Abstract
A novel virus, designated swine hepatitis E virus (swine HEV), was identified in pigs. Swine HEV crossreacts with antibody to the human HEV capsid antigen. Swine HEV is a ubiquitous agent and the majority of swine ≥3 months of age in herds from the midwestern United States were seropositive. Young pigs naturally infected by swine HEV were clinically normal but had microscopic evidence of hepatitis, and developed viremia prior to seroconversion. The entire ORFs 2 and 3 were amplified by reverse transcription–PCR from sera of naturally infected pigs. The putative capsid gene (ORF2) of swine HEV shared about 79–80% sequence identity at the nucleotide level and 90–92% identity at the amino acid level with human HEV strains. The small ORF3 of swine HEV had 83–85% nucleotide sequence identity and 77–82% amino acid identity with human HEV strains. Phylogenetic analyses showed that swine HEV is closely related to, but distinct from, human HEV strains. The discovery of swine HEV not only has implications for HEV vaccine development, diagnosis, and biology, but also raises a potential public health concern for zoonosis or xenozoonosis following xenotransplantation with pig organs.
Hepatitis E occurs predominantly in developing countries of Asia and Africa, but has also been the cause of epidemics in Mexico (1). The disease generally affects young adults and has a very high mortality rate, up to 20%, in infected pregnant women (1–4). Hepatitis E has rarely been reported in developed countries, and most of those cases have been imported (1, 4–6). The causative agent, hepatitis E virus (HEV), is transmitted primarily by the fecal–oral route, often through contaminated water (1, 4). The availability of sensitive serological tests for HEV has permitted detailed assessment of the prevalence of HEV infection (7, 8). In regions where HEV is endemic, anti-HEV have been detected in sera from convalescent individuals, as well as from the general population (1, 3). Although hepatitis E is not endemic in the United States and other developed countries, anti-HEV was found in a significant proportion, up to 28% in some areas, of healthy individuals in these countries (7, 9). It is not clear whether the anti-HEV detected in the developed countries represents infection with a nonpathogenic HEV strain or crossreactivity with a related agent.
It has been reported that anti-HEV is acquired naturally in primates and swine (1, 10), suggesting that these species have been exposed to HEV or a related agent, and that hepatitis E might be a zoonotic disease. The role of swine in HEV transmission is not clear, although domestic swine were reported to be susceptible to infection with a human HEV strain (11). Here we report the first identification of an animal strain of HEV. We designated this virus swine hepatitis E virus (swine HEV) to distinguish it from human HEV.
MATERIALS AND METHODS
Serum Samples.
Serum samples were obtained from swine of various ages in 15 commercial herds and 1 specific-pathogen-free (SPF) herd in the midwestern United States (Table 1).
Table 1.
IgG anti-HEV prevalence in swine from commercial herds in the United States
| Herd | Age | No. of swine tested | No. of swine with anti-HEV (%) |
|---|---|---|---|
| A | 6 wk | 8 | 0 (0) |
| 12 wk | 8 | 0 (0) | |
| 20 wk | 8 | 8 (100) | |
| 26 wk | 8 | 5 (63) | |
| Adult | 25 | 14 (56) | |
| B | 3–4 wk | 8 | 0 (0) |
| 5–6 wk | 8 | 0 (0) | |
| 7–8 wk | 8 | 0 (0) | |
| 13 wk | 12 | 10 (83) | |
| 6 mo | 8 | 8 (100) | |
| Adult | 17 | 16 (94) | |
| C | 2 mo | 8 | 1 (13) |
| 3 mo | 8 | 8 (100) | |
| 4 mo | 8 | 4 (50) | |
| 5 mo | 8 | 8 (100) | |
| Adult | 8 | 8 (100) | |
| D | 2 mo | 10 | 2 (20) |
| E | 6 mo | 10 | 10 (100) |
| F | 6 mo | 10 | 10 (100) |
| G | 6 mo | 10 | 10 (100) |
| H | 8 mo | 10 | 10 (100) |
| I | >1 yr | 10 | 10 (100) |
| J | 1–2 yr | 10 | 10 (100) |
| K | 1–3 yr | 10 | 10 (100) |
| L | 2 yr | 10 | 10 (100) |
| M | 2–3 yr | 10 | 10 (100) |
| N | Adult | 19 | 15 (79) |
| O | Adult | 6 | 5 (83) |
| P* | Adult | 10 | 0 (0) |
Specific-pathogen-free (SPF) swine herd.
Preparation of HEV Putative Capsid Antigen.
Insect cells were infected with recombinant baculovirus containing the putative capsid gene (ORF2) sequence of a Pakistani strain of HEV, Sar55 (8). A 55-kDa recombinant protein expressed from a recombinant baculovirus containing ORF2 was purified from insect cells (8) and used for the standard ELISA. The ORF2 recombinant protein was further purified by HPLC liquid chromatography.
Generation of Hyperimmune Swine Antibody to HEV.
Two 3-week-old SPF swine were immunized intramuscularly with 50 μg of HPLC-purified ORF2 recombinant protein mixed with Freund’s incomplete adjuvant. Booster immunizations were given at 2 and 4 weeks after the first immunization. Sera obtained before immunization and weekly for 9 weeks after immunization were used to develop an ELISA (see below).
ELISA for Anti-HEV in Swine.
The standard ELISA for anti-HEV in swine was performed essentially as described for anti-HEV in chimpanzees (8, 12), except that the secondary antibody was replaced with peroxidase-labeled goat anti-swine IgG (Kirkegaard & Perry Laboratories). All of the swine serum samples were tested in duplicate. Preimmune and hyperimmune anti-HEV positive swine sera were included as negative and positive controls, respectively.
Blocking ELISA was used to confirm the results of the standard ELISA on selected anti-HEV positive and negative serum samples. The blocking ELISA for anti-HEV in swine was performed essentially as described (13) except that the competing sera were from swine. The ORF2 protein of strain Sar55 was used for affinity purification of anti-HEV from convalescent serum of a chimpanzee exposed twice to HEV (8). The affinity-purified chimpanzee anti-HEV was conjugated with horseradish peroxidase by a custom service (ViroStat, Portland, ME) and used for the blocking ELISA (13). A serum sample was considered positive in the blocking ELISA if the OD value was reduced by ≥50% compared with the unblocked sample.
Prospective Study.
Twenty-one sows from a commercial herd were tested for IgG anti-HEV. Subsequently, 20 piglets (10 male and 10 female) were chosen from those born to seronegative sows (6 piglets) and to seropositive sows with a lower titer (6 piglets) or higher titer (8 piglets) of IgG anti-HEV. These 20 study piglets were tagged and mixed with other piglets from approximately 50 sows in the herd, and were commingled in 2 rooms in a nursery building. Piglets within a room were separated into pens by fences that allowed for nose-to-nose contacts. By the age of about 10 weeks, all piglets in the nursery building were moved to an empty finishing building that had been disinfected.
Blood samples and nasal and rectal swabs from the 20 study piglets were collected in alternate weeks from 2 weeks onward, and weekly after 14 weeks of age. The serum samples were tested for anti-HEV, and four piglets with an increasing ELISA OD value were sacrificed. Samples of 19 different tissues and organs were collected during necropsy, fixed in 10% neutral buffered formalin, and processed for routine histologic examination.
Degenerate Primers for Reverse Transcription–PCR (RT-PCR).
The sequences of 10 human HEV strains were aligned with a GeneWorks program (IntelliGenetics). Based on this alignment, two sets of degenerate primers were designed and synthesized to amplify two different regions of the capsid gene. The primer positions indicated below are relative to the published sequence of a Burmese HEV strain (14). Set one primers: external 3156 [forward, position 5687–5708, 5′-AAT(C)TATGCC(A)CAGTACCGGGTTG-3′] and 3157 (reverse, position 6395–6417, 5′-CCCTTATCCTGCTGAGCATTCTC-3′), and internal 3158 [forward, position 5972–5993, 5′-GTT(C)ATGC(T)TT(C)TGCATACATGGCT-3′] and 3159 [reverse, position 6298–6319, 5′-AGCCGACGAAATC(T)AATTCTGTC-3′]. Set two primers: external 3160 [forward, position 6578–6600, 5′-GCCGAGTAT(C)GACCAGTCCACTTA-3′] and 3161 [reverse, position 7105–7127, 5′-AT(C)AACTCCCGAGTTTTACCCACC-3′], and internal 3162 [forward, position 6645–6667, 5′-TGGTT(G)AATGTT(A)GCGACC(T)GGCGCG-3′] and 3163 [reverse, position 7063–7085, 5′-GCTCAGCGACAGTA(T)GACTGG(A)AAA-3′].
RNA Extraction and RT-PCR.
Total RNA was extracted by TriZol reagent (GIBCO/BRL) from 100 μl of serum obtained 1 or 2 weeks before seroconversion from piglets in the prospective study. Total RNA was then reverse transcribed with one of the two degenerate primers by using SuperScript II reverse transcriptase (GIBCO/BRL) at 42°C for 1 hr, and the resulting cDNA was amplified by PCR using ampliTaq Gold polymerase (Perkin–Elmer). The PCR reaction was carried out for 39 cycles of denaturation at 94°C for 1 min, annealing at 42°C for 1 min, and extension at 72°C for 2 min, followed by a nested PCR using 10 μl of the first round PCR product with a nested set of degenerate primers.
Amplification of the Entire ORFs 2 and 3 of Swine HEV.
After the first PCR fragment was amplified and sequenced, we designed two sets of primers with a swine HEV-specific primer at one end and a degenerate primer at the other end (primer sequences not shown). RT-PCR with these primers was performed essentially as described above. The entire ORFs 2 and 3 of swine HEV were amplified in this way by walking along the genome in both directions.
Sequence Analysis.
The PCR fragments were cut from 1% agarose gels and purified with a Geneclean Kit (Bio 101). Both strands were sequenced with an automated DNA sequencer. The sequences were analyzed by the geneworks program. Phylogenetic analyses were conducted with the aid of the paup software package, version 3.1.1 (David L. Swofford, Illinois Natural History Survey, Champaign).
RESULTS
Standardization of an ELISA for Swine Anti-HEV.
To establish a reliable serological test for anti-HEV in swine, we first generated hyperimmune antisera by immunizing two SPF pigs with recombinant HEV ORF2 antigen. IgG anti-HEV was first detected at 2 and 3 weeks postimmunization and reached peak ELISA titers of 10−4 at weeks 3 and 4 after immunization, respectively. The anti-HEV titer remained at 10−4 until the end of the 9-week experiment. An ELISA for swine anti-HEV was subsequently standardized by using the preimmune and hyperimmune swine sera along with 34 normal sera from swine raised in laboratory environments and from swine in an SPF herd. The ELISA cutoff value was set at 99% confidence bounds, based on the frequency distribution of the absorbance values of normal sera. The cutoff value was approximately 2.5 SD above the mean absorbance value of the normal sera.
Serological Evidence in Swine for Infection with an Agent Related to HEV.
The prevalence of anti-HEV in commercial swine herds in the midwestern United States was assessed by the standardized ELISA. Surprisingly, IgG anti-HEV was found in a majority of swine ≥3 months of age in herds in the midwestern United States, where human hepatitis E is not endemic; however, most swine ≤2 months of age were seronegative (Table 1). None of the 10 adult swine from an SPF herd was seropositive (Table 1). To further validate the serology results, we performed a blocking ELISA on selected anti-HEV positive and negative swine sera. Similar results were obtained in the blocking ELISA and the standard ELISA. The HEV antigen used in ELISA was expressed in insect cells; therefore, we also included insect cells infected with baculovirus lacking the HEV sequence as a negative antigen control and HPLC-purified HEV recombinant antigen as a positive antigen control. There was little or no reaction between the swine serum samples and the insect cells infected with the baculovirus lacking the HEV sequence. Thus, the swine anti-HEV reacted specifically with the human HEV capsid antigen in the standard ELISA and competed with anti-HEV in convalescent chimpanzee serum in the blocking ELISA. These data strongly suggested that a ubiquitous swine agent, antigenically related to human HEV, was circulating in the general swine population.
Natural Infection of Swine in a Commercial Herd.
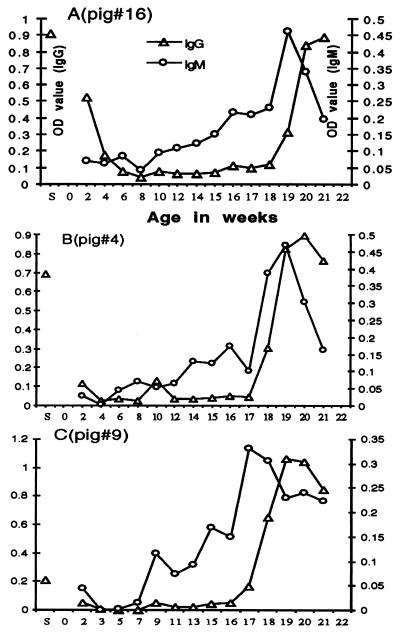
In an attempt to identify this putative HEV-related agent in pigs, a prospective study was conducted in a commercial swine herd in the midwestern United States. Consistent with our seroepidemiological results, 18 of 21 pregnant sows from this herd tested positive for anti-HEV. Piglets born to seronegative sows were seronegative at 2 weeks of age, and piglets born to seropositive sows with a lower titer of IgG anti-HEV also scored as seronegative, but had a comparatively high ELISA OD value for IgG anti-HEV (Table 2). In contrast, piglets born to seropositive sows with a higher titer of IgG anti-HEV were positive at 2 weeks of age for IgG anti-HEV (Table 2), but not for IgM anti-HEV. The level of IgG anti-HEV in seropositive piglets decreased dramatically within a few weeks after birth and had disappeared by the age of 8 or 9 weeks (Table 2, Fig. 1). Clearly, the anti-HEV detected in these newborns represented maternal antibody as evidenced by the correlation between the levels of anti-HEV in 2-week-old piglets and in their dams (Table 2), and from the fact that the anti-HEV belonged to the IgG class. However, after the maternal antibody had waned, most of the piglets developed their own antibodies to HEV. One piglet seroconverted to anti-HEV at the age of 14 weeks, followed within a few weeks by seroconversion of piglets in other pens housed in the same finishing building. The pattern of anti-HEV appearance, starting with piglets grouped near the first seropositive piglet, then followed by more distal ones, was consistent with seroconversion induced by an infectious agent (data not shown). By 21 weeks of age, 16 of the 20 study piglets had seroconverted. Two other piglets were necropsied prior to seroconversion and one piglet died of an unknown cause. The only remaining seronegative piglet had a rising ELISA OD value, but it was still below the cut-off value at the end of the 21 week study. The level of IgG anti-HEV increased steadily for several weeks after seroconversion (Fig. 1). IgM anti-HEV, indicating a newly contracted infection with this putative HEV-related agent, was also detected in all piglets that seroconverted to IgG anti-HEV. The level of IgM anti-HEV peaked about 1 week earlier than that of IgG anti-HEV and decreased rapidly over about 1–2 weeks (Fig. 1).
Table 2.
Seroconversion of piglets to anti-HEV in a commercial herd: A prospective study
| Piglet no. | Sow ELISA OD* | Piglet ELISA OD,* wk
|
Age seroconverted, wk | |
|---|---|---|---|---|
| 2 | 8 or 9 | |||
| 15 | 0.908 | 0.550 | 0.036 | † |
| 16 | 0.908 | 0.522 | 0.039 | 19 |
| 17 | 0.908 | 0.501 | 0.066 | ‡ |
| 18 | 1.011 | 0.822 | 0.103 | † |
| 19 | 1.011 | 1.264 | 0.148 | 15 |
| 20 | 1.011 | 0.979 | 0.146 | § |
| 1 | 0.692 | 0.128 | 0.024 | 18 |
| 2 | 0.692 | 0.211 | 0.071 | 21 |
| 3 | 0.692 | 0.157 | 0.052 | 20 |
| 4 | 0.692 | 0.114 | 0.026 | 18 |
| 5 | 0.431 | 0.107 | 0.065 | 18 |
| 6 | 0.431 | 0.216 | 0.059 | 21 |
| 7 | 0.424 | 0.113 | 0.073 | 16† |
| 8 | 0.424 | 0.195 | 0.093 | 15 |
| 9 | 0.209 | 0.047 | 0.050 | 18 |
| 10 | 0.209 | 0.079 | 0.090 | 19 |
| 11 | 0.209 | 0.047 | 0.039 | 19 |
| 12 | 0.245 | 0.057 | 0.061 | 14 |
| 13 | 0.245 | 0.056 | 0.038 | 16 |
| 14 | 0.245 | 0.057 | 0.012 | 20† |
ELISA cut-off value, 0.3.
Necropsied.
Remained seronegative at 21 weeks of age.
Death due to unknown cause.
Figure 1.
Seroconversion of piglets to anti-HEV. Anti-HEV responses of three representative piglets are presented. (A) Piglet born to a seropositive sow with a high titer of IgG anti-HEV. (B) Piglet born to a seropositive sow with a lower titer of IgG anti-HEV. (C) Piglet born to a seronegative sow. The ELISA OD value of IgG anti-HEV in breeder sows is indicated (S).
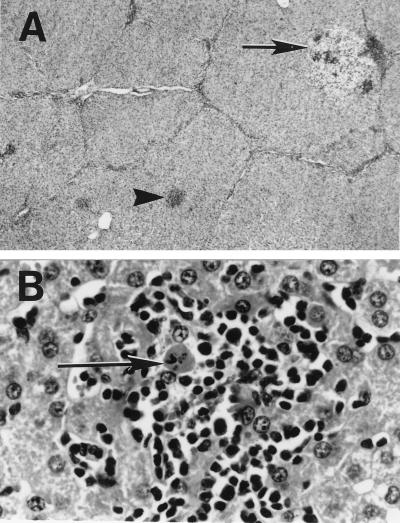
Clinical illness was not observed in the piglets. Four piglets believed to be at an early stage of infection were necropsied during the study. Except for a gross lung lesion consistent with a bacterial pneumonia in one piglet, other gross lesions were not apparent in 19 different tissues and organs examined during necropsy. Microscopically, all four piglets necropsied had evidence of hepatitis characterized by mild to moderate multifocal and periportal lymphoplasmacytic hepatitis with mild focal hepatocellular necrosis (Fig. 2). In addition, all piglets had lymphoplasmacytic enteritis, and three piglets also had mild multifocal lymphoplasmacytic interstitial nephritis. Syncytial cells were also noticed in the tonsils and Peyer’s patches of one piglet (data not shown).
Figure 2.
Liver sections from a naturally infected piglet (no. 14). (A) Multifocal lymphoplasmacytic and necrotizing hepatitis with randomly distributed foci of hepatocellular swelling and vacuolation (arrow) and foci of necrosis (arrowhead) with lymphoplasmacytic sinusoidal and periportal infiltrates (×10). (B) Foci of hepatocellular necrosis (arrow) and lymphoplasmacytic inflammation (×400). Hematoxylin/eosin staining.
Genetic Characterization of the Swine HEV.
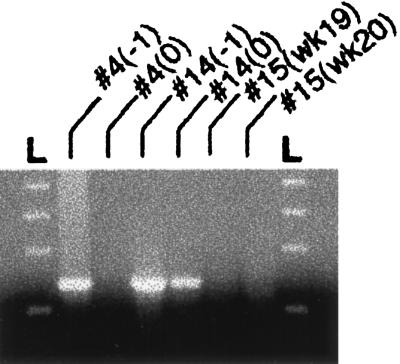
Because the swine anti-HEV reacted so strongly with the capsid protein of human HEV, it was probable that swine HEV shared nucleotide sequence similarity with human HEV. Therefore, two sets of degenerate primers derived from the HEV putative capsid gene were used to attempt the amplification of the swine HEV genome by RT-PCR of serum samples obtained 1 and 2 weeks before seroconversion. A fragment representing part of the swine HEV genome (Fig. 3) was first amplified by a nested PCR with primer set 3158 and 3159. Sequence information confirmed that this initial PCR fragment was specific for swine HEV and represented part of the ORF2.
Figure 3.
Amplification of swine HEV-specific fragment by RT-PCR. Serum samples from two piglets (nos. 4 and 14) obtained 1 week before (−1) and the week of (0) seroconversion in a prospective study were used for RT-PCR of a 344 bp fragment. Serum samples obtained at the same time (weeks 19 and 20) after birth from a seronegative piglet (no. 15) were also included. L, molecular weight marker.
Sequence Analyses of Swine HEV ORFs 2 and 3.
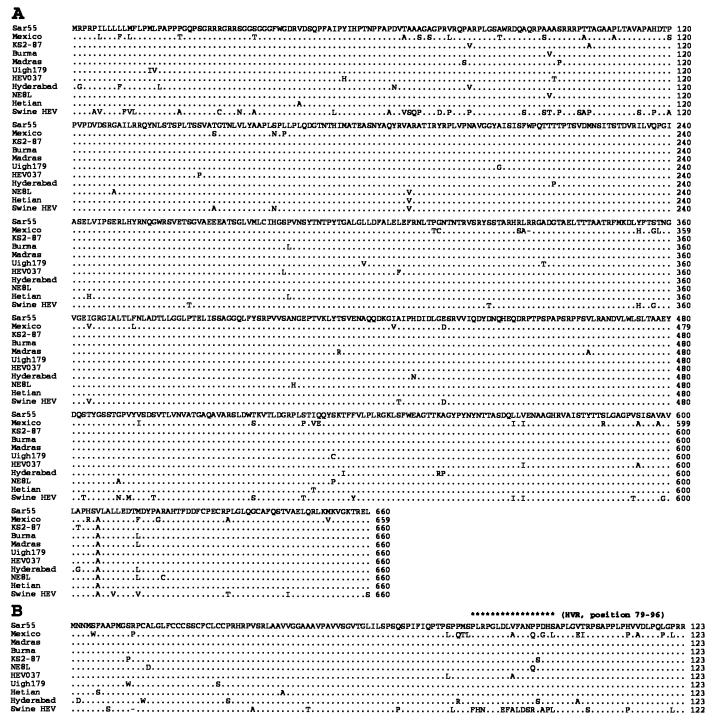
Analyses of the complete ORFs 2 and 3 sequences revealed that swine HEV is closely related to, although distinct from, human HEV strains. In the putative capsid gene (ORF2), swine HEV shares with human HEV strains about 79–80% sequence identity at the nucleotide level, and about 90–92% identity at the amino acid level (Table 3, Fig. 4). However, the relatively high amino acid identity between swine and human HEV is significantly lower than the amino acid identity (97–99%) among human HEV strains with the exception of the Mexican strain. The Mexican strain of HEV also displayed greater sequence divergence of about 92–93% amino acid identity with other human HEV strains (Table 3). However, the genetic distances between swine HEV and the Mexican strain of HEV are comparable to those between swine HEV and other human HEV strains, indicating that swine HEV is also distinct from the Mexican HEV (Table 3, Fig. 4). These data suggested that we had identified a previously unrecognized swine virus belonging to the same family as human HEV.
Table 3.
Pairwise comparison of the nucleotide and deduced amino acid sequences of ORFs 2 and 3 of the swine HEV with human HEV strains
| Virus strains | SHEV | Mexico | HEV037 | Uigh179 | Hetian | KS2-87 | Sar55 | Madras | Hyderabad | Burma | NE8L |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ORF2 | |||||||||||
| SHEV | 79 (90) | 80 (91) | 80 (91) | 80 (91) | 80 (92) | 80 (92) | 79 (92) | 79 (90) | 79 (92) | 79 (91) | |
| Mexico | 83 (79) | 81 (93) | 81 (92) | 81 (93) | 81 (93) | 81 (93) | 81 (93) | 81 (92) | 81 (93) | 81 (92) | |
| HEV037 | 84 (82) | 90 (89) | 94 (98) | 94 (98) | 94 (98) | 94 (99) | 94 (98) | 92 (97) | 94 (98) | 94 (98) | |
| Uigh179 | 84 (80) | 90 (85) | 97 (97) | 98 (98) | 98 (99) | 97 (99) | 93 (98) | 93 (97) | 94 (99) | 93 (98) | |
| Hetian | 84 (80) | 90 (85) | 97 (97) | 98 (97) | 99 (99) | 98 (99) | 93 (98) | 93 (97) | 94 (99) | 93 (98) | |
| KS2-87 | 84 (80) | 90 (85) | 97 (97) | 98 (98) | 98 (97) | 98 (99) | 93 (99) | 93 (98) | 94 (99) | 94 (98) | |
| Sar55 | 85 (82) | 91 (87) | 98 (98) | 99 (98) | 99 (98) | 99 (98) | 93 (99) | 93 (98) | 94 (99) | 93 (99) | |
| Madras | 85 (82) | 90 (87) | 98 (98) | 98 (98) | 98 (98) | 98 (98) | 99 (100) | 96 (98) | 97 (99) | 96 (98) | |
| Hyderabad | 83 (77) | 89 (84) | 95 (93) | 96 (93) | 96 (93) | 97 (95) | 97 (95) | 97 (95) | 97 (98) | 96 (97) | |
| Burma | 84 (82) | 90 (87) | 98 (98) | 98 (98) | 98 (98) | 98 (98) | 99 (100) | 99 (100) | 97 (95) | 98 (99) | |
| NE8L | 84 (81) | 90 (87) | 98 (97) | 98 (97) | 97 (97) | 97 (97) | 98 (98) | 98 (98) | 96 (93) | 99 (98) | |
| ORF3 | |||||||||||
The values in the table represent the percentage identity of nucleotide or amino acid (in parentheses) sequences
Figure 4.
Alignment of the amino acid sequences of ORFs 2 (A) and 3 (B) of swine HEV with human strains of HEV. The sequence of Sar55 strain is shown on top, and only differences are indicated. Deletions are indicated by a minus. The putative hypervariable region (HVR) in the ORF3 is indicated by asterisks. Sequences used in this alignment were Burma (14), Mexico (15), NE8L (Myanmar, ref. 16), Hyderabad (India, ref. 17), Madras (India, GenBank accession no. X99441), HEV037 (isolate from a case of fulminant hepatitis, GenBank accession no. X98292), Sar55 (Pakistan, ref. 18), KS2-87 (China, ref. 19), Hetian (China, GenBank accession no. L08816), and Uigh179 (China, ref. 20).
The small ORF3 of swine HEV had about 83–85% sequence identity at the nucleotide level with human HEV strains, but only 77–82% identity at the amino acid level (Table 3). The human HEV strains also displayed a lower percentage of identities at the amino acid level than that at the nucleotide level (Table 3). Most of the amino acid variations in ORF3 were clustered in a hypervariable region consisting of 17 amino acid residues near the carboxyl terminus (Fig. 4). In addition, the ORF3 of swine HEV had a single amino acid deletion near the amino terminus (Fig. 4).
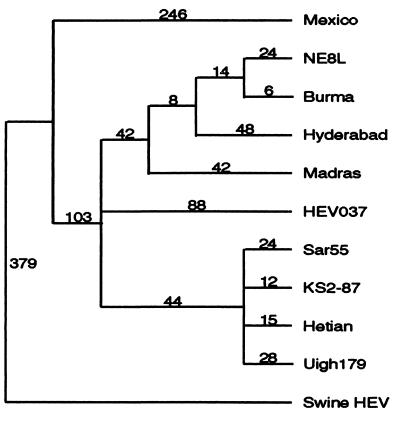
The evolutionary relationships between swine and human HEV were determined on the basis of the complete nucleotide sequences of ORFs 2 and 3. The resulting phylogenetic tree revealed that human HEV strains were represented by at least two genotypes. The first genotype was represented by the Mexican strain and the second genotype by the other human HEV strains (Fig. 5). Phylogenetically, swine HEV is unique, the most divergent of the HEV strains compared, and represents the first member of a third genotype (Fig. 5).
Figure 5.
Phylogenetic tree based on the complete nucleotide sequences of ORFs 2 and 3. The tree was constructed by maximum parsimony methods with the aid of paup software package version 3.1.1. The tree with the shortest length (most parsimonious) was found by implementing the bootstrap (1,000 replicates) with branch-and-bound search option. Branch lengths (number given above each branch) are indicated.
DISCUSSION
We have identified a novel HEV in pigs by documenting seroconversion of pigs to anti-HEV, sequence similarity to human strains of HEV, viremia just prior to seroconversion, and histologic evidence of hepatitis in the naturally infected pigs. Because the swine anti-HEV crossreacted with human HEV capsid antigen, and because the infected piglets had microscopic evidence of hepatitis during the acute stage of infection, we have tentatively designated this HEV-related agent in pigs as swine HEV.
The high prevalence of anti-HEV in commercial swine herds suggested that swine HEV is widespread in the general swine population. However, results from the prospective study showed that the naturally infected young pigs did not display clinical symptoms, although there was microscopic evidence of hepatitis, suggesting that swine HEV caused only subclinical infection in young pigs. This situation is reminiscent of hepatitis A virus infection in humans (21). Children infected with hepatitis A virus are often asymptomatic, but most infected adults show typical clinical symptoms (21). It is difficult to evaluate the outcome of natural swine HEV infection in adult pigs, however, since virtually all swine ≥3 months of age had IgG anti-HEV. Experimental infection of adult SPF swine with swine HEV will likely be necessary to answer this question.
Based on the observed similarities between swine HEV and human strains of HEV, one cannot yet determine whether swine HEV is species-specific or is circulating in the human population. Assessment of anti-HEV prevalence in pig handlers and experimental inoculation of primates with swine HEV should help address this important question. Subclinical infection of humans with swine HEV might explain the relatively high prevalence of anti-HEV in healthy individuals in the United States, where hepatitis E is not endemic (9). Development of a differential diagnostic test to distinguish between infections with swine HEV and human HEV is necessary.
The amino acid differences between swine and human HEV in the putative capsid gene are less than 10%. However, the high degree of amino acid sequence conservation in the capsid gene among human strains of HEV could argue that the differences between swine and human HEV may be of functional significance. There are well-documented examples of a single or few amino acid changes in a structural protein dramatically altering viral tropism and pathogenicity (22, 23). From the evolutionary point of view, it is not clear whether human HEV evolved from swine HEV, or vice versa, or whether swine and human HEV diverged from a common ancestor. Retrospective studies of archived serum samples from swine and humans may provide some information about the evolutionary relationship between swine and human HEV.
The possibility that swine HEV may infect humans also raises a potential public health concern for zoonosis or xenozoonosis. Xenotransplantation of pig organs has been suggested as a solution to the solid organ donor shortage for transplantations. However, xenozoonoses, the inadvertent transmission of pathogens from animal organs to human recipients, is of major concern (24). Viruses pathogenic for pigs might pose a risk to humans. However, nonpathogenic pig viruses may also become pathogenic for humans after xenotransplantation, as a result of species jumping, recombination, or adaptation in immunocompromised xenotransplantation recipients (24). Furthermore, pigs recovered from swine HEV infection might have a damaged liver (or other organ), which would limit its usefulness for xenotransplantation.
The discovery of swine HEV opens a new direction in HEV research and provides an opportunity for a better understanding of HEV pathogenesis. The similarities between swine and human HEV suggest that swine HEV infection in swine might provide an alternative animal model for HEV studies. Because swine HEV is immunologically crossreactive with human HEV and their capsid genes are very conserved, swine HEV may also prove useful as an attenuated vaccine for immunization against human hepatitis E through the “Jennerian” approach.
Acknowledgments
We would like to thank S. Vinson for providing some of the serum samples, T. Bolton for his professional help in the prospective study, and S. Tsarev for his expert help in the standardization of the ELISA. We also wish to thank D. Alling for calculating the ELISA cut-off value and D. Wong and M. Lewis for their assistance.
ABBREVIATIONS
- HEV
hepatitis E virus
- SPF
specific-pathogen-free
- RT
reverse transcription
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF011921).
References
- 1.Purcell R H. In: Fields Virology. 3rd Ed. Fields B N, Knipe D M, Howley P M, editors. Vol. 2. Philadelphia: Lippincott–Raven; 1996. pp. 2831–2843. [Google Scholar]
- 2.Wong D C, Purcell R H, Sreenivasan M A, Prasad S R, Pavri K M. Lancet. 1980;ii:876–878. doi: 10.1016/s0140-6736(80)92045-0. [DOI] [PubMed] [Google Scholar]
- 3.Arankalle V A, Tsarev S A, Chadha M S, Alling D W, Emerson S U, Banerjee K, Purcell R H. J Infect Dis. 1995;171:447–450. doi: 10.1093/infdis/171.2.447. [DOI] [PubMed] [Google Scholar]
- 4.Bradley D W. Rev Med Virol. 1992;2:19–28. [Google Scholar]
- 5.Skidmore S J, Yarbough P O, Gabor K A, Tam A W, Reyes G R. Lancet. 1991;337:1541. doi: 10.1016/0140-6736(91)93227-z. [DOI] [PubMed] [Google Scholar]
- 6.Dawson G J, Mushahwar I K, Chau K H, Gitnick G L. Lancet. 1992;340:426–427. doi: 10.1016/0140-6736(92)91507-5. [DOI] [PubMed] [Google Scholar]
- 7.Dawson G J, Chau K H, Cabal C M, Yarbough P O, Reyes G R, Mushahwar I K. J Virol Methods. 1992;38:175–186. doi: 10.1016/0166-0934(92)90180-l. [DOI] [PubMed] [Google Scholar]
- 8.Tsarev S A, Tsareva T S, Emerson S U, Kapikian A Z, Ticehurst J, London W, Purcell R H. J Infect Dis. 1993;168:369–378. doi: 10.1093/infdis/168.2.369. [DOI] [PubMed] [Google Scholar]
- 9.Thomas D L, Yarbough P O, Vlahov D, Tsarev S A, Nelson K E, Saah A J, Purcell R H. J Clin Microbiol. 1997;35:1244–1247. doi: 10.1128/jcm.35.5.1244-1247.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clayson E T, Innis B L, Myint K S A, Narupiti S, Vaughn D W, Giri S, Ranabhat P, Shrestha M P. Am J Trop Med Hyg. 1995;53:228–232. doi: 10.4269/ajtmh.1995.53.228. [DOI] [PubMed] [Google Scholar]
- 11.Balayan M S, Usmanov R K, Zamyatina D I, Karas F R. J Med Virol. 1990;32:58–59. doi: 10.1002/jmv.1890320110. [DOI] [PubMed] [Google Scholar]
- 12.Tsarev S A, Emerson S U, Tsareva T S, Yarbough P O, Lewis M, Govindarajan S, Reyes G R, Shapiro M, Purcell R H. J Infect Dis. 1993;167:1302–1306. doi: 10.1093/infdis/167.6.1302. [DOI] [PubMed] [Google Scholar]
- 13.Tsarev S A, Tsareva T S, Emerson S U, Rippy M K, Zack P, Shapiro M, Purcell R H. J Infect Dis. 1995;172:31–37. doi: 10.1093/infdis/172.1.31. [DOI] [PubMed] [Google Scholar]
- 14.Tam A W, Smith M M, Guerra M E, Huang C-C, Bradley D W, Fry K E, Reyes G R. Virology. 1991;185:120–131. doi: 10.1016/0042-6822(91)90760-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang C-C, Nguyen D, Fernandez J, Yun K Y, Fry K E, Bradley D W, Tam A W, Reyes G R. Virology. 1992;191:550–558. doi: 10.1016/0042-6822(92)90230-m. [DOI] [PubMed] [Google Scholar]
- 16.Aye T T, Uchida T, Ma X-Z, Iida F, Shikata T, Ichikawa M, Rikihisa T, Win K M. Virus Genes. 1993;7:95–110. doi: 10.1007/BF01702352. [DOI] [PubMed] [Google Scholar]
- 17.Panda S K, Nanda S K, Zafrullah M, Ansari I H, Ozdener M H, Jameel S. J Clin Microbiol. 1995;33:2653–2659. doi: 10.1128/jcm.33.10.2653-2659.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsarev S A, Emerson S U, Reyes G R, Tsareva T S, Legters L J, Malik I A, Iqbal M, Purcell R H. Proc Natl Acad Sci USA. 1992;89:559–563. doi: 10.1073/pnas.89.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yin S, Purcell R H, Emerson S U. Virus Genes. 1994;9:23–32. doi: 10.1007/BF01703432. [DOI] [PubMed] [Google Scholar]
- 20.Aye T T, Uchida T, Ma X-Z, Iida F, Shikata T, Zhuang H, Win K M. Nucleic Acids Res. 1992;20:3512. doi: 10.1093/nar/20.13.3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hollinger F B, Ticehurst J. In: Fields Virology. 2nd Ed. Fields B N, Knipe D M, editors. Vol. 1. New York: Raven; 1990. pp. 631–667. [Google Scholar]
- 22.Grieder F B, Davis N L, Aronson J F, Charles P C, Sellon D C, Suzuki K, Johnston R E. Virology. 1995;206:994–1006. doi: 10.1006/viro.1995.1022. [DOI] [PubMed] [Google Scholar]
- 23.Park B H, Matuschke B, Lavi E, Gaulton G N. J Virol. 1994;68:7516–7524. doi: 10.1128/jvi.68.11.7516-7524.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murphy F A. Science. 1996;273:746–747. doi: 10.1126/science.273.5276.746. [DOI] [PubMed] [Google Scholar]