Abstract
One-third of humans are infected with Mycobacterium tuberculosis, the causative agent of tuberculosis. Sequence analysis of two megabases in 26 structural genes or loci in strains recovered globally discovered a striking reduction of silent nucleotide substitutions compared with other human bacterial pathogens. The lack of neutral mutations in structural genes indicates that M. tuberculosis is evolutionarily young and has recently spread globally. Species diversity is largely caused by rapidly evolving insertion sequences, which means that mobile element movement is a fundamental process generating genomic variation in this pathogen. Three genetic groups of M. tuberculosis were identified based on two polymorphisms that occur at high frequency in the genes encoding catalase-peroxidase and the A subunit of gyrase. Group 1 organisms are evolutionarily old and allied with M. bovis, the cause of bovine tuberculosis. A subset of several distinct insertion sequence IS6110 subtypes of this genetic group have IS6110 integrated at the identical chromosomal insertion site, located between dnaA and dnaN in the region containing the origin of replication. Remarkably, study of ≈6,000 isolates from patients in Houston and the New York City area discovered that 47 of 48 relatively large case clusters were caused by genotypic group 1 and 2 but not group 3 organisms. The observation that the newly emergent group 3 organisms are associated with sporadic rather than clustered cases suggests that the pathogen is evolving toward a state of reduced transmissability or virulence.
One-third of the world’s population is infected with Mycobacterium tuberculosis, and 3 million human deaths annually are attributed to the organism (1, 2). Although there is a very large global pool of infected individuals and considerable chromosomal heterogeneity based on restriction fragment length polymorphism (RFLP) patterns generated by probing with mobile insertion elements (3, 4), studies of drug resistance and pathogenesis have raised the possibility that synonymous (silent) nucleotide substitutions in structural genes may be limited (5). To investigate this apparent discrepancy from the perspective of molecular population genetics, we sequenced two megabases in 26 structural genes or loci in strains of M. tuberculosis and the three closely related members of the M. tuberculosis complex (M. africanum, M. bovis, and M. microti) collected worldwide.
MATERIALS AND METHODS
Bacterial Isolates.
The study is based on a sample of 842 M. tuberculosis complex isolates recovered from diverse geographic localities. The organisms include M. tuberculosis (n = 715), M. bovis (n = 109), and M. africanum and M. microti (n = 9 each). M. tuberculosis isolates were recovered from diseased patients in the United States (five states), Latin America (Mexico, Honduras, Ecuador, Peru, Venezuela, Brazil, and Chile), Europe (Portugal, Spain, The Netherlands, Belgium, Germany, Switzerland, Italy, former Yugoslavia, and Romania), Africa (Kenya, Rwanda, Guinea, Algeria, Somalia, and Zaire), the mid-East (Yemen, Israel, Turkey, and Iran), and elsewhere (Australia, Burundi, China, India, Japan, South Korea, Mongolia, Nepal, Philippines, Sri Lanka, Tahiti, Thailand, and Vietnam) (6). This collection of M. tuberculosis isolates represents the range of insertion sequence IS6110 fingerprint diversity in the species (refs. 3 and 4 and unpublished data) and includes organisms recovered from patients with pulmonary and extrapulmonary tuberculosis. Organisms in the sample had from 0 to 21 copies of IS6110. Moreover, organisms classified into several groups based on a multiplex PCR analysis (7) were included in the analysis.
The 109 M. bovis isolates were recovered in five countries and from eight host species (6). The M. microti specimens were recovered from voles (n = 7), a pig (n = 1) in the Netherlands, and a rock hyrax (n = 1) in South Africa. Isolates of M. africanum were recovered from patients with tuberculosis living in Sierra Leone, Africa.
Species assignment of isolates of M. tuberculosis, M. africanum, M. bovis, and M. microti was based on analysis of accepted phenotypic criteria.
IS6110 RFLP Profiling.
IS6110 RFLP profiling was performed by an internationally standardized method with restriction endonuclease PvuII (3). Hybridizing bands were visualized by enhanced chemiluminescence, and banding patterns were compared with computer-assisted image analysis.
PCR Amplification and Gene Sequencing.
All or part of 26 genes were characterized (Table 1). Oligonucleotide primers used to amplify the target regions are available by request from J.M.M. Sequence data were generated with an Applied Biosystems model 373A or 377 instrument. DNA sequence data reported previously (8) were also included in this analysis. A total of 2,150,267 bp was characterized.
Table 1.
Regions of M. tuberculosis complex genes analyzed for nucleotide sequence diversity
| Gene | Size, bp | Product | No. of strains analyzed | Nucleotides sequenced, positions | No. of sites with silent variation | GenBank accession no. |
|---|---|---|---|---|---|---|
| katG | 2,170 | Catalase-peroxidase | 55 | 1,991–4,160 | 5 | X68081 |
| katG | 570 | Catalase-peroxidase | 360 | 2,881–3,330 | 2 | X68081 |
| rpoB | 350 | RNA polymerase, beta subunit | 305 | 83 –432 | 2 | L05910 |
| mabA | 700 | Fatty acid biosynthesis enzyme | 46 | 1–700 | 1 | U02492 |
| mabA | 468 | Fatty acid biosynthesis enzyme | 92 | 65–533 | 1 | U02492 |
| inhA | 805 | Enoyl reductase | 72 | 986–1,794 | 0 | U02492 |
| gyrA | 318 | DNA gyrase, A subunit | 629 | 2,384–2,701 | 6 | L27512 |
| gyrB | 351 | DNA gyrase, B subunit | 17 | 1,580–1,930 | 0 | L27512 |
| hsp65 | 366 | 65-kDa heat shock protein | 267 | 453–818 | 0 | M15467 |
| rpsL | 350 | Ribosomal protein S12 | 178 | 10–359 | 1 | L08011 |
| rrs | 1,042 | 16S rRNA | 122 | 1–1,046 | 0 | X52917 |
| aroA | 349 | 5-Enolpyruvylshikimate-3-P synthase | 8 | 714–1,063 | 0 | M62708 |
| recA | 349 | RecA protein | 9 | 1,646–1,995 | 0 | X58485 |
| ahpC | 588 | Alkylhydroperoxide reductase | 97 | 628–1,216 | 0 | U16243 |
| ahpC (upstream) | 421 | Alkylhydroperoxide reductase | 196 | 981–1,401 | 0 | U16243 |
| oxyR* | 528 | Pseudogene | 117 | 1–528 | 5 | U16243 |
| rpoV | 288 | Principal sigma factor | 48 | 2,192–2,480 | 0 | U21134 |
| 16-kDa Ag | 656 | 16-kDa antigen | 35 | 25–680 | 0 | S79751 |
| embC-A | 568 | Glycosyltransferase | 101 | 80–647 | 0 | U68480 |
| embA | 1,197 | Glycosyltransferase | 43 | 2,720–3,916 | 1 | U68480 |
| embA | 1,850 | Glycosyltransferase | 7 | 920–2,770 | 0 | U68480 |
| embB | 3,295 | Glycosyltransferase | 19 | 1–3,294 | 1 | U68480 |
| embB | 2,560 | Glycosyltransferase | 3 | 640–3,200 | 0 | U68480 |
| embB | 1,952 | Glycosyltransferase | 86 | 98–2,050 | 0 | U68480 |
| mpcA | 1,563 | Phospholipase | 30 | 438–2,000 | 2 | U49511 |
| mpcA–B | 501 | Phospholipase region | 35 | 1,748–2,249 | 0 | U49511 |
| pncA | 488 | Pyrazinamidase | 50 | 30–518 | 0 | U59967 |
| pncA | 630 | Pyrazinamidase | 131 | −82 to 558 | 2 | U59967 |
| Monooxygenase (upstream) | 340 | 29 | 11,121–11,460 | 0 | Z80108 | |
| pzaA | 1,567 | Pyrazinamidase | 30 | 54–1,620 | 0 | † |
| mdh | 1,551 | Malate dehydrogenase | 34 | 928–2,478 | 0 | † |
| ndh | 1,597 | NADH dehydrogenase | 56 | 921–2,517 | 0 | † |
| ideR | 850 | DtxR homolog | 22 | −120 to 730 | 1 | U14191 |
Ag, antigen.
oxyR is a pseudogene and, therefore, all polymorphisms are silent.
Unpublished sequence.
Determination of the Presence of IS6110 Between dnaA and dnaN.
A total of 162 strains were studied by PCR for the presence of IS6110 between dnaA and dnaN. We used PCR analysis with the following oligonucleotide primers: forward, 5′-TCCGAGATGGCCGAGCGCCG-3′; reverse, 5′-CCACCCACGACACCGCATCG-3′. Strains with the insert yielded an amplicon of ≈1,800 bp, and those without the insert had an ≈700-bp PCR product.
Assignment of Strains to Three Genotypic Groups.
Isolates of M. tuberculosis complex organisms were assigned to one of three genotypic groups based on the combinations of polymorphisms at katG codon 463 and gyrA codon 95. Group 1 has the allele combination katG codon 463 CTG (Leu) and gyrA codon 95 ACC (Thr); group 2 has katG 463 CGG (Arg) and gyrA codon 95 ACC (Thr), and group 3 organisms have katG 463 CGG (Arg) and gyrA codon 95 AGC (Ser). Polymorphism located at katG codon 463 was identified by automated DNA sequencing (9) or a PCR–RFLP strategy with restriction endonuclease NciI or MspI (10). Polymorphism occurring at gyrA codon 95 was indexed by automated DNA sequencing (11). Strains (n = 850) recovered from patients in Houston from June 1995 to February 1997 were assigned to one of the three genotypic groups. Note that only approximately 100 strains of this group of 850 were included in the sample of 842 organisms used for sequencing. Case clusters were defined as groups of five or more patients infected with the same M. tuberculosis strain, based on IS6110 pattern analysis and detailed epidemiologic contact investigation data. Because the New York City strain repository contains greater than 5,000 organisms recovered between 1991 and 1997, we used a two-pronged sampling strategy. First, random isolates representing each of the 17 IS6110 types with 20 or more organisms were analyzed. Case clusters with 20 or more patients were studied because with ≈5,000 strains of M. tuberculosis in the database, it would be a considerable undertaking to study all clusters with five or more members. Next, a random sample of 75 isolates with unique IS6110 typing patterns was selected for analysis. We also analyzed a sample of 25 isolates recovered from a statewide survey of M. tuberculosis cases in New Jersey between June 1995 and December 1996. Approximately 300 isolates were obtained during this period, and about 250 of these organisms had unique IS6110 types. The 25 isolates analyzed represented a random sample of 21 strains with unique IS6110 profiles and four organisms causing case clusters. New Jersey case clusters were defined as groups of five or more patients infected with M. tuberculosis having the same IS6110 type. The New Jersey organisms were not included in the sample of 841 strains used for the nucleotide sequence database.
RESULTS
Allelic Variation Is Largely Associated with Antibiotic Resistance.
Compilation of the two megabases of sequence data for the 26 genes revealed that greater than 95% of nucleotide substitutions caused amino acid replacements or other mutations in gene regions linked to antibiotic resistance and driven to high frequency by direct drug selection (5). Over all 26 genes examined, only 32 polymorphic nucleotide sites were identified that have not been directly associated with antibiotic resistance. Of these, 30 were synonymous (silent) changes. The 30 substitutions occurred in katG (n = 7), gyrA (n = 6), oxyR (n = 5), rpoB, mabA, pncA, and mpcA (n = 2 each), and embA, embB, ideR, and rpsL (n = 1 each). Although there was substantial chromosomal heterogeneity in RFLP patterns generated by probing with mobile elements, the striking lack of silent substitutions in M. tuberculosis complex members from global sources is unexpected given that 1 billion humans carry the pathogen and, hence, the bacterial population size worldwide must be enormous.
Allelic Variation in M. tuberculosis Complex Organisms Is Restricted Compared with Other Pathogenic Bacteria.
The level of silent nucleotide variation in M. tuberculosis complex members is greatly restricted and considerably less than observed in other pathogenic bacteria (12–27) (Table 2). Importantly, the organisms studied include other mycobacteria, and species such as Neisseria meningitidis that are strict host specialists. Thus, although members of the M. tuberculosis complex have preferred hosts, ecological host specialization per se does not account for the restricted genetic variation observed in the M. tuberculosis complex. To put the level of allelic variation in M. tuberculosis in perspective, it is even less than that in Shigella sonnei, which genetically is merely a specialized distinct pathogenic clone of Escherichia coli (29).
Table 2.
Levels of allelic polymorphism recorded in genes of pathogenic bacteria
| Organism | No. of genes or gene segments | Ds* | Relative variation† |
|---|---|---|---|
| M. tuberculosis complex | 26 | <0.01 | 1 |
| Shigella sonnei | 2 | 0.01 | 1 |
| Vibrio cholerae | 1 | 0.41 | 41 |
| Streptococcus pyogenes‡ | 3 | 1.02 | 102 |
| Neisseria meningitidis | 4 | 6.18 | 618 |
| M. avium-intracellulare | 1 | 10.10 | 1,010 |
| Escherichia coli | 11 | 11.77 | 1,177 |
| Borrelia burgdorferi | 3 | ≈20 | ≈2,000 |
| Salmonella enterica | 5 | 42.08 | 4,208 |
Average number of synonymous substitutions per 100 synonymous sites (28), calculated on the basis of sequence information published elsewhere (12-27). The value reported for S. sonnei includes both synonymous and nonsynonymous sites and is based on RFLP analysis (15).
Calculated relative to the Ds value of the M. tuberculosis complex.
Includes some unpublished data.
Identification of Three Genotypic Groups.
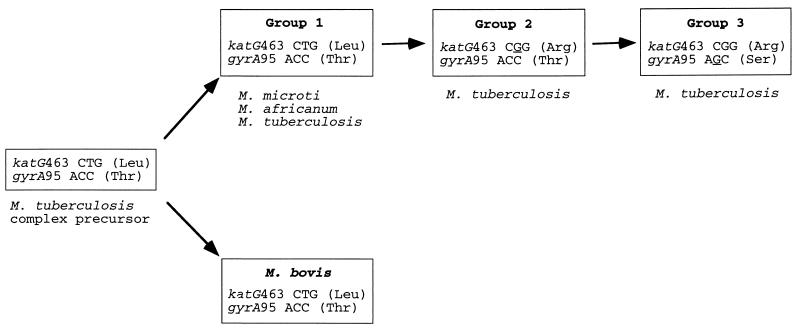
Inspection of the sequence data revealed that only the variants at katG codon 463 and gyrA codon 95 were present at high frequency. These two sites apparently do not participate in antibiotic resistance (30, 31) and, hence, were used as genetic markers that record the history of organism divergence. All members of the M. tuberculosis complex were assigned to one of three distinct genotypic groups based on the combination of polymorphisms located at these two sites (Fig. 1). All isolates of M. bovis, M. microti, and M. africanum studied had the combination of polymorphisms characteristic of genotypic group 1. In contrast, M. tuberculosis isolates fell into each of the three groups.
Figure 1.
Broad evolutionary scenario for M. tuberculosis complex organisms. The precursor of M. tuberculosis complex organisms was characterized by KatG codon 463 (Leu) and GyrA codon 95 (Thr). Strain Ravenal is a typical M. bovis isolate; New York City IS6110 type strain W and Houston IS6110 types 002, 003, 007, 015, and 033 are group 1 organisms; Erdman, Oshkosh, New York City strain C, and Houston IS6110 types 004, 006, 016, 020, and 030 are group 2; and H37Ra, H37Rv, and Houston IS6110 type 001 are group 3.
IS6110 Frequency Distribution Among Isolates of the Three Genotypic Groups.
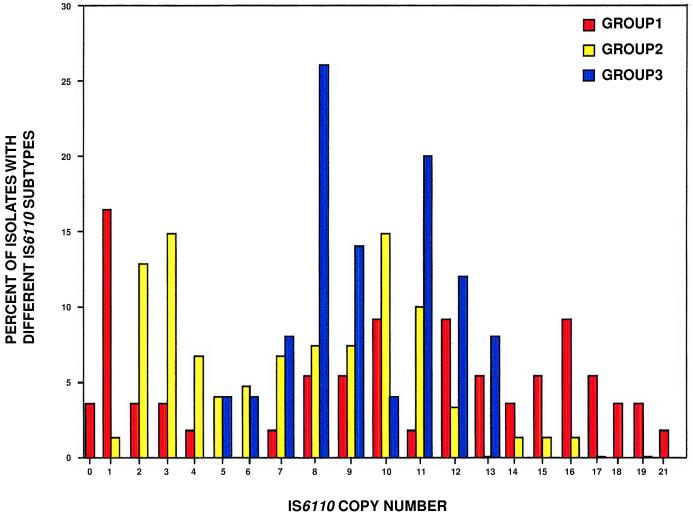
To determine the extent to which organisms assigned to the three genotypic groups have diverged in other properties, we examined the distribution of IS6110, an insertion sequence present in most M. tuberculosis isolates. The analysis was based on study of 421 organisms recovered from patients in Houston, TX, in a 13-month period. To avoid bias that would be generated by including epidemiologically associated organisms, only one isolate was included from each subtype identified by standard IS6110 typing. The frequency distribution of IS6110 copy number differed among the three genotypic groups (Fig. 2). Moreover, there was little sharing of related IS6110 profiles among groups. These data indicate that isolates of the three groups have accumulated differences in IS6110 copy number and pattern and undergone rapid chromosomal differentiation relative to silent mutations.
Figure 2.
Distribution of IS6110 copy number with respect to three genotypic groups of M. tuberculosis. The data are based on analysis of 427 isolates recovered from patients in Houston, TX. To avoid bias caused by redundant sampling of recent derivatives of epidemiologically linked isolates, only one isolate per distinct IS6110 subtype was used in the analysis. This resulted in a data set composed of 213 distinct IS6110 subtypes, of which 21% were group 1, 62% group 2, and 18% group 3.
Correlation of Site of IS6110 Insertion and Genotypic Group.
Two distinct molecular mechanisms could account for the occurrence of three genotypic groups of M. tuberculosis. One mechanism postulates that organisms assigned to each of the three genotypic groups have shared a common ancestor, and the extensive IS6110 variation recorded among members of each group arose after the divergence of the last common ancestor for each group. An alternative idea postulates that organisms have repetitively converged by independent evolutionary pathways to form the three genotypic groups. The distribution of IS6110 copy number described above is strong evidence against the convergence hypothesis. To differentiate more fully between the two hypothesis, we used PCR to examine the distribution of one randomly chosen IS6110 insertion site located between dnaA and dnaN among the three groups marked by gyrA and katG polymorphisms. IS6110 was located at the target site in 26% of 97 organisms in group 1 having distinct IS6110 profiles. In contrast, all 65 randomly chosen isolates of groups 2 and 3 tested lacked the element at this site. All 32 isolates of M. bovis, M. africanum, and M. microti studied also lacked the element at this site. The confinement of this insertion site polymorphism to a subset of group 1 M. tuberculosis further demonstrates an historical separation and genotypic differentiation of the three groups.
Additional documentation of a common ancestry of many group 1 isolates with IS6110 located between dnaA and dnaN was sought by DNA sequencing. Analysis of six randomly chosen strains with different IS6110 patterns found that all six organisms had IS6110 integrated at the identical site located 66 bp downstream from dnaA. The integration of IS6110 at precisely the same position in these group 1 members adds further strong support to the idea that the organisms have shared a common ancestor.
Genotypic Group 1 Organisms Are Ancestral to M. tuberculosis Groups 2 and 3.
In the proposed evolutionary pathway presented in Fig. 1, genotypic group 1 organisms are especially important because they link the predominantly nonhuman pathogens M. microti and M. bovis and the human specialists M. africanum and M. tuberculosis. If our evolutionary hypothesis is correct, then we expect that group 1 organisms carry a greater level of genetic diversity than group 2 and group 3 bacteria because they have had a longer time to evolve and thereby accumulate variation. Three facts support this hypothesis. (i) Group 1 organisms have the broadest range of IS6110 copy number, with 0–21 copies of this element. (ii) Group 1 organisms had 73% of all synonymous substitutions identified, despite being only about 15% of all organisms analyzed. (iii) All five variant sites detected in the pseudogene oxyR were either in M. bovis or group 1 organisms, and none were present in organisms assigned to groups 2 or 3. The neutral theory of molecular evolution predicts that allelic variation in pseudogenes exceeds that present in genes encoding protein products because functional constraints are relaxed (32). The observation of increased allelic variation in oxyR in group 1 organisms provides critical independent support of the concept that group 1 organisms are ancestral to groups 2 and 3. Also consistent with our thesis that group 1 bacteria are ancestral to groups 2 and 3 is the fact that all four organisms in our collection that lack IS6110 are group 1 members. Absence of IS6110 is the expected condition for a primitive organism from which M. tuberculosis containing this element may have arisen.
DISCUSSION
Restricted Allelic Variation.
The remarkably restricted variation in M. tuberculosis complex structural genes has important ramifications for studies of virulence and drug resistance. The lack of allelic diversity means that when amino acid polymorphisms, or regulatory region nucleotide variation are observed, there should be strong suspicion that the variation has functional consequences, such as antibiotic resistance (5). Restricted allelic diversity also means that identification of proteins with immunoprophylaxis or diagnostic utility would be of significant clinical value due to the very low probability for variation globally. Conversely, should future genomic analyses uncover highly polymorphic genes, the finding could indicate that the diversity is being driven by host immunologic responses.
The spontaneous mutation frequency of M. tuberculosis is in the range recorded for most other bacteria (33), an observation that permits us to exclude the possibility that an unusual DNA repair phenotype or replication fidelity is responsible for the lack of accumulation of neutral variation. A small population size also is unlikely to explain the limited allelic diversity given that one-third of the world’s population is infected with this bacterium. We believe that the only reasonable hypothesis to account for the lack of allelic variation is that M. tuberculosis underwent a recent evolutionary bottleneck, presumably at the time of speciation estimated to have occurred roughly 15,000 to 20,000 years ago (8).
Two main processes are driving a considerable proportion of genomic differentiation in M. tuberculosis. One mechanism responsible for creating variation is transposition of IS6110 and other mobile elements, and these events can rapidly generate new subclones (34). For example, a single multidrug-resistant organism marked by a distinct IS6110 pattern and known as strain W within years spawned at least seven variants differing by one or two copies of IS6110 (34). The level of species variation also is being profoundly increased by humans through antibiotic use. In our two-megabase data set, greater than 95% of nucleotide changes were directly associated with antibiotic resistance. All antibiotic resistance in M. tuberculosis is chromosomally mediated, unlike most pathogenic bacteria in which drug resistance is mediated by plasmid-borne genes that can be discarded when selection pressure is removed. This means that the genomic changes arising from drug selection will be removed from the species gene pool only if the resistant organisms themselves are extinguished. Given the considerable difficulties in successfully treating drug-resistant strains, and the lack of agents to eradicate latent infections, most of the resistance-associated diversity is likely to be maintained in the M. tuberculosis population.
Genotypic Group 1 and Group 2 Organisms Are Disproportionately Represented Among Clustered Cases of Tuberculosis.
One theme that has emerged from the study of bacterial population genetics is that there are frequently important biomedical and ecological correlates of population structure. Stated another way, groups of related bacterial genotypes tend to behave nonrandomly. This is especially true for species (like M. tuberculosis) in which horizontal gene transfer contributes little to the generation of genomic diversity.
The availability of extensive databases containing IS6110 typing data for ≈6,000 isolates permitted us to determine whether organisms of the three groups were equally represented among genotypes causing case clusters of tuberculosis. The Houston M. tuberculosis database has typing information for all 850 strains recovered over a 2-year period in a comprehensive population-based survey. Detailed epidemiologic data, including contact investigation, are available for all patients from whom these organisms were cultured. Interestingly, 25 of 26 case clusters involving five or more patients were caused by genotypic group 1 and 2 organisms (Table 3). This striking association was confirmed by analysis of strains recovered in metropolitan New York City. All 21 case clusters in the New York City area were also caused by genotypic group 1 and 2 organisms. Nonrandom association of group 1 and group 2 organisms with clustered cases was independent of the drug susceptibility phenotype of the strains. Critically, the dominance of group 1 and group 2 organisms as causes of case clusters was not due to the formal albeit trivial explanation that group 3 organisms do not occur in these diverse localities (Table 3).
Table 3.
Distribution of M. tuberculosis genotypic groups in Houston and the New York City area
| Place | Genotypic group*
|
|||||
|---|---|---|---|---|---|---|
| 1
|
2
|
3
|
||||
| Case clusters, no. | Unique isolates, no. | Case clusters, no. | Unique isolates, no. | Case clusters, no. | Unique isolates, no. | |
| Houston | 10 (37%) | 100 (28%) | 16 (59%) | 200 (56%) | 1 (4%)† | 55 (16%) |
| New York City area | 7 (32%) | 23 (23%) | 15 (68%) | 63 (62%) | 0 | 15 (15%) |
Group 1, allele combination katG codon 463 CTG (Leu) and gyrA codon 95 ACC (Thr); group 2, katG 463 CGG (Arg) and gyrA codon 95 ACC (Thr); group 3, katG 463 CGG (Arg) and gyrA codon 95 AGC (Ser).
Genotypic group 1 versus group 3, P ≈ 0.008; genotypic group 2 versus group 3, P ≈ 0.015; genotypic group 1 versus group 2, not significant. P values were calculated with pooled Houston and New York City area samples.
Variance in pathogen behavior is likely to be the result of two processes. Nucleotide changes may result in amino acid alteration in proteins or modification of regulatory sequences that produce increased or decreased expression of protein products. Data have been presented showing that single nucleotide or amino acid changes can significantly alter virulence of M. tuberculosis for guinea pigs and virulence in other pathogens (35, 36). A second possibility that has not yet been explored is that the site of chromosomal integration of IS6110 or other mobile elements participates. Mobile element insertion or excision from genomic sites can alter expression of genes involved in host–pathogen interactions or contribute to bacterial fitness (37, 38). Simonet et al. (37) reported that the invasin gene (inv) of Yersinia pestis (the cause of plague) is inactivated by a 708-bp IS200-like element. This single insertion prevents the pathogen from invading host cells, thereby altering the host–pathogen interaction and selecting for new virulence properties. Moreover, the site of IS1301 insertion in Neisseria meningitidis changes expression of cell surface sialic acid, a critical virulence factor in blood and brain infections (38). It is reasonable to anticipate that insights about M. tuberculosis pathogenesis will be revealed by systematic study of IS6110 integration sites and assessment of resulting functional alterations.
Although clearly more work is required to identify the molecular basis for our observation that strains of genotypic group 3 have a reduced capacity to cause large case clusters, we note that the finding has important implications for public health strategies, evolutionary biology, and virtually every aspect of M. tuberculosis investigation. Tuberculosis control strategies and transmission models have largely relied on the idea that strains or subclones are roughly equivalent in medically important biological attributes (39). Our data suggest this is not the case. Public health approaches and tuberculosis transmission models may benefit from further refinement by interpreting data in the context of the three genetic groups described herein. Advances in the study of host–pathogen interactions, genome sequences (40), antimicrobial agent resistance (5), and the genetics of host susceptibility (41) are also likely to benefit by the insights into the evolutionary history revealed by our analysis.
Acknowledgments
We thank many colleagues who supplied strains and provided critical comments on an early draft of the manuscript. In particular, R. K. Selander contributed scholarly input, and two anonymous reviewers suggested several improvements. G. Adams supplied assistance with statistical analyses. We are especially indebted to K. Davenny and A. M. Ginsberg for their support. This research was supported by Public Health Services Grants DA-09238, AI-37004, and AI-41168 to J.M.M.
ABBREVIATIONS
- RFLP
restriction fragment length polymorphism
- IS
insertion sequence
Note Added in Proof:
Analysis of an additional one megabase of sequence data from 10 genes and the DR region confirmed the broad evolutionary scenario described in Fig. 1.
References
- 1.Bloom B R, Murray C J L. Science. 1992;257:1055–1064. doi: 10.1126/science.257.5073.1055. [DOI] [PubMed] [Google Scholar]
- 2.Raviglione M C, Snider D E, Kochi A. J Am Med Assoc. 1995;273:220–226. [PubMed] [Google Scholar]
- 3.van Embden P D A, Cave M D, Crawford J T, Dale J W, Eisenach K D, Gicquel B, Hermans P W M, Martin C, McAdam R, Shinnick T M, Small P M. J Clin Microbiol. 1993;31:406–409. doi: 10.1128/jcm.31.2.406-409.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hermans P W M, Messadi F, Guebrexabher H, van Soolingen D, de Haas P E W, Heersma H, de Neeling H, Ayoub A, Portaels F, Frommel D, Zribi M, van Embden J D A. J Infect Dis. 1995;171:1504–1513. doi: 10.1093/infdis/171.6.1504. [DOI] [PubMed] [Google Scholar]
- 5.Musser J M. Clin Microbiol Rev. 1995;8:496–514. doi: 10.1128/cmr.8.4.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sreevatsan S, Escalante P, Pan X, Gillies D A, III, Siddiqui S, Khalaf C N, Kreiswirth B N, Bifani P, Adams L G, Ficht T, Perumaalla S, Cave M D, van Embden J D A, Musser J M. J Clin Microbiol. 1996;34:2007–2010. doi: 10.1128/jcm.34.8.2007-2010.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Plikaytis B B, Marden J L, Crawford J T, Woodley C L, Butler W R, Shinnick T M. J Clin Microbiol. 1994;32:1542–1546. doi: 10.1128/jcm.32.6.1542-1546.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kapur V, Whittam T S, Musser J M. J Infect Dis. 1994;170:1348–1349. doi: 10.1093/infdis/170.5.1348. [DOI] [PubMed] [Google Scholar]
- 9.Musser J M, Kapur V, Williams D L, Kreiswirth B N, van Soolingen, van Embden J D A. J Infect Dis. 1996;173:196–202. doi: 10.1093/infdis/173.1.196. [DOI] [PubMed] [Google Scholar]
- 10.Cockerill F R, III, Uhl J R, Temesgen Z, Zhang Y, Stockman L, Roberts G D, Williams D L, Kline B C. J Infect Dis. 1995;171:240–245. doi: 10.1093/infdis/171.1.240. [DOI] [PubMed] [Google Scholar]
- 11.Kapur V, Li L-L, Hamrick M R, Plikaytis B B, Shinnick T M, Telenti A, Jacobs W R, Jr, Banerjee A, Cole S, Yuen K Y, Clarridge J E, III, Kreiswirth B N, Musser J M. Arch Pathol Lab Med. 1995;119:131–138. [PubMed] [Google Scholar]
- 12.Nelson K, Whittam T S, Selander R K. Proc Natl Acad Sci USA. 1991;88:6667–6671. doi: 10.1073/pnas.88.15.6667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nelson K, Selander R K. J Bacteriol. 1992;174:6886–6895. doi: 10.1128/jb.174.21.6886-6895.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boyd E F, Nelson K, Wang F-S, Whittam T S, Selander R K. Proc Natl Acad Sci USA. 1994;91:1280–1284. doi: 10.1073/pnas.91.4.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karaolis D K R, Lan R, Reeves P R. J Clin Microbiol. 1994;32:796–802. doi: 10.1128/jcm.32.3.796-802.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karaolis D K R, Lan R, Reeves P R. J Bacteriol. 1995;177:3191–3198. doi: 10.1128/jb.177.11.3191-3198.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li J, Ochman H, Groisman E A, Boyd E F, Solomon F, Nelson K, Selander R K. Proc Natl Acad Sci USA. 1995;92:7252–7256. doi: 10.1073/pnas.92.16.7252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kapur V, Kanjilal S, Hamrick M R, Li L-L, Whittam T S, Sawyer S A, Musser J M. Mol Microbiol. 1995;16:509–519. doi: 10.1111/j.1365-2958.1995.tb02415.x. [DOI] [PubMed] [Google Scholar]
- 19.Caporale D A, Kocher T D. Mol Biol Evol. 1994;11:51–64. doi: 10.1093/oxfordjournals.molbev.a040092. [DOI] [PubMed] [Google Scholar]
- 20.Zhou J, Spratt B G. Mol Microbiol. 1992;6:2135–2146. doi: 10.1111/j.1365-2958.1992.tb01387.x. [DOI] [PubMed] [Google Scholar]
- 21.DuBose R F, Dykhuizen D E, Hartl D L. Proc Natl Acad Sci USA. 1988;85:7036–7040. doi: 10.1073/pnas.85.18.7036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bisercic M, Feutrier J Y, Reeves P R. J Bacteriol. 1991;173:3894–3900. doi: 10.1128/jb.173.12.3894-3900.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hall B G, Sharp P M. Mol Biol Evol. 1992;9:654–665. doi: 10.1093/oxfordjournals.molbev.a040751. [DOI] [PubMed] [Google Scholar]
- 24.Guttman D S, Dykhuizen D E. Science. 1994;266:1380–1383. doi: 10.1126/science.7973728. [DOI] [PubMed] [Google Scholar]
- 25.Theisen M, Borre M, Mathiesen M J, Mikkelsen B, Lebech A-M, Hansen K. J Bacteriol. 1995;177:3036–3044. doi: 10.1128/jb.177.11.3036-3044.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thampapillai G, Lan R, Reeves P. Mol Biol Evol. 1994;11:813–828. doi: 10.1093/oxfordjournals.molbev.a040165. [DOI] [PubMed] [Google Scholar]
- 27.Fiel E, Carpenter G, Spratt B G. Proc Natl Acad Sci USA. 1995;92:10535–10539. doi: 10.1073/pnas.92.23.10535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li W-H, Wu C-I, Luo C-C. Mol Biol Evol. 1986;2:150–174. doi: 10.1093/oxfordjournals.molbev.a040343. [DOI] [PubMed] [Google Scholar]
- 29.Ochman H, Whittam T S, Caugant D A, Selander R K. J Gen Microbiol. 1983;129:2715–2726. doi: 10.1099/00221287-129-9-2715. [DOI] [PubMed] [Google Scholar]
- 30.Takiff H E, Salazar L, Guerrero C, Philipp W, Huang W M, Kreiswirth B, Cole S T, Jacobs W R, Jr, Telenti A. Antimicrob Agents Chemother. 1994;38:773–780. doi: 10.1128/aac.38.4.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rouse D A, DeVito J A, Li Z, Byer H, Morris S L. Mol Microbiol. 1996;22:583–592. doi: 10.1046/j.1365-2958.1996.00133.x. [DOI] [PubMed] [Google Scholar]
- 32.Kimura M. The Neutral Theory of Molecular Evolution. Cambridge: Cambridge Univ. Press; 1983. [Google Scholar]
- 33.David H L, Newman C M. Am Rev Respir Dis. 1971;104:508–515. doi: 10.1164/arrd.1971.104.4.508. [DOI] [PubMed] [Google Scholar]
- 34.Bifani P, Plikaytis B B, Kapur V, Stockbauer K, Pan X, Lutfey S L, Moghazeh S L, Eisner W, Daniel T M, Kaplan M H, Crawford J T, Musser J M, Kreiswirth B N. J Am Med Assoc. 1996;275:452–457. [PubMed] [Google Scholar]
- 35.Collins D M, Kawakami R P, De Lisle G W, Pascopella L, Bloom B R, Jacobs W R., Jr Proc Natl Acad Sci USA. 1995;92:8036–8040. doi: 10.1073/pnas.92.17.8036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kawaoka Y, Webster R G. Microb Pathog. 1988;5:311–318. doi: 10.1016/0882-4010(88)90032-0. [DOI] [PubMed] [Google Scholar]
- 37.Simonet M, Riot B, Fortineau N, Berche P. Infect Immun. 1996;64:375–379. doi: 10.1128/iai.64.1.375-379.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hammerschmidt S, Hilse R, van Putten J P M, Gerardy-Schahn R, Unkmeir A, Frosch M. EMBO J. 1996;15:192–198. [PMC free article] [PubMed] [Google Scholar]
- 39.Blower S M, Small P C, Hopewell P C. Science. 1996;273:497–500. doi: 10.1126/science.273.5274.497. [DOI] [PubMed] [Google Scholar]
- 40.Philipp W J, Poulet S, Eiglmeier K, Pascopella, Balasubramanian V, Heym B, Bergh S, Bloom B R, Jacobs W R, Jr, Cole S T. Proc Natl Acad Sci USA. 1996;93:3132–3137. doi: 10.1073/pnas.93.7.3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Govoni G, Vidal S, Gauthier S, Skamene E, Malo D, Gros P. Infect Immun. 1996;64:2923–2929. doi: 10.1128/iai.64.8.2923-2929.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]