RNA expressed from a Rev− provirus succumbs to the effects of UPF1 depletion might seem contradictory. While negligible levels of genomic RNA do indeed enter the cytoplasm and are translated even in a Rev− background (D'Agostino et al. 1992), these results could be explained by separable processes that are regulated by UPF1 in nuclear and cytosolic compartments.
A nuclear function for UPF1 is strongly supported by our data because of the striking effects of UPF1 knockdown on nuclear-retained HIV-1 genomic RNA. The ability of UPF1 to shuttle between the nucleus and cytoplasm (Mendell et al. 2002) and its role in nonsense-mediated alternative exon skipping (Mendell et al. 2002), in exon skipping (Wang et al. 2002), in regulating nuclear Xist RNA (Ciaudo et al. 2006), and in DNA repair (Azzalin and Lingner 2006), all support nuclear roles. Our results suggest that UPF1 is needed for late gene expression stages of HIV-1, early following transcription to protect the viral mRNA from degradation. When we expressed the Rev− provirus when UPF1 was depleted, the Rev-dependent, 9-kb genomic RNA was selectively targeted, but the 2-kb RNAs that are generated by complete splicing and rapidly exported to the cytoplasm were not (Fig. 5). This selectivity for the genomic RNA strongly suggests that UPF1 exerts its protective effects on the genomic RNA when the decision for splicing is made, concomitant or upstream of the activity of Rev. CRM1-mediated nuclear export of both of these cargoes may also functionally link this host protein with the genomic RNA.
A cis-acting RNA sequence is likely necessary for the elicited effects of UPF1 depletion. The insensitivity of the 2-kb RNAs, observed most notably in the context of a Rev− provirus, could also be explained by the absence of CRS/INS in these RNAs upon complete splicing of the genomic RNA. Moreover, it has been speculated that the zinc fingers of UPF1 are implicated in binding INSs (Applequist et al. 1997) and the lack of effect of UPF1Δ20–150 on HIV-1 expression is likely due to the deletion of a zinc-finger motif in this mutant (Fig. 6). The C-terminal mutant UPF1I–1074 also did not have any effect on HIV-1 RNA. The deleted region in this mutant is speculated to be important for protein–protein or protein–RNA interactions, and these events might be important for the effects on HIV-1 (Applequist et al. 1997). These observations would be consistent with the results obtained with UPF1DE, which also has compromised RNA-binding capacity.
Based on our data, we speculate that UPF1 acts to prevent the recruitment of nuclear exosome degradative machinery (Andrulis et al. 2002) to intron-containing HIV-1 RNAs (in both Rev+ and Rev− backgrounds). On the one hand, when UPF1 is depleted from cells, intron-containing HIV-1 RNAs would be detected and rapidly cleared. On the other hand, UPF1 overexpression leads to enhanced protection of genomic RNA following transcription, and the RNA would efficiently access the cytoplasm, increasing its availability to the translation apparatus. UPF1 interacts directly with several components of the nuclear and cytoplasmic exosome (Lejeune et al. 2003), providing support for this model. The potential to correct genetic disease by targeting NMD (Kuzmiak and Maquat 2006) highlights the importance of studying UPF1 function in order to potentially home in on HIV-1 RNAs before, during, and after their export to the cytoplasm.
MATERIALS AND METHODS
siRNAs
siRNA duplexes were synthesized by QIAGEN-Xeragon. siNS is commercially available nonsilencing control duplex (QIAGEN-Xeragon). The sequence for siUPF1 (5′-AAGATGCAGTTCCGCTCCATT-3′) was reported earlier (Mendell et al. 2002). An additional siUPF1 was employed (On-Target Plus, catalog #J-011763-07, Dharmacon: 5′-GCAGCCACAUUGUAAAUCAUU-3′) having reduced off-target effects with similar efficiencies of UPF1 knockdown. The corresponding On-Target Plus siNS was also used.
Cell lines and transfections
Stable TAP- and Staufen1-TAP-expressing cell lines were created following transfection of 293T cells with the Staufen1-TAP vector generously provided by Juan Ortin (Centro Nacional de Biotecnologia, Spain) as described (Villace et al. 2004). Single clones were expanded in the presence of 600 μg/mL G418. Expression was verified by Western blot analyses and was stable for more than 22 passages. Stable control TAP-expressing cells were verified by resistance to neomycin and by PCR of genomic DNA to identify integral TAP DNA sequences. These experiments were performed in the absence and presence of transfected HIV-1 HxBRU DNA.
All other experiments employed HeLa cells. Transfection of proviruses and siRNAs was performed essentially as described (Chatel-Chaix et al. 2004) except HeLa cells were plated in six-well plates at 300,000/well for 24 h before transfection. A total of 2 μg of DNA was added per well (1 μg of proviral DNA plus 1 μg of carrier pKSII with or without siRNA RNA duplexes). siUPF1 and siNS were used at 100 nM as described (Gehring et al. 2003; Levesque et al. 2006). Proviral DNAs HxBRU and pNL4-3 were described earlier (Beriault et al. 2004). 3C1 (PTC−) and 7C3 (PTC+) HeLa cell lines were provided by Miles Wilkinson (University of Texas) (Carter et al. 1995), and NMD was assessed by Northern blotting for TCR-β transcripts. In control experiments, 100 nM cycloheximide was added 6 h before cell harvesting in order to abrogate NMD in TCR-β mRNA-expressing cell lines or to examine the dependence on translation of siUPF1-mediated effects on HIV-1 RNAs.
Total cellular RNA was isolated from cells using Trizol Reagent (Invitrogen) according to the manufacturer's instructions. The RNA was re-precipitated with ethanol before loading onto denaturing agarose/formaldehyde gels. Unspliced and spliced HIV-1 and gapdh mRNAs were identified by Northern blot analyses as described previously (Mouland et al. 2002). For the Rev− experiments, the genomic RNA was identified using a radiolabeled pol-specific cDNA probe that corresponded exactly to the riboprobe used in the FISH analyses (Levesque et al. 2006). TCR-β mRNA was identified using a cDNA probe to the second VDJ exon generated by PCR using the following primers (sense: 5′-ACACATGGAGGCTGCAGTCA; antisense: 5′-CGAAACAGTCAGTCTGGTTC) and the β433 TCR-β minigene DNA construct (generously provided by Miles Wilkinson, University of Texas). RSV mRNA was identified by Northern blot analysis. Éric A. Cohen (IRCM, Canada) provided the HxBRUΔp6 in which a stop codon was introduced following the p6 ORF (Gottlinger et al. 1991). David Ott (NIH/NCI, Frederick, MD) provided the pNL4-XX and pNL4-X proviral constructs as described (Poon et al. 2002); and the NIH AIDS Reference and Reagent Program provided the pMRev(−) contruct (Sadaie et al. 1988). Anne Gatignol (McGill University) provided the pMAL and pADA M-tropic proviral constructs. The HIV-2 ROD27 is described by Mouland et al. (2000). pC1-Flag/UPF1-Rescue (UPF1Rescue, harboring silent mutations that resists siUPF1), wild-type pC1-Flag-UPF1 (UPF1WT), and pC1-λN-UPF3 (λN/UPF3) expression constructs were described previously (Gehring et al. 2003). pC1-Flag-UPF1(VV204DI) (UPF1DI), pC1-Flag-UPF1(LECY181VRVD) (UPF1VRVD), and pC1-Flag-UPF1(TLH169RVD) (UPF1RVD) were generated in UPF1Rescue, while the ATPase mutant pC1-Flag/UPF1(DE637AA) (UPF1DE), the helicase mutants pC1-Flag/UPF1(RR857AA) (UPF1RRAA), and pC1-Flag-UPF1(R844C) (TDN), pC1-Flag/UPF1(1–1074) (UPF11–1074), and pC1-Flag/UPF1Δ20–150 (UPF1Δ20–150) were generated in UPF1WT by site-directed mutagenesis. For UPF11–1074, a stop codon was introduced at position 1075. For UPF1Δ20–150, primers were designed to delete the region in the context of wild-type UPF1. UPF1ΔN40 was generated by PCR of UPF1 with a 5′-primer starting at position 41. UPF11–1074 lacks all C-terminal phosphorylation sites that are speculated to be important for protein–protein or protein–RNA interactions; UPF1Δ20–150 is deficient for interaction with UPF2 and is missing one of the zinc fingers important for its binding to INSs on RNAs; and UPF1ΔN40 lacks part of the putative SMG5 interaction site.
Antibodies and reagents
Antisera to UPF proteins were generously supplied by Jens Lykke-Andersen (University of Colorado). Anti-p24 (CA) and anti-GAPDH antisera were purchased from Intracell and TechniScience and were described previously (Levesque et al. 2006). Anti-Flag antisera and cycloheximide were purchased from Sigma-Aldrich.
IF/FISH analyses
Immunofluorescence and fluorescence in situ hybridization were performed exactly as previously described (Levesque et al. 2006).
Colocalization analyses
For colocalization studies, laser scanning confocal microscopy (LSCM) was performed using a Zeiss Pascal LSM5 confocal microscope. Cells were mock transfected with Flag DNA or transfected with proviral HIV-1 DNA, pNL4-3, fixed at 30–36 h post-transfection, and processed for immunofluorescence as previously described (Levesque et al. 2006). Fixed cells were costained with anti-Gag p17 antisera (1:400, sheep anti-p17; from Michael Phelan, NIH AIDS Reference and Reagent Program) and rabbit anti-UPF1 antisera (1:200; from Jens Lykke-Anderson) as previously described (Levesque et al. 2006). The amount of colocalization was calculated using Manders' overlap coefficient (for entire cells) or colocalization coefficients m1 and m2 using Colocalizer Pro Software (Colocalization Research Software) as described previously (Levesque et al. 2006). The latter quantitates the contributions of each channel [(red) UPF1; (blue) Gag] to a region of interest providing the percentage of UPF1 that colocalizes with the Gag signals in the depicted region of cell is shown in the figure inset. The results shown are representative of two independently performed experiments and were calculated from more than 50 cells per condition.
Immunoprecipitation analyses
HeLa cells were transfected as described above. Cells were lysed for 30 min on ice by NP-40 lysis buffer, and Gag was immunoprecipitated as described previously using 2 mg of protein and affinity-purified mouse anti-p24 antisera (hybridoma 183-H12-5C) (Chesebro et al. 1992) from the NIH AIDS Reference and Reagent Program (Chatel-Chaix et al. 2004). Signals obtained in Western blotting of UPF1 and Gag (input and output levels) were quantitated by ImageJ software (NIH, Bethesda, MD) in order to estimate the proportion of UPF1 and Gag found in Staufen1-TAP eluates.
SUPPLEMENTAL DATA
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Miles Wilkinson and Melissa Moore for reagents and constructive discussions; Juan Ortin, Jens Lykke-Andersen, Larry Kleiman, Marv Wickens, Damian Purcell, David Ott, Anne Gatignol, Éric Cohen, Resa Sadaie, Bruce Chesebro, Hardy Chen, and Michael Phelan for antisera, reagents, and genetic clones; and the NIH AIDS Reference and Reagent Program for reagents. We thank Martin Lehmann for supplying additional images for analysis. L.A. (the first author) is a recipient of a Canadian Institutes of Health Research (CIHR) doctoral research fellowship. P.V.I. is supported by a FEBS long-term fellowship. A.J.M. is a recipient of a CIHR New Investigator Award. This work was supported by grants from the Deutsche Forschungsgemeinschaft to N.H.G. and A.E.K. (KU 563/7, KU 563/8, KU563/11) and the CIHR to A.J.M. (MOP-38111) and to A.J.M., A.E.K., and N.H.G. (OPC-83178).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.829208.
REFERENCES
- Andrulis, E.D., Werner, J., Nazarian, A., Erdjument-Bromage, H., Tempst, P., Lis, J.T. The RNA processing exosome is linked to elongating RNA polymerase II in Drosophila . Nature. 2002;420:837–841. doi: 10.1038/nature01181. [DOI] [PubMed] [Google Scholar]
- Applequist, S.E., Selg, M., Raman, C., Jack, H.M. Cloning and characterization of HUPF1, a human homolog of the Saccharomyces cerevisiae nonsense mRNA-reducing UPF1 protein. Nucleic Acids Res. 1997;25:814–821. doi: 10.1093/nar/25.4.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Askjaer, P., Jensen, T.H., Nilsson, J., Englmeier, L., Kjems, J. The specificity of the CRM1-Rev nuclear export signal interaction is mediated by RanGTP. J. Biol. Chem. 1998;273:33414–33422. doi: 10.1074/jbc.273.50.33414. [DOI] [PubMed] [Google Scholar]
- Atkin, A.L., Altamura, N., Leeds, P., Culbertson, M.R. The majority of yeast UPF1 co-localizes with polyribosomes in the cytoplasm. Mol. Biol. Cell. 1995;6:611–625. doi: 10.1091/mbc.6.5.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzalin, C.M., Lingner, J. The human RNA surveillance factor UPF1 is required for S phase progression and genome stability. Curr. Biol. 2006;16:433–439. doi: 10.1016/j.cub.2006.01.018. [DOI] [PubMed] [Google Scholar]
- Beriault, V., Clement, J.F., Levesque, K., Lebel, C., Yong, X., Chabot, B., Cohen, E.A., Cochrane, A.W., Rigby, W.F., Mouland, A.J. A late role for the association of hnRNP A2 with the HIV-1 hnRNP A2 response elements in genomic RNA, Gag, and Vpr localization. J. Biol. Chem. 2004;279:44141–44153. doi: 10.1074/jbc.M404691200. [DOI] [PubMed] [Google Scholar]
- Bhattacharya, A., Czaplinski, K., Trifillis, P., He, F., Jacobson, A., Peltz, S.W. Characterization of the biochemical properties of the human Upf1 gene product that is involved in nonsense-mediated mRNA decay. RNA. 2000;6:1226–1235. doi: 10.1017/s1355838200000546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black, A.C., Luo, J., Chun, S., Bakker, A., Fraser, J.K., Rosenblatt, J.D. Specific binding of polypyrimidine tract binding protein and hnRNP A1 to HIV-1 CRS elements. Virus Genes. 1996;12:275–285. doi: 10.1007/BF00284648. [DOI] [PubMed] [Google Scholar]
- Bohne, J., Wodrich, H., Krausslich, H.G. Splicing of human immunodeficiency virus RNA is position-dependent suggesting sequential removal of introns from the 5′ end. Nucleic Acids Res. 2005;33:825–837. doi: 10.1093/nar/gki185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhler, M., Steiner, S., Mohn, F., Paillusson, A., Mühlemann, O. EJC-independent degradation of nonsense immunoglobulin-mu mRNA depends on 3′ UTR length. Nat. Struct. Mol. Biol. 2006;13:462–464. doi: 10.1038/nsmb1081. [DOI] [PubMed] [Google Scholar]
- Carter, M.S., Doskow, J., Morris, P., Li, S., Nhim, R.P., Sandstedt, S., Wilkinson, M.F. A regulatory mechanism that detects premature nonsense codons in T-cell receptor transcripts in vivo is reversed by protein synthesis inhibitors in vitro. J. Biol. Chem. 1995;270:28995–29003. doi: 10.1074/jbc.270.48.28995. [DOI] [PubMed] [Google Scholar]
- Chamieh, H., Ballut, L., Bonneau, F., Le Hir, H. NMD factors UPF2 and UPF3 bridge UPF1 to the exon junction complex and stimulate its RNA helicase activity. Nat. Struct. Mol. Biol. 2008;15:85–93. doi: 10.1038/nsmb1330. [DOI] [PubMed] [Google Scholar]
- Chatel-Chaix, L., Clement, J.F., Martel, C., Beriault, V., Gatignol, A., DesGroseillers, L., Mouland, A.J. Identification of Staufen in the human immunodeficiency virus type 1 Gag ribonucleoprotein complex and a role in generating infectious viral particles. Mol. Cell. Biol. 2004;24:2637–2648. doi: 10.1128/MCB.24.7.2637-2648.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, Z., Muhlrad, D., Lim, M.K., Parker, R., Song, H. Structural and functional insights into the human Upf1 helicase core. EMBO J. 2007;26:253–264. doi: 10.1038/sj.emboj.7601464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesebro, B., Wehrly, K., Nishio, J., Perryman, S. Macrophage-tropic human immunodeficiency virus isolates from different patients exhibit unusual V3 envelope sequence homogeneity in comparison with T-cell-tropic isolates: Definition of critical amino acids involved in cell tropism. J. Virol. 1992;66:6547–6554. doi: 10.1128/jvi.66.11.6547-6554.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou, C.F., Mulky, A., Maitra, S., Lin, W.J., Gherzi, R., Kappes, J., Chen, C.Y. Tethering KSRP, a decay-promoting AU-rich element-binding protein, to mRNAs elicits mRNA decay. Mol. Cell. Biol. 2006;26:3695–3706. doi: 10.1128/MCB.26.10.3695-3706.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciaudo, C., Bourdet, A., Cohen-Tannoudji, M., Dietz, H.C., Rougeulle, C., Avner, P. Nuclear mRNA degradation pathway(s) are implicated in Xist regulation and X chromosome inactivation. PLoS Genet. 2006;2:e94. doi: 10.1371/journal.pgen.0020094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochrane, A.W., McNally, M.T., Mouland, A.J. The retrovirus RNA trafficking granule: From birth to maturity. Retrovirology. 2006;3:18. doi: 10.1186/1742-4690-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen, B.R. Nuclear mRNA export: Insights from virology. Trends Biochem. Sci. 2003;28:419–424. doi: 10.1016/S0968-0004(03)00142-7. [DOI] [PubMed] [Google Scholar]
- D'Agostino, D.M., Felber, B.K., Harrison, J.E., Pavlakis, G.N. The Rev protein of human immunodeficiency virus type 1 promotes polysomal association and translation of gag/pol and vpu/env mRNAs. Mol. Cell. Biol. 1992;12:1375–1386. doi: 10.1128/mcb.12.3.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks, T.M., Lykke-Andersen, J. TTP and BRF proteins nucleate processing body formation to silence mRNAs with AU-rich elements. Genes & Dev. 2007;21:719–735. doi: 10.1101/gad.1494707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring, N.H., Neu-Yilik, G., Schell, T., Hentze, M.W., Kulozik, A.E. Y14 and hUpf3b form an NMD-activating complex. Mol. Cell. 2003;11:939–949. doi: 10.1016/s1097-2765(03)00142-4. [DOI] [PubMed] [Google Scholar]
- Gottlinger, H.G., Dorfman, T., Sodroski, J.G., Haseltine, W.A. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc. Natl. Acad. Sci. 1991;88:3195–3199. doi: 10.1073/pnas.88.8.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachet, O., Ephrussi, A. Drosophila Y14 shuttles to the posterior of the oocyte and is required for oskar mRNA transport. Curr. Biol. 2001;11:1666–1674. doi: 10.1016/s0960-9822(01)00508-5. [DOI] [PubMed] [Google Scholar]
- Harger, J.W., Dinman, J.D. Evidence against a direct role for the Upf proteins in frameshifting or nonsense codon readthrough. RNA. 2004;10:1721–1729. doi: 10.1261/rna.7120504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, F., Jacobson, A. Upf1p, Nmd2p, and Upf3p regulate the decapping and exonucleolytic degradation of both nonsense-containing mRNAs and wild-type mRNAs. Mol. Cell. Biol. 2001;21:1515–1530. doi: 10.1128/MCB.21.5.1515-1530.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov, P.V., Gehring, N.H., Kunz, J.B., Hentze, M.W., Kulozik, A.E. Interactions between UPF1, eRFs, PABP and the exon junction complex suggest an integrated model for mammalian NMD pathways. EMBO J. 2008;27:736–747. doi: 10.1038/emboj.2008.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashima, I., Yamashita, A., Izumi, N., Kataoka, N., Morishita, R., Hoshino, S., Ohno, M., Dreyfuss, G., Ohno, S. Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes & Dev. 2006;20:355–367. doi: 10.1101/gad.1389006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaygun, H., Marzluff, W.F. Regulated degradation of replication-dependent histone mRNAs requires both ATR and Upf1. Nat. Struct. Mol. Biol. 2005;12:794–800. doi: 10.1038/nsmb972. [DOI] [PubMed] [Google Scholar]
- Kim, Y.K., Furic, L., Desgroseillers, L., Maquat, L.E. Mammalian Staufen1 recruits Upf1 to specific mRNA 3′UTRs so as to elicit mRNA decay. Cell. 2005;120:195–208. doi: 10.1016/j.cell.2004.11.050. [DOI] [PubMed] [Google Scholar]
- Kotsopoulou, E., Kim, V.N., Kingsman, A.J., Kingsman, S.M., Mitrophanous, K.A. A Rev-independent human immunodeficiency virus type 1 (HIV-1)-based vector that exploits a codon-optimized HIV-1 gag-pol gene. J. Virol. 2000;74:4839–4852. doi: 10.1128/jvi.74.10.4839-4852.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmiak, H.A., Maquat, L.E. Applying nonsense-mediated mRNA decay research to the clinic: Progress and challenges. Trends Mol. Med. 2006;12:306–316. doi: 10.1016/j.molmed.2006.05.005. [DOI] [PubMed] [Google Scholar]
- Lejeune, F., Li, X., Maquat, L.E. Nonsense-mediated mRNA decay in mammalian cells involves decapping, deadenylating, and exonucleolytic activities. Mol. Cell. 2003;12:675–687. doi: 10.1016/s1097-2765(03)00349-6. [DOI] [PubMed] [Google Scholar]
- Levesque, K., Halvorsen, M., Abrahamyan, L., Chatel-Chaix, L., Poupon, V., Gordon, H., DesGroseillers, L., Gatignol, A., Mouland, A.J. Trafficking of HIV-1 RNA is mediated by heterogeneous nuclear ribonucleoprotein A2 expression and impacts on viral assembly. Traffic. 2006;7:1177–1193. doi: 10.1111/j.1600-0854.2006.00461.x. [DOI] [PubMed] [Google Scholar]
- Li, S., Leonard, D., Wilkinson, M.F. T cell receptor (TCR) mini-gene mRNA expression regulated by nonsense codons: A nuclear-associated translation-like mechanism. J. Exp. Med. 1997;185:985–992. doi: 10.1084/jem.185.6.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen, J., Shu, M.D., Steitz, J.A. Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell. 2000;103:1121–1131. doi: 10.1016/s0092-8674(00)00214-2. [DOI] [PubMed] [Google Scholar]
- Macchi, P., Kroening, S., Palacios, I.M., Baldassa, S., Grunewald, B., Ambrosino, C., Goetze, B., Lupas, A., St Johnston, D., Kiebler, M. Barentsz, a new component of the Staufen-containing ribonucleoprotein particles in mammalian cells, interacts with Staufen in an RNA-dependent manner. J. Neurosci. 2003;23:5778–5788. doi: 10.1523/JNEUROSCI.23-13-05778.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malim, M.H., Hauber, J., Fenrick, R., Cullen, B.R. Immunodeficiency virus rev trans-activator modulates the expression of the viral regulatory genes. Nature. 1988;335:181–183. doi: 10.1038/335181a0. [DOI] [PubMed] [Google Scholar]
- Maquat, L.E., Li, X. Mammalian heat shock p70 and histone H4 transcripts, which derive from naturally intronless genes, are immune to nonsense-mediated decay. RNA. 2001;7:445–456. doi: 10.1017/s1355838201002229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendell, J.T., Rhys, C.M., Dietz, H.C. Separable roles for rent1/hUpf1 in altered splicing and decay of nonsense transcripts. Science. 2002;298:419–422. doi: 10.1126/science.1074428. [DOI] [PubMed] [Google Scholar]
- Mendell, J.T., Sharifi, N.A., Meyers, J.L., Martinez-Murillo, F., Dietz, H.C. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat. Genet. 2004;36:1073–1078. doi: 10.1038/ng1429. [DOI] [PubMed] [Google Scholar]
- Mohr, S.E., Dillon, S.T., Boswell, R.E. The RNA-binding protein Tsunagi interacts with Mago Nashi to establish polarity and localize oskar mRNA during Drosophila oogenesis. Genes & Dev. 2001;15:2886–2899. doi: 10.1101/gad.927001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouland, A.J., Mercier, J., Luo, M., Bernier, L., DesGroseillers, L., Cohen, E.A. The double-stranded RNA-binding protein Staufen is incorporated in human immunodeficiency virus type 1: Evidence for a role in genomic RNA encapsidation. J. Virol. 2000;74:5441–5451. doi: 10.1128/jvi.74.12.5441-5451.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouland, A.J., Coady, M., Yao, X.J., Cohen, É.A. Hypophosphorylation of poly(A) polymerase and increased polyadenylation activity are associated with human immunodeficiency virus type 1 Vpr expression. Virology. 2002;292:221–230. doi: 10.1006/viro.2001.1261. [DOI] [PubMed] [Google Scholar]
- Mouland, A.J., Cohen, É.A., DesGroseillers, L. Trafficking of HIV-1 RNA: Recent progress involving host cell RNA-binding proteins. Curr. Genomics. 2003;4:196. [Google Scholar]
- Mühlemann, O., Mock-Casagrande, C.S., Wang, J., Li, S., Custodio, N., Carmo-Fonseca, M., Wilkinson, M.F., Moore, M.J. Precursor RNAs harboring nonsense codons accumulate near the site of transcription. Mol. Cell. 2001;8:33–43. doi: 10.1016/s1097-2765(01)00288-x. [DOI] [PubMed] [Google Scholar]
- Muhlrad, D., Parker, R. Aberrant mRNAs with extended 3′ UTRs are substrates for rapid degradation by mRNA surveillance. RNA. 1999;5:1299–1307. doi: 10.1017/s1355838299990829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazarenus, T., Cedarberg, R., Bell, R., Cheatle, J., Forch, A., Haifley, A., Hou, A., Wanja Kebaara, B., Shields, C., Stoysich, K., et al. Upf1p, a highly conserved protein required for nonsense-mediated mRNA decay, interacts with the nuclear pore proteins Nup100p and Nup116p. Gene. 2005;345:199–212. doi: 10.1016/j.gene.2004.10.005. [DOI] [PubMed] [Google Scholar]
- Nott, A., Le Hir, H., Moore, M.J. Splicing enhances translation in mammalian cells: An additional function of the exon junction complex. Genes & Dev. 2004;18:210–222. doi: 10.1101/gad.1163204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal, M., Ishigaki, Y., Nagy, E., Maquat, L.E. Evidence that phosphorylation of human Upfl protein varies with intracellular location and is mediated by a wortmannin-sensitive and rapamycin-sensitive PI 3-kinase-related kinase signaling pathway. RNA. 2001;7:5–15. doi: 10.1017/s1355838201000127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios, I.M., Gatfield, D., St Johnston, D., Izaurralde, E. An eIF4AIII-containing complex required for mRNA localization and nonsense-mediated mRNA decay. Nature. 2004;427:753–757. doi: 10.1038/nature02351. [DOI] [PubMed] [Google Scholar]
- Pomerantz, R.J., Seshamma, T., Trono, D. Efficient replication of human immunodeficiency virus type 1 requires a threshold level of Rev: Potential implications for latency. J. Virol. 1992;66:1809–1813. doi: 10.1128/jvi.66.3.1809-1813.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon, D.T., Chertova, E.N., Ott, D.E. Human immunodeficiency virus type 1 preferentially encapsidates genomic RNAs that encode Pr55Gag: Functional linkage between translation and RNA packaging. Virology. 2002;293:368–378. doi: 10.1006/viro.2001.1283. [DOI] [PubMed] [Google Scholar]
- Sadaie, M.R., Benter, T., Wong-Staal, F. Site-directed mutagenesis of two trans-regulatory genes (tat-III,trs) of HIV-1. Science. 1988;239:910–913. doi: 10.1126/science.3277284. [DOI] [PubMed] [Google Scholar]
- Schwartz, S., Felber, B.K., Pavlakis, G.N. Distinct RNA sequences in the gag region of human immunodeficiency virus type 1 decrease RNA stability and inhibit expression in the absence of Rev protein. J. Virol. 1992;66:150–159. doi: 10.1128/jvi.66.1.150-159.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibuya, T., Tange, T.O., Sonenberg, N., Moore, M.J. eIF4AIII binds spliced mRNA in the exon junction complex and is essential for nonsense-mediated decay. Nat. Struct. Mol. Biol. 2004;11:346–351. doi: 10.1038/nsmb750. [DOI] [PubMed] [Google Scholar]
- Sun, X., Perlick, H.A., Dietz, H.C., Maquat, L.E. A mutated human homologue to yeast Upf1 protein has a dominant-negative effect on the decay of nonsense-containing mRNAs in mammalian cells. Proc. Natl. Acad. Sci. 1998;95:10009–10014. doi: 10.1073/pnas.95.17.10009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villace, P., Marion, R.M., Ortin, J. The composition of Staufen-containing RNA granules from human cells indicates their role in the regulated transport and translation of messenger RNAs. Nucleic Acids Res. 2004;32:2411–2420. doi: 10.1093/nar/gkh552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J., Hamilton, J.I., Carter, M.S., Li, S., Wilkinson, M.F. Alternatively spliced TCR mRNA induced by disruption of reading frame. Science. 2002;297:108–110. doi: 10.1126/science.1069757. [DOI] [PubMed] [Google Scholar]
- Wilkinson, M.F. A new function for nonsense-mediated mRNA-decay factors. Trends Genet. 2005;21:143–148. doi: 10.1016/j.tig.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Zhang, J., Sun, X., Qian, Y., Maquat, L.E. Intron function in the nonsense-mediated decay of β-globin mRNA: Indications that pre-mRNA splicing in the nucleus can influence mRNA translation in the cytoplasm. RNA. 1998;4:801–815. doi: 10.1017/s1355838298971849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolotukhin, A.S., Michalowski, D., Bear, J., Smulevitch, S.V., Traish, A.M., Peng, R., Patton, J., Shatsky, I.N., Felber, B.K. PSF acts through the human immunodeficiency virus type 1 mRNA instability elements to regulate virus expression. Mol. Cell. Biol. 2003;23:6618–6630. doi: 10.1128/MCB.23.18.6618-6630.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]