Abstract
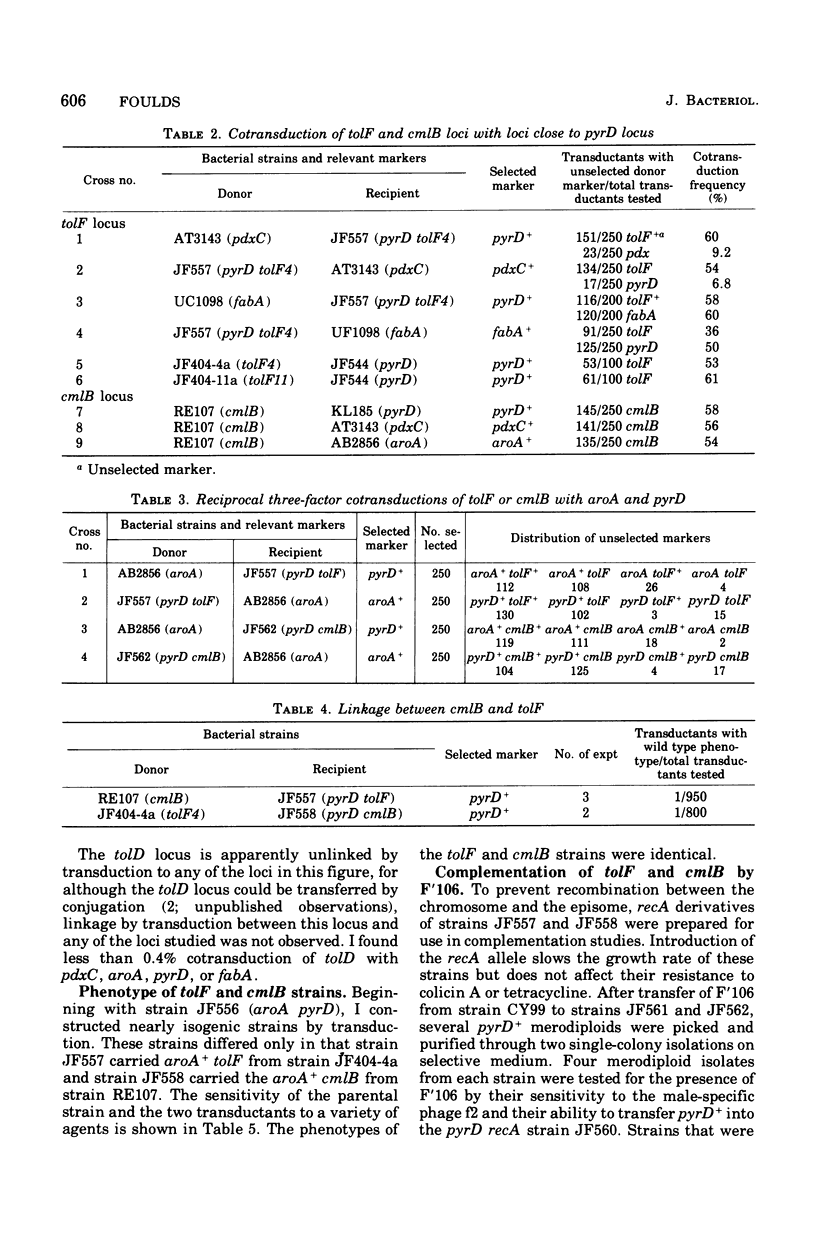
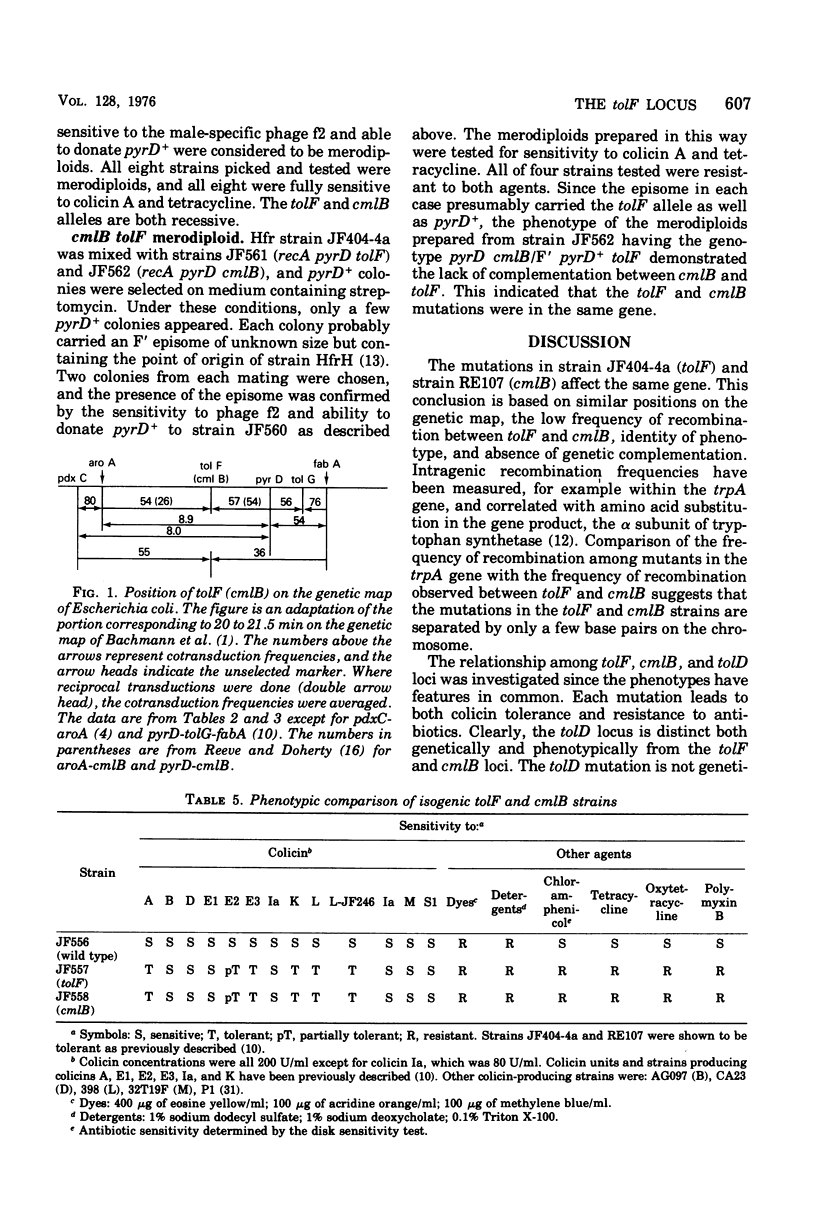
The tentative map position on the Escherichia coli chromosome of the tolF locus, determining tolerance to colicins A, E2, E3, K, and L, has been confirmed by three-point transduction. It lies between the aroA and pyrD loci at about 21 min on the linkage map of Bachmann et al. (1976). The cmlB locus, determining increased resistance to the antibiotics chloramphenicol and tetracycline, also lies in this region (Reeve, 1966). Phenotypic and genetic comparison of isogenic strains that carry a mutation in either the tolF or cmlB locus makes it likely that these loci are closely related or identical. The tolD locus determining tolerance to colicins E2 and E3 as well as increased resistance to antibiotics has been reported to be located close to the aroA locus as a result of conjugation experiments (Eriksson-Grennberg et al. 1965). However, tolD did not cotransduce with any of several loci in this region, indicating that the mutation is not located within the region of the genetic map corresponding to approximately 19 to 22.5 min.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bachmann B. J., Low K. B., Taylor A. L. Recalibrated linkage map of Escherichia coli K-12. Bacteriol Rev. 1976 Mar;40(1):116–167. doi: 10.1128/br.40.1.116-167.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burman L. G., Nordström K. Colicin tolerance induced by ampicillin or mutation to ampicillin resistance in a strain of Escherichia coli K-12. J Bacteriol. 1971 Apr;106(1):1–13. doi: 10.1128/jb.106.1.1-13.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronan J. E., Jr, Silbert D. F., Wulff D. L. Mapping of the fabA locus for unsaturated fatty acid biosynthesis in Escherichia coli. J Bacteriol. 1972 Oct;112(1):206–211. doi: 10.1128/jb.112.1.206-211.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies J. K., Reeves P. Genetics of resistance to colicins in Escherichia coli K-12: cross-resistance among colicins of group A. J Bacteriol. 1975 Jul;123(1):102–117. doi: 10.1128/jb.123.1.102-117.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Haan P. G., Hoekstra W. P., Verhoef C., Felix H. S. Recombination in Escherichia coli. 3. Mapping by the gradient of transmission. Mutat Res. 1969 Nov-Dec;8(3):505–512. doi: 10.1016/0027-5107(69)90067-0. [DOI] [PubMed] [Google Scholar]
- Eriksson-Grennberg K. G., Boman H. G., Jansson J. A., Thorén S. Resistance of Escherichia coli to Penicillins I. Genetic Study of Some Ampicillin-Resistant Mutants. J Bacteriol. 1965 Jul;90(1):54–62. doi: 10.1128/jb.90.1.54-62.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulds J., Barrett C. Characterization of Escherichia coli mutants tolerant to bacteriocin JF246: two new classes of tolerant mutants. J Bacteriol. 1973 Nov;116(2):885–892. doi: 10.1128/jb.116.2.885-892.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulds J. Chromosomal location of the tolG locus for tolerance to bacteriocin JF246 in Escherichia coli K-12. J Bacteriol. 1974 Mar;117(3):1354–1355. doi: 10.1128/jb.117.3.1354-1355.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulds J. Purification and partial characterization of a bacteriocin from Serratia marcescens. J Bacteriol. 1972 Jun;110(3):1001–1009. doi: 10.1128/jb.110.3.1001-1009.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HELINSKI D. R., YANOFSKY C. Correspondence between genetic data and the position of amino acid alteration in a proein. Proc Natl Acad Sci U S A. 1962 Feb;48:173–183. doi: 10.1073/pnas.48.2.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low B. Formation of merodiploids in matings with a class of Rec- recipient strains of Escherichia coli K12. Proc Natl Acad Sci U S A. 1968 May;60(1):160–167. doi: 10.1073/pnas.60.1.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeve E. C. Characteristics of some single-step mutants to chloramphenicol resistance in Escherichia coli K12 and their interactions with R-factor genes. Genet Res. 1966 Apr;7(2):281–286. doi: 10.1017/s0016672300009708. [DOI] [PubMed] [Google Scholar]
- Reeve E. C., Doherty P. Linkage relationships of two genes causing partial resistance to chloramphenicol in Escherichia coli. J Bacteriol. 1968 Oct;96(4):1450–1451. doi: 10.1128/jb.96.4.1450-1451.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeve E. C. Genetic analysis of some mutations causing resistance to tetracycline in Escherichia coli K12. Genet Res. 1968 Jun;11(3):303–309. doi: 10.1017/s0016672300011484. [DOI] [PubMed] [Google Scholar]
- Rossi J. J., Berg C. M. Differential recovery of auxotrophs after penicillin enrichment in Escherichia coli. J Bacteriol. 1971 May;106(2):297–300. doi: 10.1128/jb.106.2.297-300.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STACEY K. A., SIMSON E. IMPROVED METHOD FOR THE ISOLATION OF THYMINE-REQUIRING MUTANTS OF ESCHERICHIA COLI. J Bacteriol. 1965 Aug;90:554–555. doi: 10.1128/jb.90.2.554-555.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnaitman C. A. Outer membrane proteins of Escherichia coli. 3. Evidence that the major protein of Escherichia coli O111 outer membrane consists of four distinct polypeptide species. J Bacteriol. 1974 May;118(2):442–453. doi: 10.1128/jb.118.2.442-453.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signer E. R. Interaction of prophages at the att80 site with the chromosome of Escherichia coli. J Mol Biol. 1966 Jan;15(1):243–255. doi: 10.1016/s0022-2836(66)80224-3. [DOI] [PubMed] [Google Scholar]



