Abstract
The Salmonella enterica serotype Typhimurium (S. typhimurium) genome contains a large repertoire of putative fimbrial operons that remain poorly characterized because they are not expressed in vitro. In this study, insertions that induced expression of the putative stdABCD fimbrial operon were identified from a random bank of transposon mutants by screening with immuno-magnetic particles for ligand expression (SIMPLE). Transposon insertions upstream of csgC and lrhA or within dam, setB and STM4463 (renamed rosE) resulted in expression of StdA and its assembly into fimbrial filaments on the cell surface. RosE is a novel negative regulator of Std fimbrial expression as indicated by its repression of a std::lacZ reporter construct and by binding of the purified protein to a DNA region upstream of the stdA start codon. Expression of Std fimbriae in the rosE mutant resulted in increased attachment of S. typhimurium to human colonic epithelial cell lines (T-84 and CaCo-2). A rosE mutant exhibited a reduced ability to compete with virulent S. typhimurium for colonization of murine organs, while no defect was observed when both competing strains carried a stdAB deletion. These data suggest that a tight control of Std fimbrial expression mediated by RosE is required during host pathogen interaction.
Introduction
Laboratory-grown cultures of Salmonella enterica serotype Typhimurium (S. typhimurium) commonly elaborate only type 1 fimbriae (Duguid et al., 1966) encoded by the fim operon (Clegg et al., 1987) and thin curled fimbriae (also known as thin aggregative fimbriae or curli) (Grund and Weber, 1988; Stolpe et al., 1994) encoded by the csg (agf) gene cluster (Romling et al., 1998). Two other S. typhimurium fimbrial gene clusters, termed pef (Friedrich et al., 1993) and lpf (Bäumler and Heffron, 1995), induce expression of fimbriae when the cloned genes are introduced into Escherichia coli. However, nucleotide sequencing identified nine additional putative fimbrial operons in the S. typhimurium genome, termed bcf (Tsolis et al., 1999), stf (Emmerth et al., 1999; Morrow et al., 1999), saf (Folkesson et al., 1999), stb, stc, std, sth, sti and stj (McClelland et al., 2001). The fact that nine putative fimbrial operons were only identified by sequence analysis in this genetically well-characterized organism reflects the tight control of their expression in vitro, which is detectable neither by Western blot (Humphries et al., 2005) nor by flow cytometry (Humphries et al., 2003) after growth of S. typhimurium under standard laboratory conditions. This lack of in vitro expression has prevented a thorough functional characterization of the encoded adhesins.
The std operon was initially identified during sequence analysis of the human-adapted S. enterica serotype Typhi strain CT18 (Townsend et al., 2001). Many of the putative fimbrial operons identified in the genomes of human-adapted Salmonella serotypes, including S. enterica serotypes Paratyphi A and Typhi (strains CT18 and Ty2), contain pseudogenes (i.e. genes carrying frameshift mutations or stop codons) (Parkhill et al., 2001; Deng et al., 2003; McClelland et al., 2004). Interestingly, only two fimbrial operons, tcf and std, share the following characteristics: they are present and apparently intact in all three available genomes of human-adapted Salmonella serotypes (Andrews-Polymenis and Bäumler, 2006). DNA–DNA hybridization studies show that the majority of Salmonella serotypes investigated contain orthologues of the std operon (Townsend et al., 2001; Porwollik et al., 2002; 2004; Chan et al., 2003; Anjum et al., 2005). Sequence comparison of its usher protein identifies the std operon as a member of the π-fimbriae, a group including well-characterized virulence factors such as pyelonephritis-associated (P) fimbriae of E. coli and the Mannose-resistant/Proteus-like (MR/P) fimbriae of Proteus mirabilis (Nuccio and Bäumler, 2007). Deletion of the std operon in S. typhimurium causes a competitive disadvantage during intestinal persistence in a mouse model (Weening et al., 2005). Mice infected with S. typhimurium seroconvert to StdA, which provides indirect evidence for in vivo expression of the std operon (Humphries et al., 2005). Gene expression profiling of an S. typhimurium dam mutant recently demonstrated that transcription of the std operon is repressed by Dam methylation (Balbontin et al., 2006). However, as the elaboration of surface structures has not been demonstrated in S. typhimurium, std must still be considered a putative fimbrial operon.
In this study we used a genetic screen to identify novel genes silencing expression of the std operon in vitro and demonstrated for the first time expression of the encoded fimbriae in S. typhimurium. Our results represent an important first step in characterizing Std fimbriae and lead the way for future functional studies on the remaining putative fimbrial operons present in the S. typhimurium genome.
Results
Identification of S. tyhimurium mutants expressing StdA using immuno-magnetic particles for ligand expression (SIMPLE)
To identify novel regulatory genes controlling expression of StdA in vitro, we generated mutant libraries in an S. typhimurium fim mutant (AJB4) using the transposons Mud-Cam (Elliott, 1993) and T-POP (Rappleye and Roth, 1997). Approximately 18 500 Mud-Cam mutants were generated, approximately 40% of which resulted from homologous recombination of the transposon into the hisD gene (data not shown), because the transposon was transduced from a donor strain (TE3461) carrying a hisD::Mud-Cam insertion. Thus, the Mud-Cam mutant bank contained an estimated 11 000 random insertion mutants. In addition, a bank containing approximately 9000 random T-POP insertion mutants was generated in an S. typhimurium fim mutant (AJB4). To this end, a plasmid encoding a Tn10 transposase with broad target specificity (pNK2880) was introduced into the S. typhimurium fim mutant. The T-POP transposon was delivered into this strain by transduction from an S. typhimurium donor (TH3923) (Lee et al., 2007) carrying the transposon on an E. coli F′ plasmid. A derivative of S. typhimurium isolate SR11 was used for this procedure, because this strain can be transduced with P22 to chloramphenicol (Mud-Cam) or tetracycline (T-POP) resistance, but is resistant to P22-mediated lysis, thus avoiding a selection for phage-resistant mutants during culture of mutant pools.
The immuno-magnetic particles for ligand expression (SIMPLE) approach (Nuccio et al., 2007) is based on the idea that incubation of a mutant bank with magnetic particles coated with antiserum raised against a fimbrial protein can be used to enrich for fimbriated mutants by immuno-magnetic separation (Nuccio et al., 2007). The Mud-Cam and T-POP mutant banks were divided into 16 and 7 pools, respectively, and each pool was used to inoculate a static Luria–Bertani (LB) broth (pH 7) culture with (for T-POP) or without (for Mud-Cam) tetracyline. Each pool was incubated with magnetic particles coated with anti-StdA serum; particle-bound bacteria were re-suspended in broth and used to inoculate a second static broth culture to repeat the enrichment procedure. After two enrichment cycles, a sample from each pool was grown statically in LB broth and expression of StdA by bacteria in the pool was assessed by Western blot (Fig. 1). Four T-POP pools and five Mud-Cam pools were positive for StdA expression, suggesting that enrichment for a mutant expressing this protein had occurred. Twenty bacterial colonies from each positive pool were grown statically in LB broth to identify individual transposon mutants expressing StdA. One StdA-expressing mutant from each pool was selected, a lysate was prepared using phage KB1 int (because SR11 derivatives are resistant to P22-mediated lysis) and the transposon insertion was transduced into an S. typhimurium fim mutant (AJB4). Transduction yielded four T-POP mutants (CD6R, CD7R, CD12R and CD15R) and five Mud-Cam mutants (CD2R, CD8R, CD10R, DB2R and DB3R) that expressed StdA in LB broth.
Fig. 1.
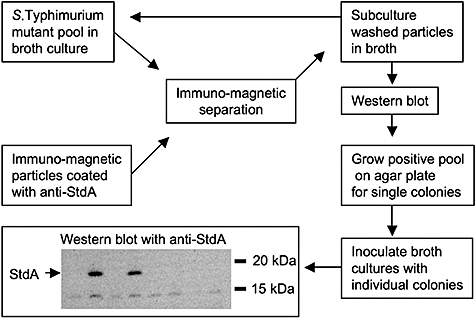
Flow chart of the SIMPLE approach used to identify S. typhimurium mutants expressing StdA. The bottom panel shows a Western blot detected with rabbit anti-StdA serum. Each lane represents a culture inoculated with a single colony from a mutant pool after two rounds of enrichment with immuno-magnetic particles coated with anti-StdA serum. The molecular mass of standard proteins is indicated on the right.
Identification of transposon insertion sites
DNA regions flanking the T-POP transposon insertion sites in strains CD2R, CD6R, CD7R, CD8R, CD12R, CD10R, CD15R, DB2R and DB3R were cloned by inverse polymerase chain reaction (PCR) and the respective nucleotide sequences were determined. Four Mud-Cam insertion sites were either within or immediately upstream of the dam open reading frame, at base pair positions +803 (CD2R), +36 (DB3R), +10 (DB2R) or −40 (CD10R) relative to the start codon. One T-POP insertion was located in damX, the open reading frame located directly upstream of dam, at base pair position +20 (CD12R) relative to the start codon (Fig. 2). The deoxyadenosine methylase (Dam) encoded by the dam gene has previously been implicated in controlling expression of fimbriae in E. coli (Blyn et al., 1990; van der Woude and Low, 1994) and S. typhimurium (Nicholson and Low, 2000).
Fig. 2.

Transposon insertion sites in S. typhimurium mutants expressing StdA.
A. Arrows indicate the size and orientation of genes in the S. typhimurium genome. The location of transposon insertions is indicated above each genetic map. Arrowheads indicate the orientation of the tet promoter of T-POP (CD12R, CD6R, CD7R, CD15R).
B. Comparison of the yjgG–yjgK intergenic regions of S. typhimurium strain LT2 (top) and E. coli strain K12 (bottom). Closed arrows indicate the size and orientation of genes present in the S. typhimurium genome but absent from E. coli K12. Open arrows indicate genes present in S. typhimurium and E. coli. Grey arrows indicate genes present in E. coli K12 but absent in S. typhimurium.
The T-POP insertion in CD6R was located in the csgA csgC intergenic region, at base pair position −60 relative to the csgC start codon. The csg gene cluster is involved in the biosynthesis of thin curled fimbriae in S. typhimurium (Romling et al., 1998); however, no function in fimbrial biosynthesis has been assigned to the csgC open reading frame. One T-POP insertion (CD15R) was found in the setB gene, at base pair position +1145 relative to the start codon. The setB gene encodes a sugar transporter (Liu et al., 1999) and mutations have been associated with pleiotrophic phenotypes, including a delay in chromosome segregation (Espeli et al., 2003).
One T-POP mutant (CD7R) did not express StdA after growth in LB broth in the absence of tetracycline (Fig. 3), suggesting that the tetA promoter of T-POP may drive expression of a positive regulatory element in this mutant. The T-POP insertion in CD7R was located in the yfbQ open reading frame, at base pair position +600 relative to the start codon. Mutant CD7R carried the T-POP insertion in an orientation in which the lrhA gene is located downstream of the tetA promoter (Fig. 2). LrhA is a negative regulator of flagellar, chemotactic, motility and type 1 fimbrial gene expression in E. coli (Lehnen et al., 2002; Blumer et al., 2005).
Fig. 3.
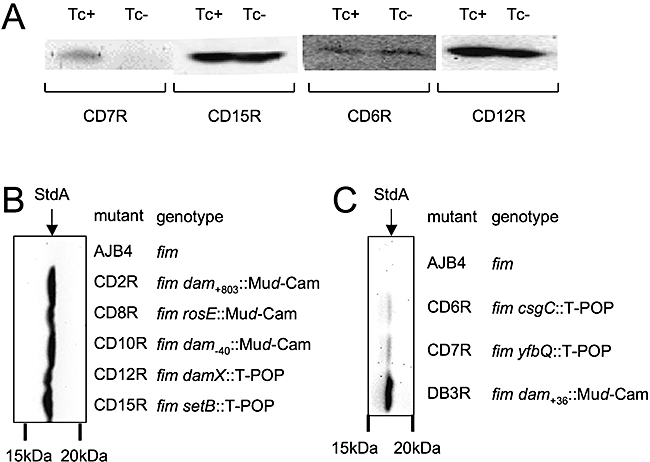
Expression of StdA by S. typhimurium mutants.
A. Expression of StdA detected by Western blot in T-POP insertion mutants (CD7R, CD15R, CD6R and CD12R) grown in the presence (Tc+) or absence (Tc−) of tetracycline.
B. Expression of StdA (arrow) detected after short (20 min) exposure of a Western blot. Bacterial strains analysed are indicated on the right. The molecular mass of standard proteins is indicated at the bottom.
C. Expression of StdA (arrow) detected after overnight exposure of a Western blot. Bacterial strains analysed are indicated on the right. The molecular mass of standard proteins is indicated at the bottom.
One Mud-Cam insertion was located upstream of open reading frame STM4463 (from hereon referred to as rosE), at base pair position −40 relative to the start codon. The protein encoded by rosE had homology to transcriptional regulators and showed the highest sequence identity to the ArgR (29%) (Lim et al., 1987) and Fur (16%) (Schäffer et al., 1985) proteins of E. coli. The rosE gene was located on an approximately 5 kb islet that was found to be present in all sequenced genomes of Salmonella serotypes and in genomes of uropathogenic E. coli, but absent from other sequenced E. coli genomes (Fig. 2). Open reading frames on this islet encode a putative arginine deiminase system composed of arginine deiminase (STM4467), carbamate kinase (STM4466), ornithine carbamyltransferase (STM4465) and an arginine-ornithine antiporter (STM4464). Similar arginine deiminase operons have been described in both Gram-positive and Gram-negative bacteria where they are involved in catabolizing arginine to ornithine, ammonia and carbon dioxide with the concomitant production of ATP (Luthi et al., 1986; 1990; Maghnouj et al., 1998).
StdA expression in S. typhimurium mutants is associated with enhanced transcription from the std promoter
Analysis of StdA expression divided mutants identified by SIMPLE into those exhibiting a strong signal by Western blot minutes after exposure (Fig. 3B) and those in which a signal was only detectable after prolonged exposure of Western blots (Fig. 3C). Mutants with high levels of StdA expression included rosE::Mud-Cam (CD8R), setB::Mud-Cam (CD15R), dam+803::Mud-Cam (CD2R), dam−40::Mud-Cam (CD10R), dam+10::Mud-Cam (DB2R) and damX::T-POP (CD12R). The damX::T-POP mutant (CD12R) expressed StdA when cultures were inoculated from glycerol stocks, but not when cultures were inoculated from a colony grown on LB agar plates, which may result from selection for compensatory mutations (however, this was not further analysed). None of the other mutants exhibited this phenotype. Mutants with lower levels of StdA expression included yfbQ::T-POP (CD7R) and csgC::T-POP (CD6R), while the dam+36::Mud-Cam mutant (DB3R) exhibited an intermediate level of expression (Fig. 3C).
To determine whether expression of StdA in individual mutants was accompanied with increased expression of the std operon, the promoter region upstream of stdA (Pstd) was introduced into vector pUJ10 oriented such that it controlled expression of a promoterless lacZ gene. The resulting plasmid (pDC57) was introduced into each of the mutants and their isogenic parent (AJB4). The parent strain AJB4(pDC57) expressed only low levels of β-galactosidase activity (approximately 2 Miller units on average) while all the T-POP or Mud-Cam insertions characterized in this study exhibited a significantly increased expression of the Pstd::lacZ reporter construct. The lowest levels of β-galactosidase expression were detected in the yfbQ::T-POP mutant (fivefold induction) and the csgC::T-POP mutant (sixfold induction) (Fig. 4), which also exhibited the weakest expression of StdA by Western blot (Fig. 3C). A stronger induction of the Pstd::lacZ reporter construct was detected in the setB::Mud-Cam mutant (ninefold induction), the rosE::Mud-Cam mutant (11-fold induction), the damX::T-POP mutant (26-fold induction) and the dam mutants. Expression of StdA was detected after short exposure of Western blots in all mutants in which β-galactosidase expression from the Pstd::lacZ reporter construct was increased ninefold or higher compared with the parental strain (Fig. 4). These data suggest that expression of StdA in the T-POP or Mud-Cam mutants identified in this study were at least in part due to increased expression from the Pstd promoter. However, expression of the Pstd::lacZ reporter fusion below the threshold level observed in the setB::Mud-Cam mutant resulted in a dramatic reduction of StdA expression detected by Western blot (Fig. 4) to levels only detectable after prolonged exposure (Fig. 3C). Due to the relatively low level of StdA expression detected in the yfbQ::T-POP mutant (CD7R) and the csgC::T-POP mutant (CD6R) (Fig. 3C), we did not further analyse the respective mutations.
Fig. 4.
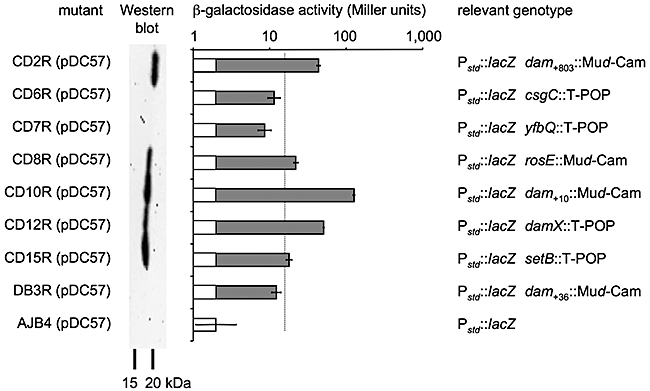
Transcription of the stdA gene in S. typhimurium strains detected with a plasmid-based (pDC57) lacZ transcriptional fusion (Pstd::lacZ). Bacterial strains analysed are indicated on the right. The second panel from the left shows a Western blot of each strain detected with anti-StdA serum. The molecular mass of standard proteins is indicated at the bottom. The β-galactosidase activities detected in each strain are shown at the centre. Each bar represents the average from three independent experiments ± standard deviation. The genotype of each strain is given on the right.
StdA assembles into fimbrial filaments on the surface of S. typhimurium that mediate attachment to human intestinal epithelial cells
Mutants that strongly expressed StdA were further analysed by electron microscopy to determine whether they elaborated fimbriae on their surface. When bacteria were cultured statically at 37°C in LB broth no fimbrial filaments were detected on the surface of the parental S. typhimurium fim mutant (AJB4) (data not shown), which is consistent with previous observations that type 1 fimbriae, but not thin-curled fimbriae, are expressed under this growth condition (Grund and Weber, 1988). As expected, introduction of the dam, setB and rosE mutations into a ΔstdAB derivative of AJB4 (CD21) did not result in expression of StdA (data not shown). However, surface appendages were visualized by negative staining on the surface of the dam (CD2R), setB (CD15R) and rosE (CD8R) mutants identified by SIMPLE (Fig. 5A). The identity of these surface structures was further investigated by immunogold labelling with anti-StdA serum. Gold particles labelled filamentous surface structures on the surfaces of the S. typhimurium dam+803::Mud-Cam (CD2R), rosE::Mud-Cam (CD8R) and setB::T-POP (CD15R) mutants, demonstrating that StdA was incorporated into fimbrial structures in these strains (Fig. 5B–D). In contrast, labelling with rabbit anti-StdA serum and goat anti-rabbit gold conjugate did not result in deposition of gold particles on the surface of the parental S. typhimurium fim mutant (AJB4) (data not shown). Immunoelectron microscopy thus provided the first evidence that the std operon encodes fimbriae in S. typhimurium.
Fig. 5.
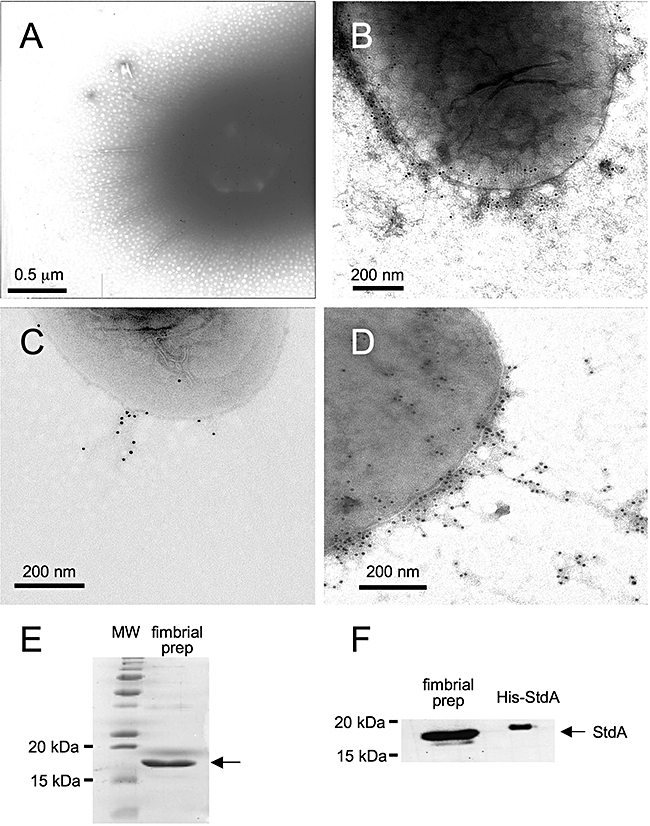
Expression of fimbriae on the surface of S. typhimurium transposon mutants investigated by electron microscopy and purification of fimbriae.
A. Expression of fimbrial filaments on the surface of a dam+803::Mud-Cam mutant (CD2R) was visualized by negative staining. B–D. Immunoelectron microscopy detecting expression of StdA on the surface of a dam+803::Mud-Cam mutant (CD2R) (B), a rosE::Mud-Cam mutant (CD8R) (C) and a setB::T-POP mutant (CD15R) (D). E and F. Coomassie stain (E) and Western blot (F) of a fimbrial preparation (fimbrial prep) obtained from an S. typhimurium fim fliC fljB dam+803::Mud-Cam mutant. (E) The Coomassie-stained gel shows standard proteins (MW, left lane) and a fimbrial preparation (right lane). The molecular mass of relevant standard proteins is indicated on the left. The arrow indicates a single major protein band present in the fimbrial preparation. (F) The Western blot shows the fimbrial preparation (left lane) and purified His–StdA fusion protein (right lane). Note that due to its 6xHis tag, the His–StdA protein has a slightly greater molecular mass than native StdA (arrow). The molecular mass of standard proteins is indicated on the left.
Fimbriae were purified from an S. typhimurium fim fliC fljB dam+803::Mud-Cam mutant (to prevent contamination with flagella and type 1 fimbriae) after their removal from the surface by mechanical shearing. Separation of proteins by SDS-PAGE followed by Coomassie blue staining revealed that the fimbrial preparation contained a single protein band with an apparent molecular mass that was similar to that predicted for the mature StdA protein (16 kDa) (Fig. 5E). In addition to a fimbrial usher (StdB) and a chaperone (StdC), the std operon encodes another putative subunit (StdD) whose predicted molecular mass was considerably larger than that of StdA. Western blot analysis showed that the protein band present in the fimbrial extract reacted with anti-StdA antiserum (Fig. 5F). Collectively, these data suggest that StdA is the major subunit of fimbriae encoded by the std operon.
To investigate the function of fimbriae encoded by the std operon, we studied bacterial adhesion to two human colonic epithelial cell lines, T-84 cells and CaCo-2 cells. Compared with its parent (AJB4), the rosE::Mud-Cam mutant (CD8R) was recovered in significantly higher numbers from both CaCo-2 cells and T84 cells (Fig. 6A and B). This increased recovery was due to the presence of std, because deletion of this fimbrial operon abolished adherence of the rosE::Mud-Cam mutant (CD20). Western blot analysis of the bacterial inoculum confirmed that only adherent bacterial cultures expressed StdA (Fig. 6C). These data demonstrated that fimbriae encoded by the std operon mediate attachment to human colonic epithelial cells.
Fig. 6.

Adherence of S. typhimurium strains to human colonic epithelial cells.
A and B. Adherence to T84-cells (A) or CaCo-2 cells (B) was determined at 4°C to prevent bacterial invasion. Data are shown as averages of cell-associated bacteria ± standard deviation. Statistical significance of differences is indicated by brackets. C. Expression of StdA in the bacterial inoculum was investigated by Western blot with anti-StdA serum.
RosE is a negative regulator of StdA expression
Complementation of dam mutations was not attempted because both gene inactivation and introduction of the cloned gene can result in the same phenotype (e.g. increased spontaneous mutation rates in S. typhimurium) (Torreblanca and Casadesus, 1996). To complement the rosE::Mud-Cam and setB::T-POP mutations, both open reading frames were PCR amplified along with their ribosome binding sites, but without their promoter sequences, and cloned in vector pBAD30 behind the E. coli arabinose promoter (PBAD). The resulting plasmids (pCD58 and pCD59 respectively) were introduced into the respective S. typhimurium mutants and strains were cultured in the presence of glucose (to repress expression from PBAD) or in the presence of arabinose (to induce expression from PBAD). Complementation of the setB::T-POP mutant (CD15R) with the cloned setB gene (pCD59) resulted in reduced StdA expression detected by Western blot. Similarly, introduction of the cloned rosE gene (pCD58) into the rosE::Mud-Cam (CD8R) mutant resulted in repression of StdA expression (Fig. 7). These data suggested that inactivation of rosE and setB were responsible for increased expression of StdA strains CD8R and CD15R respectively.
Fig. 7.
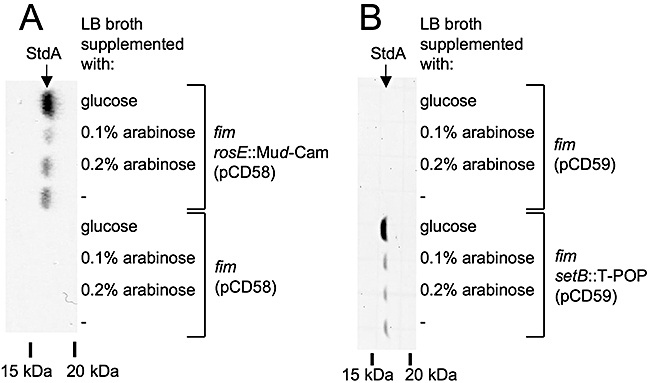
Complementation of S. typhimurium mutants expressing StdA.
A. Complementation of the CD8R mutant (fim rosE::Mud-Cam) (top) and its parent AJB4 (fim) (bottom) with the rosE gene cloned under control of the arabinose promoter (pCD58). A Western blot detected with anti-StdA serum is shown on the left. The molecular mass of standard proteins is indicated at the bottom. The presence or absence of glucose or arabinose is indicated on the right.
B. Complementation of the CD15R mutant (fim setB::T-POP) (bottom) and its parent AJB4 (fim) (top) with the setB gene cloned under control of the arabinose promoter (pCD59). A Western blot detected with anti-StdA serum is shown on the left. The molecular mass of standard proteins is indicated at the bottom. The presence or absence of glucose or arabinose is indicated on the right.
As RosE showed homology to transcriptional regulators, we investigated whether this protein would bind to a DNA region upstream of the stdA open reading frame. To this end, a fusion protein between RosE and a histidine tag (His–RosE) was constructed and purified. Purified His–RosE protein was tested for its ability to bind the stdA upstream DNA region using an electrophoretic mobility shift assay (EMSA). Addition of increasing concentrations of His–RosE to a biotin-labelled PCR product containing nucleotides −225 to −375 relative to the stdA start codon (Fig. 8A, region 2) resulted in appearance of a band with higher molecular weight (Fig. 8B). In contrast, His–RosE did not result in an electrophoretic mobility shift of a biotin-labelled DNA region comprising nucleotides −275 to −126 relative to the stdA start codon (Fig. 8A, region 1). The electrophoretic mobility shift of biotin-labelled DNA region 2 could be inhibited by pre-incubation of His–RosE with unlabelled DNA region 2, indicating specific binding of His–RosE to the stdA upstream DNA region (Fig. 8C).
Fig. 8.

Electrophoretic mobility shift assay (EMSA) of DNA regions located upstream of the stdA start codon using purified His–RosE protein.
A. The locations of DNA fragments (brackets) used for the EMSA (region 1 and region 2) are shown relative to the stdA open reading frame (filled arrow). The positions of three Dam methylation sites (GATC) present in the putative std promoter region are indicated.
B. EMSA of biotin-labelled DNA (region 1 or region 2) and different concentrations of His–RosE. Shifted DNA fragments are indicated by an arrow.
C. Competitive EMSA of biotin-labelled DNA (region 2, 35 pg well−1) in the presence (+) or absence (−) of purified His–RosE protein (70 pmol well−1) and the indicated amounts of unlabelled competitor DNA (region 2). Shifted DNA fragments are indicated by an arrow.
Collectively, our data showed that RosE bound to a DNA region upstream of the stdA start codon (Fig. 8), repressed transcription from the stdA promoter (Fig. 4) and prevented expression (Fig. 7) and assembly of StdA into fimbrial filaments (Fig. 5). These results identified RosE as a transcriptional regulator of Std fimbrial expression in S. typhimurium.
RosE-mediated repression of the std operon is required for full infectivity of S. typhimurium
Next we wanted to determine whether constitutive expression of the std operon in a rosE mutant would alter the course of infection in mice. To this end, the rosE::Mud-Cam insertion was introduced into the S. typhimurium wild type (SR11) to give rise to strain CD30. The S. typhimurium wild type was marked with a mutation in phoN (CD31), encoding alkaline phosphatase. Inactivation of phoN abolishes the ability to cleave 5-bromo-4-chloro-3-indolyl phosphate (XP). Growth on LB agar plates supplemented with XP thus provided an easy means to distinguish between mutants carrying a mutation in rosE (PhoN+ blue colonies) and CD31 (PhoN- white colonies). Inactivation of phoN does not reduce the ability of S. typhimurium to colonize organs or faeces of mice during competitive infections (Kingsley et al., 2003; Weening et al., 2005). A group of six genetically resistant mice (CBA/J) was inoculated with a 1:1 mixture of the S. typhimurium phoN mutant (CD31) and a rosE mutant (CD30). Recovery of bacteria from the faeces showed that the rosE mutant was recovered at significantly reduced numbers at 7 days after infection (Fig. 9A). Determination of the bacterial tissue load at 10 days after infection revealed a significant competitive disadvantage of the rosE mutant for colonizing caecal contents, caecal wall and the spleen (Fig. 9B).
Fig. 9.
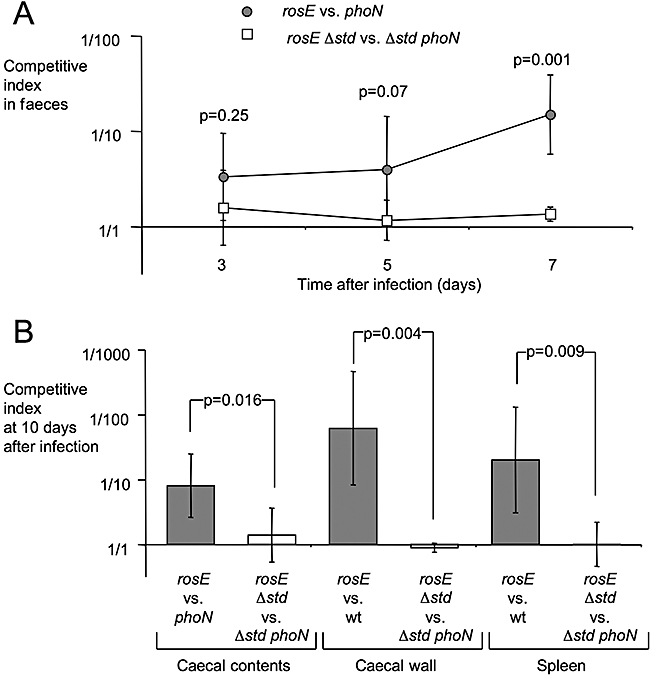
Effect of a mutation in rosE on the ability of S. typhimurium to colonize genetically resistant (CBA/J) mice.
A. Mice were infected with a 1:1 mixture of the S. typhimurium phoN mutant and a rosE mutant (grey circles) or with a 1:1 mixture of std phoN mutant and a std rosE mutant (open squares) and bacteria were recovered from faeces over time. Data are shown as geometric means of competitive indices ± standard deviation. Statistical significance of differences between both competitive infection experiments are indicated by P-values.
B. Mice were infected with a 1:1 mixture of the S. typhimurium phoN mutant and a rosE mutant (grey bars) or with a 1:1 mixture of std phoN mutant and a std rosE mutant (open bars) and bacteria were recovered from organs at 10 days after infection. Data are shown as geometric means of competitive indices ± standard deviation. Statistical significance of differences between both competitive infection experiments are indicated by P-values.
We next determined whether reduced recovery of the rosE mutant from the faeces and organs of mice was due to uncontrolled expression of the std operon or due to altered expression of other genes that may be regulated by RosE. We reasoned that deletion of the std fimbrial biosynthesis genes from both competing strains would eliminate a competitive defect only if recovery of the rosE mutant was due to uncontrolled expression of the std operon. The std operon was deleted from the S. typhimurium phoN mutant (CD31) and the rosE mutant (CD30) to give rise to strains CD32 and CD33 respectively. A group of six genetically resistant mice (CBA/J) was inoculated with a 1:1 mixture of the std phoN mutant (CD32) and the std rosE mutant (CD33). Recovery of bacteria from the faeces and organs of mice showed that the std rosE mutant (CD33) was fully able to compete with the std phoN mutant (CD32) for colonization of mice (Fig. 9). These data provided evidence that RosE-mediated repression of the std operon is required for full virulence of S. typhimurium in mice.
Discussion
Sequence analysis has identified multiple putative fimbrial operons in the S. typhimurium genome that remain poorly characterized (Emmerth et al., 1999; Folkesson et al., 1999; Morrow et al., 1999; Tsolis et al., 1999; McClelland et al., 2001). We have recently developed a new method, termed SIMPLE, that can be used to isolate fimbriated S. typhimurium mutants from a random transposon library (Nuccio et al., 2007). Here we have applied this method for the first time to a putative fimbrial operon, stdABCD, which was identified in the S. typhimurium genome by whole genome sequencing. The SIMPLE method proved to be a powerful approach for isolating mutants expressing fimbriae encoded by the std operon in vitro. The isolation of such mutants represents an important first step in the characterization of fimbriae encoded by the std operon and opened the way for functional studies on their binding specificity. Specifically, in vitro expression of the std operon enabled us to demonstrate that the encoded fimbriae mediate attachment of S. typhimurium to human colonic epithelial cell lines (i.e. T-84 and CaCo-2).
While this work was in progress, Balbontin et al. (2006) demonstrated that transcription of the std operon is repressed by Dam methylation in S. typhimurium. Our data confirmed and extended this observation by showing that mutations in dam resulted in expression of StdA filaments on the bacterial cell surface. The Dam protein methylates adenine residues located within the palindromic sequence GATC (Razin and Friedman, 1981). Inspection of the std promoter region reveals three GATC sites arranged in the motif 5′-acGATCa-(N6)-tcGATCgt-atcGATCta-3′ located upstream of a putative stdA promoter (Balbontin et al., 2006) (211 bp upstream of the stdA start codon). A similar arrangement of GATC sites and flanking regions is found in the promoter region of the E. coli agn43 gene, which contains the sequence 5′-acGATCa-(N12)-tgGATCgt-(N4)-atcGATCga-3′ located downstream of the transcriptional start site (32 bp upstream of the agn43 start codon) (Waldron et al., 2002; Wallecha et al., 2002). Dam-mediated methylation of these GATC sites is part of a phase variation mechanism controlling expression of antigen 43, an adhesin of the autotransporter family (Henderson and Owen, 1999; Haagmans and van der Woude, 2000). However, while E. coli dam mutants do not express antigen 43, S. typhimurium dam mutants did express StdA, which illustrated that regulation of Std fimbriae differed from that of the E. coli autotransporter.
A novel transcriptional regulator of std expression identified in this study is RosE, a protein with homology to the arginine repressor ArgR of E. coli (Lim et al., 1987). Analysis of a ProsE::lacZ reporter construct and complementation of a rosE-40::Mud-Cam mutant (CD8R) with the cloned rosE gene suggested that RosE repressed transcription of the std operon. Furthermore, EMSAs showed that RosE bound a DNA region located between base pairs −375 and −225 relative to the stdA start codon. For comparison, the GATC sites implicated in Dam-mediated repression of stdA expression (Balbontin et al., 2006) are located between base pairs −239 and −214 relative to the stdA start codon. Collectively, these data suggested that similar to its homologue ArgR, the RosE protein functioned as a transcriptional repressor. The rosE gene is located on a genetic islet that is well conserved among the genus Salmonella, thus making it likely that co-ordination of gene expression by RosE is a general feature of Salmonella serotypes.
While our data show that RosE prevented StdA expression in vitro, characterization of an S. typhimuriumΔstdAB mutant in the mouse model suggests that RosE-mediated suppression must be relieved under some conditions in vivo. The S. typhimuriumΔstdAB mutant has a competitive defect in colonizing the caecum of mice and in being shed with the faeces (Weening et al., 2005), suggesting that fimbriae encoded by the std operon are expressed in the intestinal lumen. StdA seroconversion of mice infected with S. typhimurium provides further indirect evidence for in vivo expression of fimbriae encoded by the std operon (Humphries et al., 2005). Interestingly, we found that a mutation in rosE resulted in a competitive defect of S. typhimurium during colonization of mice. This competitive colonization defect was no longer observed when std biosynthesis genes were deleted from both competing strains. These data suggest that a mutation in rosE resulted in attenuated mouse virulence of S. typhimurium because fimbriae encoded by the std operon were expressed in vivo at an inappropriate time or location. Deletion of the std operon results in a competitive defect of S. typhimurium in colonizing the caecum of mice, while colonization of the spleen is not altered (Weening et al., 2005). In contrast, the uncontrolled expression of the std operon in a rosE mutant reduced the ability of S. typhimurium to colonize both intestinal sites (i.e. the caecum) and extra-intestinal sites (i.e. the spleen). Collectively, these data illustrate that a tight control of std fimbrial expression is critical during host pathogen interaction in vivo.
Experimental procedures
Bacterial strains, media and growth conditions
Salmonella enterica serotype Typhimurium strains used in this study are shown in Table 1. An S. typhimurium fim fliC fljB dam+803::Mud-Cam mutant was generated by transducing the fliC and fljB mutations of S. typhimurium strain EHW26 (Raffatellu et al., 2005) into strain CD2R. E. coli strains DH5α MCR (Gibco BRL) and TOP10 (invitrogen) were used for cloning experiments. Insertions in the phoN gene were introduced into S. typhimurium strains by P22-mediated transduction from strain AJB715.
Table 1.
S. typhimurium strains used in this study.
| Strain | Genotype | Source or reference |
|---|---|---|
| SR11 | Xylose fermenting mutant of wild-type isolate BA2 | Schneider and Zinder (1956) |
| ADH17 | SR11, ΔstdAB::Km | Humphries et al. (2003) |
| CD30 | SR11, rosE::Mud-Cam | This study |
| CD31 | SR11, phoN::Km | This study |
| CD32 | SR11, phoN::Km ΔstdAB::Km | This study |
| CD33 | SR11, rosE::Mud-Cam ΔstdAB::Km | This study |
| AJB4 | SR11, nalidixic acid resistant, fim | Bäumler et al. (1996) |
| CD6R | AJB4, csgC::T-pop | This study |
| CD7R | AJB4, yfbQ::T-pop | This study |
| CD8R | AJB4, rosE::Mud-Cam | This study |
| CD10R | AJB4, dam−40::Mud-Cam | This study |
| CD12R | AJB4, damX::Mud-Cam | This study |
| CD15R | AJB4, setB::T-pop | This study |
| DB3R | AJB4, dam+36::Mud-Cam | This study |
| CD20 | AJB4, rosE::Mud-Cam ΔstdAB::Km | This study |
| CD21 | AJB4, ΔstdAB::Km | This study |
| CD2R | AJB4, dam+803::Mud-Cam | This study |
| CD3 | CD2R, fliC fljB | This study |
| ATCC14028 | Wild-type isolate from cattle | American Type Culture Collection |
| IR715 | Nalidixic acid-resistant derivative of ATCC14028 | Stojiljkovic et al. (1995) |
| EHW26 | IR715, fliC::Tn10 fljB5001::MudJ | Raffatellu et al. (2005) |
| AJB715 | IR715, phoN::Km | Kingsley et al. (2003) |
| LT2 | ATCC 700720 | Lilleengen (1948) |
| TE3461 | LT2, hisD9953::Mud-Cam hisA9944::Mud-1 | Elliott (1993) |
| TH3923 | LT2, pJS28(CarbR, P22-9+)/F′114ts Lac+zzf-20::Tn10[tetA::MudP](TcS) zzf-3823::Tn10dTc[del-25](T-POP)/leuA414 hsdSB Fels2− | Rappleye and Roth (1997) |
All strains were cultured aerobically or statically at 37°C in LB broth (10 g l−1 tryptone, 5 g l−1 yeast extract, 5 g l−1 NaCl). For the SIMPLE approach, bacteria were cultured in LB buffered to pH 7 with 100 mM 2-(N-morpholino)ethanesulphonic acid (MES) (Nicholson and Low, 2000). When appropriate, antibiotics were added at the following concentrations (mg l−1): kanamycin (Km), 100; nalidixic acid (Nal), 50; carbenicillin (Carb), 100; tetracycline (Tc), 20. To detect expression of beta-galactosidase on agar plates, 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) was added at a concentration of 40 mg l−1. For detection of alkaline phosphatase (PhoN) activity in S. typhimurium 30 mg l−1 XP was added to LB agar plates.
Genetic techniques
Generalized transducing phages P22 HT int-105 and KB1 int were used to generate lysates of S. typhimurium as previously described (Miller, 1972). Transductants were streaked for single colonies on Evans blue Uridine (EBU) agar (Bochner, 1984) and phage-free colonies were cross-streaked against P22 H5 (for P22 HT int-105), or KB1 int (for KB1) to confirm phage sensitivity.
Mud-Cam insertion mutants of S. typhimurium strain AJB4 were isolated using the method of Hughes and Roth (1988). In brief, a P22 HT int-105 lysate of TE3461 (Elliott, 1993) was used to transduce S. typhimurium to chloramphenicol resistance. Mud-Cam insertions were selected on LB+Cm plates (approximately 1000–2000 mutants per plate) and pooled by flooding with 5 ml of LB broth and re-suspending colonies.
Tn10dTc[del-25](T-POP) insertion mutants of S. typhimurium AJB4 were generated as described previously (Rappleye and Roth, 1997), using a P22 lysate grown on TH3923 to transduce into S. typhimurium AJB4 carrying plasmid pNK2880 (Rappleye and Roth, 1997; Lee et al., 2007). Transductants were selected on LB+Tc agar (approximately 1000–2000 mutants per plate) and pooled by flooding with 5 ml of LB broth and re-suspending colonies.
SIMPLE protocol
SIMPLE was performed as described previously (Nuccio et al., 2007). In brief, 100 μl aliquots of BioMag Protein G particles (Qiagen) were washed three times in 750 μl of TN buffer (0.1 M Tris-HCl, 0.15 M NaCl, pH 7.5) and incubated for 1 h at room temperature in 100 μl of pre-absorbed anti-StdA serum on an automatic roller. The particles were washed as described above and incubated at room temperature for half an hour with constant inversion on an automatic roller in 650 μl of Particle-Blocking (PB) buffer (TN buffer + 1% casein, prepared fresh). Bacteria (500 μl of a culture grown statically at 37°C for 2 days in LB-MES pH 7) were added and the tubes were incubated for 1 h at room temperature with constant inversion. The particles were washed three times with PB buffer by gentle inversion and re-suspended in 1 ml of PBS (to generate serial 10-fold dilutions that were spread on agar plates) or in the appropriate growth medium (to incubate bacteria statically at 37°C for 2 days and use the resulting culture to repeat the above protocol). For each mutant pool, 10 single colonies from a plate were analysed by Western blot using anti-StdA serum.
Western blotting
The polyclonal rabbit anti-StdA serum (Humphries et al., 2003) was diluted 1:5 in PBS supplemented with 0.2% sodium azide and pre-absorbed with ADH17 as described previously (Gruber and Zingales, 1995). For Western blot analysis, 10 μl containing approximately 2 × 108 colony-forming units (cfu) re-suspended in PBS was mixed with an equal volume of sodium dodecyl sulphate (SDS)-polyacrylamide gel electrophoresis (PAGE) loading buffer and boiled for 10 min. These whole-cell lysates were separated by 15% SDS-PAGE, transferred to Immobilon-P (Millipore) membranes using a Trans-Blot semi-dry transfer cell (Bio-Rad) and incubated with anti-StdA serum diluted 1:500 (final). Binding of anti-StdA serum was detected with Goat anti-Rabbit-Alkaline Phosphatase conjugate and the Immun-Star chemiluminescent substrate (Bio-Rad). Bands were visualized with a BioSpectrumAC Imaging System (UVP).
Cloning of transposon flanking DNA
DNA regions adjacent to the transposon insertion sites were amplified by an inverse PCR as described previously (Bäumler et al., 1994). Genomic DNA was digested with either AluI or TaqI and ligated with T4 DNA Ligase. DNA flanking Mud-Cam insertions was amplified using primer 5′-CCGAATAATCCAATGTCCTCCCGGT-3′ in combination with either 5′-AGTGCGCAATAACTTGCTCTCGTTC-3′ (for TaqI-digested DNA) or 5′-CGAAAAACAAAAACACTGCAAATCATTTCAATAAC-3′ (for AluI-digested DNA). DNA flanking T-POP insertions was amplified using primers 5′-CGCTTTTCCCGAGATCATATG-3′ and TPOP-AluTaq, 5′-GCACTTGTCTCCTGTTTACTCC-3′ (for AluI-digested DNA) and PCR SuperMix HiFi (Invitrogen). PCR products were gel-purified (QIAEX II gel purification kit, Qiagen) and cloned into pCR2.1 with the TOPO TA cloning kit (Invitrogen). The inserts of the resulting plasmids were sequenced by SeqWright (Houston, TX).
Complementation
The rosE gene was PCR amplified with the primers 5′-GAGCTCTAAGGTGCATTTATGAAGGA-3′ and 5′-AAGCTTACTCATCGCAAACGGTTCTTA-3′, cloned into pCR2.1 and the insert excised with SacI and HindIII and cloned into vector pBAD30 (Guzman et al., 1995) to give rise to plasmid pCD58. The setB gene was PCR amplified with the primers 5′-GAATTCCGTAAACTCCGCCTCTCTTCACAC-3′ and 5′-AAGCTTGCTGAAATGTGTCGAAGAGTAAA-3′, cloned into pCR2.1 and the insert excised with EcoRI and HindIII and cloned into vector pBAD30 to give rise to plasmid pCD59. The plasmids were introduced into S. typhimurium strains by electroporation. The resulting strains were grown overnight at 37°C in LB+Carb supplemented with 10 mM glucose. This overnight culture was used to inoculate (1:100) LB+Carb or LB+Carb supplemented with 0.2% of glucose, 0.1% arabinose or 0.2% arabinose. Cultures were grown at 37°C statically to an OD600 of 0.4–0.6 and StdA expression assessed by Western blot with rabbit anti-StdA serum (Humphries et al., 2003).
Electron microscopy
For microscopy, bacteria were grown for 2 days in a static culture, washed twice in PBS and re-suspended in EM grade water (EM Science) at a titre of approximately 1 × 109 cfu ml−1. Bacteria were allowed to attach to a formvar/carbon-coated grid (EM Science) for 2 min. For negative staining of fimbriae, the grids were incubated for 1 min in 1% uranyl-acetate (UA). For immunogold labelling the grids were incubated for 20 min with rabbit anti-StdA serum diluted 1/250 in PBS containing 1% bovine serum albumin (BSA). Grids were washed five times for 1 min in PBS containing 1% BSA. Grids were then incubated for 20 min in goat anti-rabbit 10 nm gold conjugate (EM Science) diluted 1/20 in PBS containing 1% BSA. Grids were washed three times for 1 min in PBS containing 1% BSA and three times for 1 min in EM grade water. Grids were incubated for 1 min with 1% UA before they were analysed by electron microscopy.
Purification of fimbriae
An S. typhimurium fim fliC fljB dam+803::Mud-Cam mutant was grown statically in 2 l of LB broth at 37°C overnight, harvested by centrifugation and re-suspended in 10 ml of 0.5 mM Tris 75 mM NaCl. StdA fimbriae were separated from the cells by mechanical shearing in a blender for three 1 min periods, after which cells and cellular debris were removed by centrifugation (3500 r.p.m. 30 min, 4°C). The supernatant was collected and passed through a 0.45 μm filter (Millipore), and (NH4)2SO4 (60% final concentration) was added to precipitate the fimbriae. Precipitated fimbriae were recovered by centrifugation (14 000 r.p.m. 30 min, 4°C). The pellet was re-suspended in 50 μl of sterile water and was analysed by SDS-PAGE and Western blot.
Tissue culture experiments
The colorectal carcinoma cell lines T84 (ATCC CCL-248) and Caco-2 (HTB-37) were obtained from the American Type Culture Collection. T84 cells were routinely cultured in Dulbecco's modified Eagle's medium (DMEM)-F12 medium (Gibco), containing 1.2 g l−1 sodium bicarbonate, 2.5 mM l-glutamine, 15 mM HEPES and 0.5 mM sodium pyruvate (Gibco), supplemented with 10% fetal calf serum (FCS). Caco-2 cells were maintained in Minimum essential medium (Eagle) (Gibco) containing 2.5 mM l-glutamine, 0.1 mM non-essential amino acids, 1 mM sodium pyruvate, Earle's balanced salt solution (BSS) and 15% FCS. For assays, T84 cells were seeded in 24-well plates at a density of approximately 1 × 105 cells per well or in six-well plates at a density of 5 × 105 cells per well and incubated for 48 h. Caco-2 cells were grown to confluency in 24-well plates for 5 days. Prior to adherence assays, epithelial cells were washed with ice-cold PBS. Ice-cold medium was added, cells were infected with bacteria (105 cfu well−1) and allow to adhere for 1 h at 4°C (to prevent invasion). Non-adherent bacteria were removed by five washes with PBS and adherent bacteria re-suspended in PBS containing 1% (v/v) Triton X-100. Serial 10-fold dilutions were spread on LB agar plates to determine the number of cell-associated bacteria per well.
β-Galactosidase assay
The promoter region of StdA was PCR amplified with the primers 5′-TCTAGACCTGAACTTTCCATCGAA-3′ and 5′-CCCGGGATATCCCCCAGCCTGCTG-3′. The PCR product was cloned into pCR2.1 (Invitrogen), the insert excised with XbaI and EcoRI and cloned into vector pUJ10 (deLorenzo et al., 1990) to give rise to plasmid pCD57. Plasmid pCD57 was introduced into S. typhimurium strains by electroporation. For β-galactosidase measurements, LB+Carb was inoculated 1:100 from an overnight culture and bacteria were grown to an optical density at 600 nm (OD600) of 0.4–0.6. The enzymatic activity of β-galactosidase in each culture was determined using standard methodology (Miller, 1972).
Purification of His–RosE
An N-terminal 6xHis–RosE fusion protein was constructed using the cloning vector pQE30 (Qiagen). The rosE gene was PCR amplified using the primers 5′-GGATCCATGAAGGAATACGATGATTAC-3′ and 5′-GAGCTCTTATGAATTTAAATTCATTTTAAG-3′ and the product was cloned into pCR2.1 (Invitrogen). The insert was then cloned into the BamHI and SacI sites of pQE30 to give rise to plasmid pCD62. Plasmid pCD62 was electroporated into E. coli TOP10 and the resulting strain was grown overnight at 37°C in LB media supplemented with 10 mM glucose and carbenicillin (100 mg l−1) to maintain the plasmid. To induce the production of His–RosE, cells were grown to mid-log phase before IPTG was added to a final concentration of 2 mM. Cells were harvested by centrifugation after a 4 h incubation. His–RosE was purified by nickel-affinity chromatography (Ni-NTA, Invitrogen) according to the manufacturer's protocol, but after cell fragments had been removed from cell lysates by centrifugation, the supernatant was centrifuged at 15000 r.p.m. for 30 min to remove denatured protein before being applied to the affinity column. The fractions containing His–RosE were immediately aliquotted and frozen at −80°C in 5% sucrose. The His–RosE concentration was determined by measuring the absorbance at 280 nm.
Electrophoretic mobility shift assay
Fragments of the region upstream of the stdA open reading frame were generated by PCR amplification using the primer sets 5′-CCTGAACTTTCCATCGAAAAA-3′, 5′-TAAAAAGTATTTCTTTGAT-3′ (region 1) and 5′-TCGATGATTATGATCGTAAT-3′, 5′-ATGGAAAAGCAAAACATA-3′ (region 2) which were biotinylated at the 5′ end (MWG Bioscences, High Point, NC). The PCR products were gel-purified in a vertical acrylamide gel. The vertical acrylamide gel was prepared by adding 0.1 ml of 50× Tris-acetate EDTA (242 g of Tris, 57.1 ml of glacial acetic acid, 37.2 g of Na2EDTA·2H2O in 1 l of distilled H2O), 0.66 ml of acrylamide/bis-acrylamide (37.5:1, Roche), 6 μl of TEMED and 30 μl of ammonium persulphate to 5.33 ml of distilled H2O. After electrophoresis, fragments were excised and eluted by electrophoresis in dialysis tubes for 15 min at 100 V. The DNA concentration was determined by spectrophotometry. The binding assays were performed in a total volume of 20 μl containing the following: 1 μg ml−1 Poly(dI-dC), 50% glycerol, 1% non-ylphenyl-polyethylene glycol (NP-40), 1.05 M KCl, 10 mM TRIS (pH 7.5), 1 mM DTT, 100 mM MgCl2, 200 mM EDTA, 35 pg of biotinylated DNA, various concentrations of His–RosE and unlabelled competitor DNA as required. Competitor DNA was generated by PCR amplification using the primers listed above without the biotin label. Binding reactions were allowed to incubate for 20 min at room temperature prior to adding loading buffer and separating fragments on a pre-run 5% acrylamide gel in Tris-borate EDTA buffer (Bio-Rad Precast Gels). Gels were run at 4°C at 100 V until the bromophenol blue dye front had migrated approximately three-fourths down the length of the gel. The DNA–protein complexes were transferred to a nylon membrane (Roche Applied Science) using a semi-dry blotter (Bio-Rad) at 15 V for 30 min. The DNA was UV cross-linked to the membrane, and biotinylated DNA on the membrane was detected using the LighShiftTM Chemiluminescent EMSA Kit (Pierce Biotechnology, Rockford, IL) according to the manufacturer's protocol.
Animal experiments
Eight- to 12-week-old CBA/J (Jackson Laboratory) mice were used throughout this study. Bacteria were routinely cultured as standing overnight cultures prior to infection. In all experiments the bacterial titre of the inoculum was determined by spreading serial 10-fold dilutions on agar plates containing appropriate antibiotics and determining cfu. For competitive infection experiments, groups of five mice were infected by oral gavage with a 1:1 mixture of mutant and isogenic parent at a dose of approximately 5 × 108 cfu per mouse. Faecal pellets were homogenized in 1 ml of PBS. The limit of detection was approximately 0.08 cfu per milligram of faeces. Caecum, caecal contents and spleen were harvested and homogenized in 5 ml of PBS pH 7.4. Dilutions of faecal pellets and homogenized organs were spread on LB agar plates containing the appropriate antibiotics. Agar plates were supplemented with XP to distinguished between colonies expressing PhoN and colonies that were PhoN negative. Data were normalized and competitive indices calculated by dividing the output ratio (cfu mutant/cfu wild type) by the input ratio (cfu mutant/cfu wild type). Competitive indices were converted logarithmically prior to the calculation of averages and statistical analysis. A Student's t-test was used to determine whether differences were statistically significant.
Acknowledgments
We would like to thank Sebastian Winter for helpful discussions and Maarten de Jong for help with the EMSA. We are grateful to Dr Casadesus for sharing results prior to publication. We would also like to thank Grete Adamson for her help with electron microscopy. Work in A.J.B.'s laboratory was supported by USDA/NRICGP Grant No. 2002-35204-12247 and Public Health Service Grants AI040124, AI044170 and AI065534.
References
- Andrews-Polymenis H, Bäumler AJ. Pathogenomics of Salmonella species. In: Hacker J, Dobrindt U, editors. Pathogenomics. Weinheim: Wiley-VCH; 2006. pp. 109–124. [Google Scholar]
- Anjum MF, Marooney C, Fookes M, Baker S, Dougan G, Ivens A, Woodward MJ. Identification of core and variable components of the Salmonella enterica subspecies I genome by microarray. Infect Immun. 2005;73:7894–7905. doi: 10.1128/IAI.73.12.7894-7905.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balbontin R, Rowley G, Pucciarelli MG, Lopez-Garrido J, Wormstone Y, Lucchini S, et al. DNA adenine methylation regulates virulence gene expression in Salmonella enterica serovar Typhimurium. J Bacteriol. 2006;188:8160–8168. doi: 10.1128/JB.00847-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäumler AJ, Heffron F. Identification and sequence analysis of lpfABCDE, a putative fimbrial operon of Salmonella typhimurium. J Bacteriol. 1995;177:2087–2097. doi: 10.1128/jb.177.8.2087-2097.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäumler AJ, Kusters JG, Stojiljkovic I, Heffron F. Salmonella typhimurium loci involved in survival within macrophages. Infect Immun. 1994;62:1623–1630. doi: 10.1128/iai.62.5.1623-1630.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäumler AJ, Tsolis RM, Heffron F. Contribution of fimbrial operons to attachment to and invasion of epithelial cell lines by Salmonella typhimurium. Infect Immun. 1996;64:1862–1865. doi: 10.1128/iai.64.5.1862-1865.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumer C, Kleefeld A, Lehnen D, Heintz M, Dobrindt U, Nagy G, et al. Regulation of type 1 fimbriae synthesis and biofilm formation by the transcriptional regulator LrhA of Escherichia coli. Microbiology. 2005;151:3287–3298. doi: 10.1099/mic.0.28098-0. [DOI] [PubMed] [Google Scholar]
- Blyn LB, Braaten BA, Low DA. Regulation of pap pilin phase variation by a mechanism involving differential dam methylation states. EMBO J. 1990;9:4045–4054. doi: 10.1002/j.1460-2075.1990.tb07626.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochner BR. Curing bacterial cells of lysogenic viruses by using UCB indicator plates. Biotechniques. 1984;2:234–240. [Google Scholar]
- Chan K, Baker S, Kim CC, Detweiler CS, Dougan G, Falkow S. Genomic comparison of Salmonella enterica serovars and Salmonella bongori by use of an S. enterica serovar typhimurium DNA microarray. J Bacteriol. 2003;185:553–563. doi: 10.1128/JB.185.2.553-563.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clegg S, Purcell BK, Pruckler J. Characterization of genes encoding type 1 fimbriae of Klebsiella pneumoniae, Salmonella typhimurium, and Serratia marcescens. Infect Immun. 1987;55:281–287. doi: 10.1128/iai.55.2.281-287.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Liou SR, Plunkett G, Mayhew GF, Rose DJ, Burland V, et al. Comparative genomics of Salmonella enterica serovar Typhi strains Ty2 and CT18. J Bacteriol. 2003;185:2330–2337. doi: 10.1128/JB.185.7.2330-2337.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duguid JP, Anderson ES, Campbell I. Fimbriae and adhesive properties in Salmonellae. J Pathol Bacteriol. 1966;92:107–138. doi: 10.1002/path.1700920113. [DOI] [PubMed] [Google Scholar]
- Elliott T. Transport of 5-aminolevulinic acid by the dipeptide permease in Salmonella typhimurium. J Bacteriol. 1993;175:325–331. doi: 10.1128/jb.175.2.325-331.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmerth M, Goebel W, Miller SI, Hueck CJ. Genomic subtraction identifies Salmonella typhimurium prophages, F-related plasmid sequences, and a novel fimbrial operon, stf, which are absent in Salmonella typhi. J Bacteriol. 1999;181:5652–5661. doi: 10.1128/jb.181.18.5652-5661.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espeli O, Nurse P, Levine C, Lee C, Marians KJ. SetB: an integral membrane protein that affects chromosome segregation in Escherichia coli. Mol Microbiol. 2003;50:495–509. doi: 10.1046/j.1365-2958.2003.03736.x. [DOI] [PubMed] [Google Scholar]
- Folkesson A, Advani A, Sukupolvi S, Pfeifer JD, Normark S, Lofdahl S. Multiple insertions of fimbrial operons correlate with the evolution of Salmonella serovars responsible for human disease. Mol Microbiol. 1999;33:612–622. doi: 10.1046/j.1365-2958.1999.01508.x. [DOI] [PubMed] [Google Scholar]
- Friedrich MJ, Kinsey NE, Vila J, Kadner RJ. Nucleotide sequence of a 13.9 kb segment of the 90 kb virulence plasmid of Salmonella typhimurium: the presence of fimbrial biosynthetic genes. Mol Microbiol. 1993;8:543–558. doi: 10.1111/j.1365-2958.1993.tb01599.x. [DOI] [PubMed] [Google Scholar]
- Gruber A, Zingales B. Alternative method to remove antibacterial antibodies from antisera used for screening of expression libraries. Biotechniques. 1995;19:28–30. [PubMed] [Google Scholar]
- Grund S, Weber A. A new type of fimbriae on Salmonella typhimurium. Zentralbl Veterinarmed [B] 1988;35:779–782. doi: 10.1111/j.1439-0450.1988.tb00560.x. [DOI] [PubMed] [Google Scholar]
- Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haagmans W, van der Woude M. Phase variation of Ag43 in Escherichia coli: Dam-dependent methylation abrogates OxyR binding and OxyR-mediated repression of transcription. Mol Microbiol. 2000;35:877–887. doi: 10.1046/j.1365-2958.2000.01762.x. [DOI] [PubMed] [Google Scholar]
- Henderson IR, Owen P. The major phase-variable outer membrane protein of Escherichia coli structurally resembles the immunoglobulin A1 protease class of exported protein and is regulated by a novel mechanism involving Dam and oxyR. J Bacteriol. 1999;181:2132–2141. doi: 10.1128/jb.181.7.2132-2141.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes KT, Roth JR. Transitory cis complementation: a method for providing transposition functions to defective transposons. Genetics. 1988;119:9–12. doi: 10.1093/genetics/119.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries AD, Raffatellu M, Winter S, Weening EH, Kingsley RA, Droleskey R, et al. The use of flow cytometry to detect expression of subunits encoded by 11 Salmonella enterica serotype Typhimurium fimbrial operons. Mol Microbiol. 2003;48:1357–1376. doi: 10.1046/j.1365-2958.2003.03507.x. [DOI] [PubMed] [Google Scholar]
- Humphries AD, DeRidder S, Bäumler AJ. Salmonella enterica serotype Typhimurium fimbrial proteins serve as antigens during infection of mice. Infect Immun. 2005;73:5329–5338. doi: 10.1128/IAI.73.9.5329-5338.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsley RA, Humphries AD, Weening EH, De Zoete MR, Winter S, Papaconstantinopoulou A, et al. Molecular and phenotypic analysis of the CS54 island of Salmonella enterica serotype typhimurium: identification of intestinal colonization and persistence determinants. Infect Immun. 2003;71:629–640. doi: 10.1128/IAI.71.2.629-640.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Wozniak C, Karlinsey JE, Hughes KT. Genomic screening for regulatory genes using the T-POP transposon. Methods Enzymol. 2007;421:159–167. doi: 10.1016/S0076-6879(06)21014-0. [DOI] [PubMed] [Google Scholar]
- Lehnen D, Blumer C, Polen T, Wackwitz B, Wendisch VF, Unden G. LrhA as a new transcriptional key regulator of flagella, motility and chemotaxis genes in Escherichia coli. Mol Microbiol. 2002;45:521–532. doi: 10.1046/j.1365-2958.2002.03032.x. [DOI] [PubMed] [Google Scholar]
- Lilleengen K. Typing of Salmonella typhimurium by means of bacteriophage. Acta Pathol Microbiol Scand Suppl. 1948;77:2–125. [Google Scholar]
- Lim DB, Oppenheim JD, Eckhardt T, Maas WK. Nucleotide sequence of the argR gene of Escherichia coli K-12 and isolation of its product, the arginine repressor. Proc Natl Acad Sci USA. 1987;84:6697–6701. doi: 10.1073/pnas.84.19.6697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JY, Miller PF, Willard J, Olson ER. Functional and biochemical characterization of Escherichia coli sugar efflux transporters. J Biol Chem. 1999;274:22977–22984. doi: 10.1074/jbc.274.33.22977. [DOI] [PubMed] [Google Scholar]
- deLorenzo V, Herrero M, Jakubzik U, Timmis KN. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J Bacteriol. 1990;172:6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luthi E, Mercenier A, Haas D. The arcABC operon required for fermentative growth of Pseudomonas aeruginosa on arginine: Tn5-751-assisted cloning and localization of structural genes. J Gen Microbiol. 1986;132:2667–2675. doi: 10.1099/00221287-132-10-2667. [DOI] [PubMed] [Google Scholar]
- Luthi E, Baur H, Gamper M, Brunner F, Villeval D, Mercenier A, Haas D. The arc operon for anaerobic arginine catabolism in Pseudomonas aeruginosa contains an additional gene, arcD, encoding a membrane protein. Gene. 1990;87:37–43. doi: 10.1016/0378-1119(90)90493-b. [DOI] [PubMed] [Google Scholar]
- McClelland MKE, Sanderson J, Spieth SW, Clifton P, Latreille L, Courtney S, et al. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature. 2001;413:852–856. doi: 10.1038/35101614. [DOI] [PubMed] [Google Scholar]
- McClelland MKE, Sanderson SW, Clifton P, Latreille S, Porwollik A, Sabo R, et al. Comparison of genome degradation in Paratyphi A and Typhi, human-restricted serovars of Salmonella enterica that cause typhoid. Nat Genet. 2004;36:1268–1274. doi: 10.1038/ng1470. [DOI] [PubMed] [Google Scholar]
- Maghnouj A, de Sousa Cabral TF, Stalon V, Vander Wauven C. The arcABDC gene cluster, encoding the arginine deiminase pathway of Bacillus licheniformis, and its activation by the arginine repressor argR. J Bacteriol. 1998;180:6468–6475. doi: 10.1128/jb.180.24.6468-6475.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JH. Experiments in Molecular Genetics. New York: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- Morrow BJ, Graham JE, Curtiss R., 3rd Genomic subtractive hybridization and selective capture of transcribed sequences identify a novel Salmonella typhimurium fimbrial operon and putative transcriptional regulator that are absent from the Salmonella typhi genome. Infect Immun. 1999;67:5106–5116. doi: 10.1128/iai.67.10.5106-5116.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson B, Low D. DNA methylation-dependent regulation of pef expression in Salmonella typhimurium. Mol Microbiol. 2000;35:728–742. doi: 10.1046/j.1365-2958.2000.01743.x. [DOI] [PubMed] [Google Scholar]
- Nuccio SP, Bäumler AJ. Evolution of the chaperone/usher assembly pathway: fimbrial classification goes greek. Microbiol Mol Biol Rev. 2007;71:551–575. doi: 10.1128/MMBR.00014-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuccio SP, Chessa D, Weening EH, Raffatellu M, Clegg S, Bäumler AJ. SIMPLE approach for isolating mutants expressing fimbriae. Appl Environ Microbiol. 2007;73:4455–4462. doi: 10.1128/AEM.00148-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhill J, Dougan G, James KD, Thomson NR, Pickard D, Wain J, et al. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature. 2001;413:848–852. doi: 10.1038/35101607. [DOI] [PubMed] [Google Scholar]
- Porwollik S, Wong RM, McClelland M. Evolutionary genomics of Salmonella: gene acquisitions revealed by microarray analysis. Proc Natl Acad Sci USA. 2002;99:8956–8961. doi: 10.1073/pnas.122153699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porwollik S, Boyd EF, Choy C, Cheng P, Florea L, Proctor E, McClelland M. Characterization of Salmonella enterica subspecies I genovars by use of microarrays. J Bacteriol. 2004;186:5883–5898. doi: 10.1128/JB.186.17.5883-5898.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffatellu M, Chessa D, Wilson RP, Dusold R, Rubino S, Bäumler AJ. The Vi capsular antigen of Salmonella enterica serotype Typhi reduces Toll-like receptor-dependent interleukin-8 expression in the intestinal mucosa. Infect Immun. 2005;73:3367–3374. doi: 10.1128/IAI.73.6.3367-3374.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappleye CA, Roth JR. A Tn10 derivative (T-POP) for isolation of insertions with conditional (tetracycline-dependent) phenotypes. J Bacteriol. 1997;179:5827–5834. doi: 10.1128/jb.179.18.5827-5834.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razin A, Friedman J. DNA methylation and its possible biological roles. Prog Nucleic Acid Res Mol Biol. 1981;25:33–52. doi: 10.1016/s0079-6603(08)60482-1. [DOI] [PubMed] [Google Scholar]
- Romling U, Bian Z, Hammar M, Sierralta WD, Normark S. Curli fibers are highly conserved between Salmonella typhimurium and Escherichia coli with respect to operon structure and regulation. J Bacteriol. 1998;180:722–731. doi: 10.1128/jb.180.3.722-731.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäffer S, Hantke K, Braun V. Nucleotide sequence of the iron regulatory gene. Fur Mol Gen Genet. 1985;200:110–113. doi: 10.1007/BF00383321. [DOI] [PubMed] [Google Scholar]
- Schneider HA, Zinder ND. Nutrition of the host and natural resistance to infection. V. An improved assay employing genetic markers in the double strain inoculation test. J Exp Med. 1956;103:207–223. doi: 10.1084/jem.103.2.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojiljkovic I, Bäumler AJ, Heffron F. Ethanolamine utilization in Salmonella typhimurium: nucleotide sequence, protein expression and mutational analysis of the cchA cchB eutE eutJ eutG eutH gene cluster. J Bacteriol. 1995;177:1357–1366. doi: 10.1128/jb.177.5.1357-1366.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolpe H, Grund S, Schroder W. Purification and partial characterization of type 3 fimbriae from Salmonella typhimurium var. copenhagen. Zentralbl Bakteriol. 1994;281:8–15. doi: 10.1016/s0934-8840(11)80631-6. [DOI] [PubMed] [Google Scholar]
- Torreblanca J, Casadesus J. DNA adenine methylase mutants of Salmonella typhimurium and a novel dam-regulated locus. Genetics. 1996;144:15–26. doi: 10.1093/genetics/144.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend SM, Kramer NE, Edwards R, Baker S, Hamlin N, Simmonds M, et al. Salmonella enterica serotype Typhi possesses a unique repertoire of fimbrial gene sequences. Infect Immun. 2001;69:2894–2901. doi: 10.1128/IAI.69.5.2894-2901.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsolis RM, Townsend SM, Miao EA, Miller SI, Ficht TA, Adams LG, Bäumler AJ. Identification of a putative Salmonella enterica serotype typhimurium host range factor with homology to IpaH and YopM by signature-tagged mutagenesis. Infect Immun. 1999;67:6385–6393. doi: 10.1128/iai.67.12.6385-6393.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldron DE, Owen P, Dorman CJ. Competitive interaction of the OxyR DNA-binding protein and the Dam methylase at the antigen 43 gene regulatory region in Escherichia coli. Mol Microbiol. 2002;44:509–520. doi: 10.1046/j.1365-2958.2002.02905.x. [DOI] [PubMed] [Google Scholar]
- Wallecha A, Munster V, Correnti J, Chan T, van der Woude M. Dam- and OxyR-dependent phase variation of agn43: essential elements and evidence for a new role of DNA methylation. J Bacteriol. 2002;184:3338–3347. doi: 10.1128/JB.184.12.3338-3347.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weening EH, Barker JD, Laarakker MC, Humphries AD, Tsolis RM, Bäumler AJ. The Salmonella enterica serotype Typhimurium lpf, bcf, stb, stc, std, and sth fimbrial operons are required for intestinal persistence in mice. Infect Immun. 2005;73:3358–3366. doi: 10.1128/IAI.73.6.3358-3366.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Woude MW, Low DA. Leucine-responsive regulatory protein and deoxyadenosine methylase control the phase variation and expression of the sfa and daa pili operons in Escherichia coli. Mol Microbiol. 1994;11:605–618. doi: 10.1111/j.1365-2958.1994.tb00340.x. [DOI] [PubMed] [Google Scholar]


