Abstract
Cells respond to DNA double-strand breaks (DSBs) and uncapped telomeres by recruiting checkpoint and repair factors to the site of lesions. Single-stranded DNA (ssDNA) is an important intermediate in the repair of DSBs and is produced also at uncapped telomeres. Here, we provide evidence that binding of the checkpoint protein Rad9, through its Tudor domain, to methylated histone H3-K79 inhibits resection at DSBs and uncapped telomeres. Loss of DOT1 or mutations in RAD9 influence a Rad50-dependent nuclease, leading to more rapid accumulation of ssDNA, and faster activation of the critical checkpoint kinase, Mec1. Moreover, deletion of RAD9 or DOT1 partially bypasses the requirement for CDK1 in DSB resection. Interestingly, Dot1 contributes to checkpoint activation in response to low levels of telomere uncapping but is not essential with high levels of uncapping. We suggest that both Rad9 and histone H3 methylation allow transmission of the damage signal to checkpoint kinases, and keep resection of damaged DNA under control influencing, both positively and negatively, checkpoint cascades and contributing to a tightly controlled response to DNA damage.
Keywords: chromatin, DNA damage checkpoint, DNA repair, histone methylation, telomeres
Introduction
Eukaryotic cells evolved a complex system to protect the genome from spontaneous and exogenous DNA damage. A central role is played by DNA damage checkpoint pathways, signal transduction cascades coordinating DNA replication, repair and recombination with cell cycle progression. The defining feature of an active checkpoint is the arrest of cell proliferation at the G1/S or G2/M transitions, or the slowing down of DNA replication (Elledge, 1996; Nyberg et al, 2002). Many details of the DNA damage checkpoint have been established using genetic and biochemical approaches in budding and fission yeast; the basic checkpoint response has been shown to be conserved in other eukaryotes (Longhese et al, 1998; Melo and Toczyski, 2002; Rouse and Jackson, 2002; Lydall and Whitehall, 2005). In budding yeast, it has been useful to examine the roles of checkpoint proteins at uncapped telomeres and unrepaired double-strand breaks (DSBs). Several repair, recombination and checkpoint factors are recruited at a DSB site, according to a well-established order (Lisby et al, 2004). DSB ends are initially processed to generate long 3′ single-stranded DNA (ssDNA) tails. The Mre11, Rad50, Xrs2 (MRX) complex is involved in this process, as mutations in the corresponding genes reduce the resection rate (White and Haber, 1990; Ivanov et al, 1994; Lee et al, 1998). However, MRX is not likely to be the nuclease itself: point mutations within Mre11 catalytic site do not affect resection of DSB ends (Moreau et al, 1999; Lee et al, 2002; Llorente and Symington, 2004). The nature of the nuclease(s) involved in DSB processing has been elusive; moreover, we lack information on the regulatory mechanisms governing DNA end resection (see Harrison and Haber, 2006 for a recent review). It has been recently shown that, both in human cells and in Schizosaccharomyces pombe, CtIP, a partner of the MRN complex, is required for efficient resection of DSB ends (Limbo et al, 2007; Sartori et al, 2007). In budding yeast, CtIP seems to correspond to Sae2, which also has a positive function in DSB processing (Clerici et al, 2005; Sartori et al, 2007). Recent studies show that CDK1 kinase is important for the generation of ssDNA tails; in fact, inhibition of CDK1 strongly interferes with resection (Aylon et al, 2004; Ira et al, 2004). ssDNA is bound by the RPA heterotrimer, generating a structure important for recruiting checkpoint factors (Kornbluth et al, 1992; Zou and Elledge, 2003; Zou et al, 2003; Majka et al, 2006).
PI3-like kinases have essential functions in checkpoint signal transduction in all eukaryotes. In budding yeast, the Mec1/Ddc2 checkpoint protein kinase is stimulated both by binding to RPA-coated ssDNA and by the checkpoint sliding clamp (Rad17, Mec3 and Ddc1) (Zou and Elledge, 2003; Zou et al, 2003; Majka et al, 2006). Mec1 phosphorylates several targets, among these Ddc2, Ddc1, Rad9 and the protein kinases Chk1 and Rad53. Rad9 is a checkpoint adaptor molecule, linking the upstream Mec1 kinase with downstream Rad53 and Chk1 kinases, and it is essential for checkpoint function (Sanchez et al, 1996; Gardner et al, 1999; Blankley and Lydall, 2004). It is thought that Mec1-dependent phosphorylation of Rad9 recruits and catalyses Rad53 activation (Gilbert et al, 2001; Sweeney et al, 2005).
As well as being essential for cell cycle arrest, Rad9 contributes to DNA damage metabolism because Rad9 inhibits the accumulation of ssDNA at uncapped telomeres (Lydall and Weinert, 1995). This effect of Rad9 is not simply checkpoint signal transduction dependent, because other checkpoint proteins, such as Rad24, are also required to signal cell cycle arrest at uncapped telomeres and yet have the opposite resection phenotype (Lydall and Weinert, 1995). However, so far, no biochemical mechanism by which Rad9 inhibits resection at uncapped telomeres has been discovered. In addition, it was not known whether Rad9 inhibits ssDNA accumulation at other types of lesion, such as DSBs.
Budding yeast Rad9 interacts with the methylated K79 residue of histone H3, through the Rad9 Tudor domain, and this interaction regulates Rad9 function after DNA is damaged (Giannattasio et al, 2005; Wysocki et al, 2005; Grenon et al, 2007). Similar results were reported for S. pombe Crb2, where the binding target seems to be methylated H4-K20 (Sanders et al, 2004; Du et al, 2006), whereas for human 53BP1 binding to both residues has been reported (Huyen et al, 2004; Botuyan et al, 2006). Loss of methylation of the K79 residue of histone H3 impairs Rad9 phosphorylation and activation of Rad53, after DNA damage in G1 cells. Furthermore, a rad9Y798Q point mutation within the Tudor domain prevents Rad9 binding to chromatin and Rad9 hyper-phosphorylation after DNA damage (Giannattasio et al, 2005; Wysocki et al, 2005; Grenon et al, 2007; Hammet et al, 2007, and Supplementary Figure 1). The simplest explanation for these data is that methylated histone H3-K79 is involved in recruiting Rad9 to damaged chromosomes and that this contributes to Rad9 hyper-phosphorylation and checkpoint activation.
As H3-K79 appears to be constitutively methylated by the Dot1 methyltransferase in undamaged cells (90% of H3 is methylated at K79; van Leeuwen et al, 2002), it has been proposed that the critical event for Rad9 recruitment may be a DNA-damage-induced change in the status of chromatin, allowing exposure of this methylated residue (Huyen et al, 2004). Another intriguing option would be that Rad9 is always weakly bound to methylated H3-K79; upon damage Rad9 oligomerization may cause its accumulation at the sites of lesion. Moreover, post-translational modifications may induce changes in Rad9-binding mode and allow it to interact with other partners (Du et al, 2006; Hammet et al, 2007).
Here, we produce evidence that the interaction between histone H3-K79 and Rad9 inhibits accumulation of ssDNA at DSBs and at uncapped telomeres. This mechanism, requiring methylation of histone H3 and the Tudor domain of Rad9, regulates resection and appears to represent a strategy that coordinates cell cycle arrest with nuclease progression, thus limiting the amount of ssDNA generated during the cellular response to DNA damage.
Results
Methylation of H3-K79 controls Mec1 kinase activation after DNA damage
Dot1 is required for the G1/S DNA damage checkpoint (Giannattasio et al, 2005; Wysocki et al, 2005). To investigate the effect of the loss of DOT1 on Mec1 kinase activity directly, we analysed the phosphorylation of its proximal target Ddc2, after DNA damage. Ddc2 is a stable partner of Mec1 and is directly phosphorylated by Mec1 in vivo and in vitro (Paciotti et al, 2000; Rouse and Jackson, 2000; Wakayama et al, 2001). Cells were arrested in G1 to avoid complications due to cell cycle-dependent phosphorylation of Ddc2 during the S/G2 phases (Paciotti et al, 2000). Analogous experiments in G2-arrested cells gave comparable results (Supplementary Figure 2). Surprisingly, in time-course analyses, we observed an increase in the phosphorylated form of Ddc2 in dot1Δ cells. G1-arrested wild-type (WT) and dot1Δ cells, expressing HA-tagged Ddc2, were treated with zeocine, which induces DSBs, and Ddc2 phosphorylation was evaluated at different times after the treatment. We consistently found that dot1Δ cells showed a hyper-modification of Ddc2, after induction of DSBs (Figure 1A and B). To verify that the increase in Ddc2 phosphorylation was due to Mec1 and not to another kinase, we compared Ddc2 phosphorylation in WT, dot1Δ, mec1-1 and dot1Δ mec1-1 mutants. Figure 1C shows that all the phospho-Ddc2 signal, detectable in WT and dot1Δ cells, disappears when Mec1 is defective, demonstrating that the increase in Ddc2 phosphorylation observed in dot1Δ cells is indeed due to the Mec1 kinase. These results suggest that more Mec1–Ddc2 kinase complexes can be activated after DSB induction, in the absence of Dot1, and hence H3-K79 methylation.
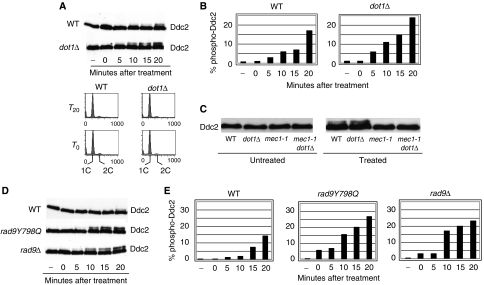
Figure 1.
Loss of DOT1 or RAD9 leads to Mec1 kinase hyper-activation after induction of DSBs. (A) WT (YLL683.8/3b) and dot1Δ (YFL403/10b) cells carrying an HA-tagged version of DDC2 at its chromosomal locus were arrested in G1 and treated with 50 μg/ml zeocine to induce DSBs. At the indicated times (–: untreated cells) samples were taken, protein extracts were prepared and the phosphorylation-dependent mobility shift of Ddc2 in SDS–PAGE was monitored by western blotting with 12CA5 antibodies (top panels). FACS profiles of the cultures (bottom panels) show that both cultures did not escape the G1 block throughout the experiment. (B) Quantification of the percentage of phosphorylated form relative to the total Ddc2 protein (from (A)). (C) Ddc2 phosphorylation was monitored in WT (YFL693), dot1Δ (YFL694), mec1-1 (YFL219/9b), mec1-1 dot1Δ (YFL571.1) cells after treatment with zeocine, as in (A). (D) Western blots of Ddc2 in WT (YLL683.8/3b), rad9Y798Q (YFL502) and rad9Δ (YFL407/5a) cells, at different times after induction of DSBs. (E) Quantification of the bands from (D).
Failure to recruit Rad9 to histone H3 leads to an increase in Mec1 activation
Previous reports suggested that Dot1-dependent methylation of H3-K79 is critical for docking Rad9 to damaged chromatin (Wysocki et al, 2005; Toh et al, 2006). To test whether impairment of Rad9 recruitment to histone H3 would, similarly to a dot1Δ mutation, lead to an increase in Mec1 activity, we introduced a RAD9 allele carrying a Y798Q point mutation in its Tudor domain; this mutation prevents Rad9 recruitment to damaged DNA, damage-dependent phosphorylation of Rad9 and the activation of Rad53 (Wysocki et al, 2005; Grenon et al, 2007, and Supplementary Figure 1). The kinetics of Mec1 activation after DSB induction was analysed in rad9Y798Q and rad9Δ cells. Figure 1D shows that, similarly to what was found in a dot1Δ strain, cells completely lacking Rad9 or expressing rad9Y798Q exhibit faster Ddc2 phosphorylation after DSB induction; quantification of the phospho-Ddc2 form confirmed the observation (Figure 1E). Taken together, these results strongly suggest that loss of Rad9 binding to methylated H3-K79 leads to a faster and more robust activation of Mec1 kinase in response to DSBs.
A robust DNA damage checkpoint is not triggered by DSBs themselves, but rather by processed DNA ends, containing long stretches of ssDNA, which recruit Mec1–Ddc2 kinase complexes (White and Haber, 1990; Lydall et al, 1996; Lee et al, 1998; Usui et al, 2001; Harrison and Haber, 2006). Therefore, the Mec1 hyper-activation detected in dot1Δ and rad9 mutants could be explained if DSBs were more rapidly processed to ssDNA when Rad9 does not bind methylated H3.
Dot1 and Rad9 limit resection of DNA DSB ends
To test the hypothesis that more rapid activation of Mec1 kinase results from a faster production of ssDNA intermediates in dot1Δ, rad9Y798Q and rad9Δ cells, we investigated the kinetics of ssDNA formation after a single unrepairable DSB in these mutants, using an inducible HO endonuclease. Cells were arrested in G2, to prevent cell cycle-dependent effects on resection, and samples were collected at various time points after induction of the nuclease. ssDNA regions in genomic DNA were revealed by the loss of restriction sites distal to the HO-cut site, leading to the accumulation of uncut DNA fragments that were detected with a strand-specific probe, after alkaline electrophoresis (White and Haber, 1990; Shroff et al, 2004; Clerici et al, 2006. See Figure 2B for a map of the MAT locus and the location of probe used in these experiments). The kinetics of appearance of longer DNA fragments suggests that dot1Δ, rad9Y798Q and rad9Δ cells all showed more rapid resection than WT cells (Figure 2A). This finding was confirmed using a different assay where resection leads to the disappearance of a specific DNA restriction fragment in Southern blots (see Figures 5 and 6). These data suggest that the impairment of Rad9 binding to methylated H3-K79, as seen in rad9Δ, rad9Y798Q, dot1Δ and rad9Y798Q dot1Δ (not shown), leads to faster resection at an HO-induced DSB.
Figure 2.
Resection of a DSB is faster in dot1Δ, rad9Y798Q and rad9Δ cells. (A) WT (JKM179), dot1Δ (YFL399), rad9Y798Q (YFL504) and rad9Δ (YFL419) cells, carrying a unique HO-cut site at the MAT locus and expressing the HO endonuclease under the inducible GAL1 promoter, were grown in presence of lactate and arrested with nocodazole. HO was induced with galactose 2%. Genomic DNA, extracted from samples collected at the indicated times, was digested with SspI and separated on alkaline denaturating gels. Resection was monitored by Southern blotting using a ribo-probe specific for the 3′strand. (B) Scheme of the MAT locus. The figure shows the positions of the HO-cut site, and of the probe (asterisk) used for the experiments shown in (A). The black vertical bars indicate the SspI sites. The products of the digestion of differently resected molecules are shown above the scheme.
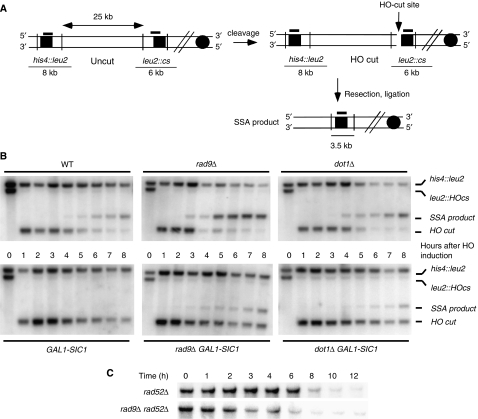
Figure 5.
Loss of Rad9 bypasses CDK1 requirement for SSA and resection. (A) Map of the YMV80 Chr III region, containing the HO-cut site. Vertical bars show the relevant KpnI sites. The thick lines above the map indicate the positions where the probe hybridizes. After HO cleavage, DNA is resected. When the left and right leu2 sequences have been converted to ssDNA, repair by SSA can take place and can be monitored by the appearance of a SSA product in a Southern blot. (B) Exponentially growing YEP+raffinose cell cultures of WT (YMV80) and isogenic rad9Δ (Y31), GAL1-SIC1 (Y20), rad9Δ GAL1-SIC1 (Y293), dot1Δ (YFL736) and dot1Δ GAL-SIC1 (YFL738) strains, carrying an HO-cut site and a gal-inducible HO gene, were arrested with nocodazole; galactose was added at time zero. KpnI-digested DNA, prepared from cells collected at the indicated times, was analysed by Southern blotting with a LEU2 probe. Two fragments, 8 and 6 kb long (his4∷leu2, leu2∷HOcs) are evident in the absence of HO cut, whereas the HO-induced DSB causes the disappearance of the 6-kb species and the formation of a 2.5-kb fragment (HO-cut fragment). Repair by SSA converts such fragment to a repair product of 3.5-kb (SSA product). (C) Speed of resection was evaluated in rad52Δ (YMV037) and rad52Δ rad9Δ (YMV038) cells under similar conditions by monitoring the disappearance of the his4∷leu2 signal.
Figure 6.
Loss of RAD9 or DOT1 increase resection at DSBs bypassing the requirement for Cdk1. The graphs represent quantifications of data shown in Figure 5. (A) Quantification of the his4∷leu2 band, measuring resection at the distal site. The intensity of each band was normalized with respect to loading. The signal present before HO cutting was set to 100%. (B) Quantification of the SSA product band. ▪ rad9Δ; □ rad9Δ GAL-SIC1; • dot1Δ; ○ dot1Δ GAL-SIC1; ♦ WT; ⋄ WT GAL-SIC1.
Dot1 and Rad9 control DNA processing at uncapped telomeres
The results reported so far suggest that a complex containing methylated H3 and Rad9 on damaged DNA inhibits ssDNA accumulation at DSBs. Consistent with this interpretation, earlier studies showed that Rad9 inhibits the accumulation of ssDNA at uncapped telomeres (Lydall and Weinert, 1995; Zubko et al, 2004). To assess the role of methylated H3-K79, and its relationship with Rad9, at uncapped telomeres, we analysed both DNA processing and checkpoint activation in a cdc13-1 mutant background. At temperatures higher than 26°C, cdc13-1 cells accumulate ssDNA and block cell division at the G2/M checkpoint (Garvik et al, 1995).
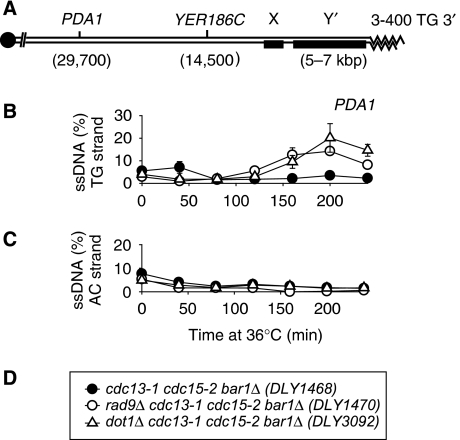
We first examined whether, as found above at DSBs, Dot1 inhibited resection at uncapped telomeres. We used QAOS (quantitative amplification of ssDNA) (Booth et al, 2001; Zubko et al, 2006) to measure the accumulation of ssDNA in synchronous cultures of dot1Δ cdc13-1 strains. A bar1Δ mutation was present in strains to ensure efficient G1 cell cycle arrest with alpha factor and a cdc15-2 mutation was present to ensure that checkpoint-deficient cells did not initiate more than one round of DNA replication during the course of an experiment because at 36°C cdc15-2 mutants arrest in late anaphase (Zubko et al, 2006). We measured ssDNA at the PDA1 locus, which lies about 30 kb away from the end of ChV-R (Figure 3A), because ssDNA does not accumulate at this locus in RAD+ strains but does accumulate if Rad9 function is compromised (Figure 3B). As shown in Figure 3B, dot1Δ cdc13-1 cells, similarly to rad9Δ cdc13-1 strains, had significantly increased levels of ssDNA at PDA1, relative to cdc13-1 strains suggesting that Dot1, similar to Rad9, protects subtelomeric DNA from nucleolytic degradation in cdc13-1 mutants.
Figure 3.
Dot1 protects subtelomeric DNA from degradation in cdc13-1 mutants. (A) Schematic of ChrVR telomere. (B) Accumulation of ssDNA on the TG strand at PDA1. (C) Accumulation of ssDNA on the AC strand at PDA1. (D) Key to (B, C). In (B, C), ssDNA was measured by QAOS. A single representative experiment is shown with error bars indicating the error of the mean of three independent measurements of the same DNA samples, in most cases the error bars are small and are hidden by the symbols.
Dot1 affects checkpoint activation in response to telomere uncapping
As Dot1, the H3-K79 methylase, is important for activating Rad9 in response to DSBs (Giannattasio et al, 2005; Wysocki et al, 2005; Toh et al, 2006), we tested the checkpoint role of Dot1 after telomere uncapping. Impairing checkpoint pathways, for example rad9Δ, partially suppresses the temperature sensitivity of cdc13-1 strains (Weinert et al, 1994; Zubko et al, 2004). Figure 4A shows that Dot1 inhibits growth of cells with uncapped telomeres because dot1Δ cdc13-1 strains grow better than cdc13-1 strains at 26.5°C. We note that rad9Δ cdc13-1 strains grow better than dot1Δ cdc13-1 strains. To test whether Dot1 growth inhibition of cdc13-1 cells is mediated by the interaction of Rad9-Tudor domain with H3-K79me, we analysed the effect of the rad9Y798Q mutation. Even though we routinely observed that rad9Y798Q cdc13-1 strains grew better than dot1Δ cdc13-1 strains, rad9Y798Q cdc13-1 mutants behave most similarly to dot1Δ cdc13-1 cells and grow better than cdc13-1 cells but less well than cdc13-1 rad9Δ cells (Figure 4A). This can be explained if the rad9Y798Q point mutation affects the structure of Rad9 or its interaction with other proteins. Moreover, loss of DOT1 causes redistribution of the SIR factors and this could also influence the vitality of cdc13-1 cells. The epistatic relationships between these mutations are shown in Supplementary Figure 3. Taken together, these data suggest that the role of Dot1 in responding to telomere uncapping is mediated by the Tudor domain of Rad9. However, our finding that rad9Y798Q cdc13-1 and dot1Δ cdc13-1 mutants do not grow as well as cdc13-1 rad9Δ strains, at semi-permissive temperature, suggests the existence of a Rad9-dependent mechanism acting independently of the H3-K79me/Rad9 Tudor domain at uncapped telomeres.
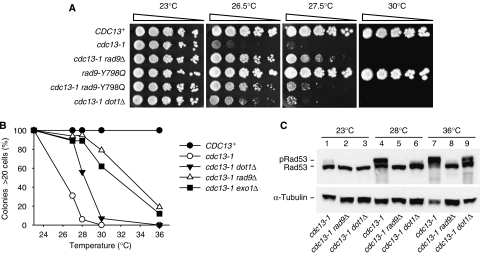
Figure 4.
Dot1 contributes to checkpoint-dependent arrest of cdc13-1 mutants. (A) Five-fold dilutions of strains with indicated phenotypes were spotted onto YEPD plates and grown for 3 days at the temperatures shown. Strains were CDC13+ (DLY640), cdc13-1 (DLY1195), cdc13-1 rad9Δ (DLY1256), rad9-Y798Q (YFL502), cdc13-1 rad9-Y798Q (DLY3083) and cdc13-1 dot1Δ (DLY2880). (B) Single cells were spread onto agar plates and colony formation of CDC13+ (DLY640), cdc13-1 (DLY1195), cdc13-1 dot1Δ (DLY2881) and cdc13-1 rad9Δ (DLY1256) cells was monitored after 20 h at temperatures shown (see Supplementary Figure 4). Number of colonies containing more than 20 cells is plotted against temperature. (C) Cultures of cdc13-1 (DLY1195), cdc13-1 rad9Δ (DLY1256) and cdc13-1 dot1Δ (DLY2881) were grown at the indicated temperatures for 5 h and Rad53 levels in protein extracts were immunoblotted with anti-Rad53 antibodies or anti-tubulin antibodies, as a loading control.
Our observations suggest a role for H3-K79 methylation in checkpoint activation following telomere uncapping. To address this directly, single cells were monitored for their ability to form colonies at different temperatures (Figure 4B, and Supplementary Figure 4). Figure 4B plots the fraction of colonies that contain more than 20 cells for checkpoint and nuclease-deficient cdc13-1 strains over a range of temperatures. The higher the temperature the smaller the colony size for both checkpoint-proficient and -deficient cells. At temperatures higher than 26°C, checkpoint proficient cdc13-1 cells divide slowly and form smaller colonies than checkpoint- or nuclease-deficient strains (Figure 4B). At high levels of telomere uncapping, for example, 30°C and 36°C, the colonies of dot1Δ cdc13-1 strains are of similar size as those of cdc13-1. At lower levels of uncapping, for example, 27°C and 28°C, dot1Δ cdc13-1 cells form larger colonies than cdc13-1 strains (Figure 4B). However, dot1Δ cdc13-1 colonies are smaller than cdc13-1 rad9Δ cells at 27°C (Supplementary Figure 4). This, and the spots tests shown in Figure 4A, suggest that Dot1 has a partial function in checkpoint activation, a function that is only detectable at low levels of DNA damage. Consistent with this interpretation, we saw complete and efficient cell cycle arrest of dot1Δ cdc13-1 mutants in the first cycle after G1 release in the ssDNA measurement experiment shown in Figure 3 (data not shown).
The role of Dot1 in checkpoint activation at intermediate temperatures was confirmed by measuring phosphorylation of Rad53. At 23°C, telomeres in cdc13-1 strains are capped and Rad53 is largely hypo-phosphorylated (Figure 4C, lanes 1–3). In response to low levels of telomere uncapping (28°C), Rad53 is hyper-phosphorylated in a Dot1- and Rad9-dependent manner (Figure 4C, lanes 4–6). However, at higher levels of telomere uncapping (36°C) a significant fraction of Rad53 is hyper-phosphorylated in dot1Δ cdc13-1 strains (Figure 4C, lane 9). These observations suggest that at low levels of telomere uncapping, Dot1 has a more important function in Rad9-dependent checkpoint activation that it does at higher levels of uncapping, when presumably alternative mechanisms ensure that Rad9 is activated.
Loss of Rad9 or Dot1 partially bypasses the requirement for CDK1 in DSB resection
The molecular mechanisms controlling DNA end resection at DSBs and uncapped telomeres are poorly understood. Recent reports showed that CDK1 activity is necessary to obtain effective resection, both at DSBs and uncapped telomeres (Aylon et al, 2004; Ira et al, 2004; Vodenicharov and Wellinger, 2006). Indeed, inhibition of CDK1, by overexpression of the inhibitor Sic1, leads to a notable reduction of 5′–3′ processing of DSB ends (Aylon et al, 2004; Ira et al, 2004, and Figure 5B). To understand the relationships between the Rad9-dependent and the CDK1-dependent mechanisms that regulate resection, we analysed the processing of a DSB in rad9Δ and dot1Δ cells after inhibition of CDK1 activity. We used a yeast strain where a site-specific DSB can be repaired by single-strand annealing (SSA) between two short regions of homology flanking the cut site (Figure 5A) (Sugawara et al, 2000; Vaze et al, 2002). We chose this system for analysis of resection because in this context we can follow simultaneously processing at sites close and far away from the DSB; moreover, we can monitor the appearance of the repair product, which being dependent upon the generation of ssDNA is also a measure of the velocity of resection. At various times after induction of the HO cut and after the concomitant induction of CDK1 inhibitor Sic1, we monitored both DNA end processing and DSB repair by SSA, through Southern blot analysis. Our data show that loss of RAD9 or DOT1 accelerates the appearance of the band corresponding to the repaired chromosome (the SSA product in Figure 5B); the repair product is clearly detectable 5 h after HO induction in WT cells, whereas it is already visible at the 3 h time point in the rad9Δ strain (Figures 5B and 6B). This confirms that absence of RAD9 stimulates DSB resection and SSA and the results with dot1Δ argue that this effect partially depends upon H3-K79 methylation. Interestingly, whereas SIC1 overexpression almost completely inhibits the generation of the SSA product in a WT background, loss of RAD9 or DOT1 significantly reduces the effect due to the inhibition of CDK1 (Figures 5B and 6B). This may suggest that the CDK1-dependent stimulation of resection could exert a function by overcoming the inhibitory effect of Rad9 bound to H3-K79me.
As SSA-based DNA repair depends upon annealing of two complementary ssDNA, faster SSA product generation in this experiment agrees with the faster resection observed in the experiments described above (Figures 1 and 2), but a contribution by a more efficient recombination mechanism could not be excluded. To this aim, we tested the effect of RAD9 deletion in the absence of the critical recombination protein Rad52. SSA is reduced >95% in the absence of Rad52. Figure 5C shows that resection in rad52Δ rad9Δ double mutant cells is accelerated, compared with rad52Δ cells. This is also confirmed by measuring resection of the his4∷leu2 fragment in Figure 5B (see quantifications in Figure 6A) and by monitoring resection of the HO-cut fragment in a yeast strain where the break cannot be repaired by SSA (Supplementary Figure 5). These results suggest that loss of RAD9 most likely accelerates SSA DNA repair through a faster accumulation of ssDNA.
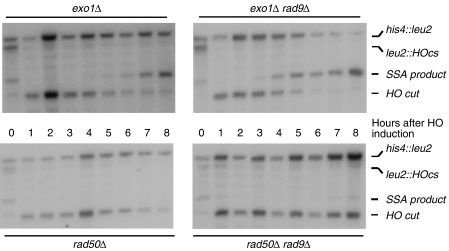
The identity of all the nucleases resecting DSB ends and uncapped telomeres has not yet been fully elucidated, but previous reports implicated the MRX complex and Exo1 in this process (Llorente and Symington, 2004; Harrison and Haber, 2006) as well as undefined nucleases, ExoX, ExoY (Zubko et al, 2004; Harrison and Haber, 2006). To help determine how Rad9 regulates resection at DSBs, we examined cells lacking Exo1 or Rad50. Figure 7 shows that loss of Rad9 causes an accelerated resection in the absence of Exo1, where the SSA product appears at the 3 h time point in a rad9Δ background and at the 6 h time point in a RAD9 background. These results suggest that Exo1 is not required for the rapid rates of resection observed in rad9Δ strains. On the other hand, if RAD50 is missing, there is no strong effect of the loss of RAD9, suggesting that Rad9 inhibits, at least in part, a RAD50-dependent nuclease (Figure 7). Similar conclusions can be drawn analysing resection at the distal site (Supplementary Figure 6).
Figure 7.
Fast resection in the absence of Rad9 depends upon RAD50. SSA was analysed in exo1Δ (Y28), exo1Δ rad9Δ (YFL802), rad50Δ (YFL827) and rad50Δ rad9Δ (YFL809) using Southern blotting as described in Figure 5.
Discussion
DNA damage checkpoint pathways are powerful intracellular signal transduction cascades that, following genomic lesions, inhibit cell cycle progression and coordinate DNA replication, repair and recombination with cell division. DNA damage checkpoint pathways are understood to be based on kinase-dependent signal transduction cascades stimulated by damaged DNA (Longhese et al, 1998; Lydall and Whitehall, 2005); in this context, Rad9, the first checkpoint protein to be identified, has been assigned a mediator function, necessary to link the upstream kinase, Mec1, with activation of the downstream kinases Rad53 and Chk1 (Gilbert et al, 2001; Blankley and Lydall, 2004; Sweeney et al, 2005).
Rad9 has recently been shown to bind to methylated K79 residue of histone H3 through the Rad9 Tudor domain and this interaction is important for the checkpoint role of Rad9 (Giannattasio et al, 2005; Wysocki et al, 2005). The finding that other Tudor domain checkpoint mediator proteins bind methylated histones suggests functional evolutionary conservation of the interactions between checkpoint proteins and modified histones; in fact, mammalian 53BP1 and S. pombe Crb2p interact with methylated H4-K20 (Sanders et al, 2004; Botuyan et al, 2006; Du et al, 2006). We show here that Dot1, the H3-K79 methylase, and the Tudor domain of Rad9, also have a negative feedback function in the checkpoint response to DSBs, where they inhibit processing of DSB ends and activation of the Mec1 kinase. By analysing the extent of phosphorylation of Ddc2, the most proximal target of Mec1, after induction of DSBs, we observed that loss of methylation of H3-K79 leads to more rapid activation of Mec1 and to an increase in the level of phosphorylated species. Methylation of histone H3 in nucleosomes, similar to histone tail acetylation, could alter chromatin structure directly or influence susceptibility to nucleases, affecting Mec1 activation. On the other hand, Dot1-dependent methylation of H3 could help in the recruitment of other factors, such as Rad9, that may influence Mec1 activity. Indeed, a mutation affecting Rad9 Tudor domain or a deletion of RAD9, also caused hyper-activation of Mec1, strongly suggesting that H3-K79 interaction with Rad9 is important to limit Mec1 activation. Moreover, although both the Rad9 Tudor domain and Dot1 contribute to the checkpoint response at low levels of telomere uncapping, they are not necessary at high levels of telomere uncapping.
Much of what we know on the mechanisms involved in activation of Mec1 derives from studying the response to site-specific DSBs. The DNA ends generated by the cleavage are processed by nucleolytic activities, which leave long tails of ssDNA (see Harrison and Haber, 2006 for a review). RPA-covered ssDNA appears to be a prerequisite for Mec1 activation (Zou and Elledge, 2003; Zou et al, 2003). Therefore, we evaluated the effect of Dot1 and Rad9 on the amount of ssDNA generated after a site-specific DSB. Our data show that various genetic manipulations expected to cause loss of Rad9 binding to histone H3 lead to an increase in the speed of resection both at DSBs and uncapped telomeres, suggesting that chromatin-bound Rad9 could represent a functional or physical barrier to exonucleolytic processing of DSBs and uncapped telomeres.
How does the Rad9 H3-K79 interaction affect ssDNA accumulation at telomeres and DSBs? The question is difficult to address at this stage because important details about the mechanisms by which ssDNA is generated are lacking. For example, the nucleases responsible for generating ssDNA have not been fully defined yet, although there are roles for Exo1, MRX and other nuclease activities (Llorente and Symington, 2004; Zubko et al, 2004; Harrison and Haber, 2006). Moreover, the requirement for CDK1 activity in DSB processing suggests that CDK1 may target a protein involved in resection (Aylon et al, 2004; Ira et al, 2004). In S. pombe the Rad9 orthologue, Crb2, mediates the effect of CDK1 on DSB repair (Caspari et al, 2002). Moreover, phosphorylation by CDK1 allows Crb2 to bind close to DSBs through the interaction with Cut5, the orthologue of Dpb11, and this suffices for activating the DNA damage response to DSB (Du et al, 2006). To better define the mechanisms controlling nucleolytic processing of DSBs, we investigated the effect of CDK1 inhibition on resection in the absence of RAD9. Our data show that deletion of RAD9 makes resection much less sensitive to CDK1 inhibition; a similar, albeit reduced, effect is detected in the absence of Dot1, suggesting that binding to methylated H3-K79 is a critical regulatory process. A likely explanation for this result is that one major requirement for CDK1 in DSB processing relies on the removal of Rad9-dependent inhibition of resection, even though a direct effect on some nuclease or other factors cannot be excluded. It is interesting that Rad9 has been shown to be phosphorylated by CDK1 (Ubersax et al, 2003; Grenon et al, 2007) and this may modulate chromatin accessibility by nucleases.
Intriguingly, the inhibitory effect of Rad9 on resection is most evident when monitoring the disappearance of a DNA fragment far away from a DSB or uncapped telomere, compared with regions closer to the primary lesion (Supplementary Figure 7, and Zubko et al, 2004). These observations, together with the residual inhibition by GAL-SIC1 in rad9Δ cells (Figure 5B), suggest that a different CDK1 target could be involved in controlling resection initiation, whereas Rad9 may be limiting the speed and amount of DNA processing. This hypothesis would be consistent with recent findings, reporting an important role for CtIP in the control of DSB resection (Limbo et al, 2007; Sartori et al, 2007).
Further attempts to define the mechanism confirm that Exo1- and Rad50-dependent nucleases participate in resecting DSBs and suggest that the observed Rad9-dependent inhibition affects in part or in whole RAD50-dependent nuclease activity. In contrast, at uncapped telomeres MRX is not responsible for generating ssDNA, this function can be ascribed to unidentified nucleases (ExoX, ExoY) (Foster et al, 2006). Importantly, high levels of ssDNA accumulation after telomere uncapping in exo1Δ rad9Δ double mutants indicate that Rad9-dependent inhibition of ssDNA production may be a general aspect of the DNA damage response (Zubko et al, 2004). Notably, the only transcriptional change caused by deletion of DOT1, and therefore induced by loss of H3-K79 methylation, is overexpression of Y′ repeats, which are found at the telomeres (van Leeuwen et al, 2002). It has been postulated that this is because, in the absence of H3-K79 methylation, the Sir2 histone deacetylase moves from its normal location at telomeres to spread more evenly around the genome. This movement correlates with a loss of the heterochromatic status at telomeres and may be responsible, in part, for the enhanced resection at uncapped telomeres in dot1 and rad9 mutants.
Figure 8 summarizes possible mechanisms by which the Rad9 H3-K79 interaction may affect ssDNA accumulation at DSBs and telomeres: Rad9, when bound to chromatin, may represent a direct structural impediment to nuclease activity or it may promote the formation of a chromatin structure that inhibits exonucleolytic processing of DNA. This could be, in part or completely, due to the interaction of the Tudor domain of Rad9 with methylated H3. But other interactions between Rad9 and chromatin may also contribute to inhibiting nuclease activity; for example, the BRCT domain of Rad9 may interact with phosphorylated checkpoint proteins, or histones, at sites of DNA damage and inhibit nuclease activity (see Du et al, 2006; Hammet et al, 2007). Consistent with the idea of Rad9 affecting nuclease activity by more than one chromatin interaction, yeast strains defective in H3-K79 methylation are not as defective at inhibiting resection as rad9Δ mutants.
Figure 8.
Potential mechanisms for the role of Rad9 in resection inhibition in response to DSBs and uncapped telomeres. Rad9 bound to the methylated H3-K79 interferes with the action of the nuclease(s) or generates a non permissive chromatin configuration.
The eukaryotic DNA damage response requires the coordinated interactions between many molecular players, including damaged DNA, checkpoint proteins, chromatin, chromatin modifiers, double-stranded DNA, ssDNA, RPA, clamps, clamp loaders and kinases. Consequently, DNA damage responses are powerful intracellular pathways, which are potentially harmful for the cells; in fact, they may cause inappropriate cell cycle arrest and amplification of DNA damage if their regulation is lost. Previous work showed that chromatin modification is necessary for generating high levels of ssDNA (van Attikum et al, 2004). We now report the opposite role for a chromatin modification. Our findings suggest that interactions between the checkpoint protein Rad9 and methylated histone H3 inhibits ssDNA accumulation at DSBs and in response to uncapped telomeres. Tudor domains are conserved across evolution, as are methylated histone residues and we propose that Tudor domain/histone interactions may regulate resection also in other eukaryotic cell types.
Materials and methods
Strains and plasmids
Strains are listed in Supplementary Table 1. YFL399, YFL504 and YFL419 were derived from JKM179. To construct strains, standard genetic procedures of transformation and tetrad analysis were followed (Adams et al, 1998). YFL502 and YFL504 were obtained by integration of EcoRI-digested plasmid pFL37.1 at the RAD9 locus. Pop-out events were selected on FOA plates. The Y798Q mutation in Rad9 Tudor domain was checked by PCR. Y31, Y20, Y293, YMV037, YMV038, YFL736, YFL738, YFL827, YFL809, Y28 and YFL802 derive from YMV80. Deletions and tag fusions were generated by the one-step PCR system (Longtine et al, 1998). Serial dilution and maximum permissive temperature analysis were performed as described (Maringele and Lydall, 2002).
Plasmid pFL36.1 was obtained by cloning a RAD9-3HA fragment into XhoI-NotI-digested pRS306. pFL37.1 was obtained introducing the Tudor domain mutation by site-specific mutagenesis in pFL36.1.
Microcolony assays
Cells were inoculated into 2 ml YEPD, grown overnight at 23°C until they reached saturation. The next morning, cells were sonicated briefly and plated. Plates were incubated at the indicated temperature for 20 h, photographed using a Leica DC 300F microscope and the number of cells present in 20 colonies was counted/estimated. The experiment was repeated four times, and a representative experiment is shown.
G1 block and treatment with genotoxic agents
Cells were grown in YEPD medium at 28°C to a 5 × 106 cells/ml and arrested with α-factor (10 μg/ml). Arrested cells (30 ml (untreated)) were mock treated and the rest of the culture was treated with zeocine (50 μg/ml). Treated cells (30 ml) were spun and resuspended in 20% trichloroacetic acid (TCA) for protein extract preparation at 0, 5, 10, 15 and 20 min after treatment. Cell cycle profiles were analysed by standard flow cytometry.
SDS–PAGE and western blot
TCA protein extract was prepared (Muzi-Falconi et al, 1993) and separated by SDS–PAGE. Western blotting was performed with anti-Rad53, anti-HA (12CA5) or anti-tubulin antibodies using standard techniques. Quantification was obtained with a Typhoon after incubation with fluorescent secondary antibodies.
Analysis of Rad53 phosphorylation levels in cdc13-1 cells
Saturated cultures grown at 23°C were diluted to 8 × 106 cells/ml and allowed to double to 1.6 × 107 cells before incubation at 28°C or 36°C for a further 5 h. A control culture was grown in parallel at 23°C. Cells were harvested, washed in H2O and proteins were extracted with 10% TCA and solubilized in SDS–PAGE sample buffer (Blankley and Lydall, 2004). Protein samples were analysed by immunoblotting with anti-Rad53 antibody (DL58, kind gift from D Durocher).
ssDNA measurements at telomeres
ssDNA was isolated from cultures and quantified using the QAOS assay (Booth et al, 2001) as recently described (Zubko et al, 2006).
Measurement of DNA resection and SSA at DSBs
Cells grown in YEP lactate 3% medium at 28°C to a concentration of 5 × 106 cells/ml were arrested with nocodazole (20 μg/ml). A DSB was produced by adding 2% galactose and inducing the expression of the HO endonuclease. The maintenance of the arrest was confirmed by FACS analysis and monitoring of nuclear division. Genomic DNA was isolated at intervals, and the loss of the 5′ ends of the HO-cleaved MAT locus was determined by Southern blotting (Lee et al, 1998; Vaze et al, 2002; Clerici et al, 2005). All the experiments have been repeated at least three times. In the corresponding figures, one representative example is shown with its quantification.
Supplementary Material
Supplementary Figures
Acknowledgments
C Santocanale, D Durocher, K Gull, D Stern, M Clerici and MP Longhese are acknowledged for materials and reagents. M Giannattasio, F Puddu and members of DL's lab are thanked for discussions. This work was supported by grants from AIRC, Fondazione Cariplo, the European Union FP6 Integrated Project DNA repair and MIUR (to MM-F and PP). DL and VS were supported by the Wellcome Trust (075294) and CRUK (C23629/A7951). The financial support of Telethon-Italy (grant no. GGP030406 to MM-F) and research support from NIH grants GM20056 and GM61766 to JEH are gratefully acknowledged.
References
- Adams A, Kaiser C, Cold Spring Harbor Laboratory (1998) Methods in Yeast Genetics: a Cold Spring Harbor Laboratory Course Manual. Plainview, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Aylon Y, Liefshitz B, Kupiec M (2004) The CDK regulates repair of double-strand breaks by homologous recombination during the cell cycle. EMBO J 23: 4868–4875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blankley RT, Lydall D (2004) A domain of Rad9 specifically required for activation of Chk1 in budding yeast. J Cell Sci 117: 601–608 [DOI] [PubMed] [Google Scholar]
- Booth C, Griffith E, Brady G, Lydall D (2001) Quantitative amplification of single-stranded DNA (QAOS) demonstrates that cdc13-1 mutants generate ssDNA in a telomere to centromere direction. Nucleic Acids Res 29: 4414–4422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botuyan MV, Lee J, Ward IM, Kim JE, Thompson JR, Chen J, Mer G (2006) Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell 127: 1361–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspari T, Murray JM, Carr AM (2002) Cdc2-cyclin B kinase activity links Crb2 and Rqh1-topoisomerase III. Genes Dev 16: 1195–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerici M, Mantiero D, Lucchini G, Longhese MP (2005) The Saccharomyces cerevisiae Sae2 protein promotes resection and bridging of double strand break ends. J Biol Chem 280: 38631–38638 [DOI] [PubMed] [Google Scholar]
- Clerici M, Mantiero D, Lucchini G, Longhese MP (2006) The Saccharomyces cerevisiae Sae2 protein negatively regulates DNA damage checkpoint signalling. EMBO Rep 7: 212–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du LL, Nakamura TM, Russell P (2006) Histone modification-dependent and -independent pathways for recruitment of checkpoint protein Crb2 to double-strand breaks. Genes Dev 20: 1583–1596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elledge SJ (1996) Cell cycle checkpoints: preventing an identity crisis. Science 274: 1664–1672 [DOI] [PubMed] [Google Scholar]
- Foster SS, Zubko MK, Guillard S, Lydall D (2006) MRX protects telomeric DNA at uncapped telomeres of budding yeast cdc13-1 mutants. DNA Repair (Amst) 5: 840–851 [DOI] [PubMed] [Google Scholar]
- Gardner R, Putnam CW, Weinert T (1999) RAD53, DUN1 and PDS1 define two parallel G2/M checkpoint pathways in budding yeast. EMBO J 18: 3173–3185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garvik B, Carson M, Hartwell L (1995) Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol Cell Biol 15: 6128–6138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannattasio M, Lazzaro F, Plevani P, Muzi-Falconi M (2005) The DNA damage checkpoint response requires histone H2B ubiquitination by Rad6-Bre1 and H3 methylation by Dot1. J Biol Chem 280: 9879–9886 [DOI] [PubMed] [Google Scholar]
- Gilbert CS, Green CM, Lowndes NF (2001) Budding yeast Rad9 is an ATP-dependent Rad53 activating machine. Mol Cell 8: 129–136 [DOI] [PubMed] [Google Scholar]
- Grenon M, Costelloe T, Jimeno S, O'Shaughnessy A, Fitzgerald J, Zgheib O, Degerth L, Lowndes NF (2007) Docking onto chromatin via the Saccharomyces cerevisiae Rad9 Tudor domain. Yeast 24: 105–119 [DOI] [PubMed] [Google Scholar]
- Hammet A, Magill C, Helerhorst J, Jackson SP (2007) Rad9 BRCT domain interaction with phosphorylated H2AX regulates the G1 checkpoint in budding yeast. EMBO Rep 8: 851–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison JC, Haber JE (2006) Surviving the breakup: the DNA damage checkpoint. Annu Rev Genet 40: 209–235 [DOI] [PubMed] [Google Scholar]
- Huyen Y, Zgheib O, Ditullio RA Jr, Gorgoulis VG, Zacharatos P, Petty TJ, Sheston EA, Mellert HS, Stavridi ES, Halazonetis TD (2004) Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature 432: 406–411 [DOI] [PubMed] [Google Scholar]
- Ira G, Pellicioli A, Balijja A, Wang X, Fiorani S, Carotenuto W, Liberi G, Bressan D, Wan L, Hollingsworth NM, Haber JE, Foiani M (2004) DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature 431: 1011–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov EL, Sugawara N, White CI, Fabre F, Haber JE (1994) Mutations in XRS2 and RAD50 delay but do not prevent mating-type switching in Saccharomyces cerevisiae. Mol Cell Biol 14: 3414–3425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornbluth S, Smythe C, Newport JW (1992) In vitro cell cycle arrest induced by using artificial DNA templates. Mol Cell Biol 12: 3216–3223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SE, Bressan DA, Petrini JH, Haber JE (2002) Complementation between N-terminal Saccharomyces cerevisiae mre11 alleles in DNA repair and telomere length maintenance. DNA Repair (Amst) 1: 27–40 [DOI] [PubMed] [Google Scholar]
- Lee SE, Moore JK, Holmes A, Umezu K, Kolodner RD, Haber JE (1998) Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell 94: 399–409 [DOI] [PubMed] [Google Scholar]
- Limbo O, Chahwan C, Yamada Y, de Bruin RA, Wittenberg C, Russell P (2007) Ctp1 is a cell-cycle-regulated protein that functions with Mre11 complex to control double-strand break repair by homologous recombination. Mol Cell 28: 134–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby M, Barlow JH, Burgess RC, Rothstein R (2004) Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell 118: 699–713 [DOI] [PubMed] [Google Scholar]
- Llorente B, Symington LS (2004) The Mre11 nuclease is not required for 5′ to 3′ resection at multiple HO-induced double-strand breaks. Mol Cell Biol 24: 9682–9694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longhese MP, Foiani M, Muzi-Falconi M, Lucchini G, Plevani P (1998) DNA damage checkpoint in budding yeast. EMBO J 17: 5525–5528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961 [DOI] [PubMed] [Google Scholar]
- Lydall D, Nikolsky Y, Bishop DK, Weinert T (1996) A meiotic recombination checkpoint controlled by mitotic checkpoint genes. Nature 383: 840–843 [DOI] [PubMed] [Google Scholar]
- Lydall D, Weinert T (1995) Yeast checkpoint genes in DNA damage processing: implications for repair and arrest. Science 270: 1488–1491 [DOI] [PubMed] [Google Scholar]
- Lydall D, Whitehall S (2005) Chromatin and the DNA damage response. DNA Repair (Amst) 4: 1195–1207 [DOI] [PubMed] [Google Scholar]
- Majka J, Niedziela-Majka A, Burgers PM (2006) The checkpoint clamp activates Mec1 kinase during initiation of the DNA damage checkpoint. Mol Cell 24: 891–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maringele L, Lydall D (2002) EXO1-dependent single-stranded DNA at telomeres activates subsets of DNA damage and spindle checkpoint pathways in budding yeast yku70Delta mutants. Genes Dev 16: 1919–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo J, Toczyski D (2002) A unified view of the DNA-damage checkpoint. Curr Opin Cell Biol 14: 237–245 [DOI] [PubMed] [Google Scholar]
- Moreau S, Ferguson JR, Symington LS (1999) The nuclease activity of Mre11 is required for meiosis but not for mating type switching, end joining, or telomere maintenance. Mol Cell Biol 19: 556–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzi-Falconi M, Piseri A, Ferrari M, Lucchini G, Plevani P, Foiani M (1993) De novo synthesis of budding yeast DNA polymerase alpha and POL1 transcription at the G1/S boundary are not required for entrance into S phase. Proc Natl Acad Sci USA 90: 10519–10523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyberg KA, Michelson RJ, Putnam CW, Weinert TA (2002) Toward maintaining the genome: DNA damage and replication checkpoints. Annu Rev Genet 36: 617–656 [DOI] [PubMed] [Google Scholar]
- Paciotti V, Clerici M, Lucchini G, Longhese MP (2000) The checkpoint protein Ddc2, functionally related to S. pombe Rad26, interacts with Mec1 and is regulated by Mec1-dependent phosphorylation in budding yeast. Genes Dev 14: 2046–2059 [PMC free article] [PubMed] [Google Scholar]
- Rouse J, Jackson SP (2000) LCD1: an essential gene involved in checkpoint control and regulation of the MEC1 signalling pathway in Saccharomyces cerevisiae. EMBO J 19: 5801–5812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouse J, Jackson SP (2002) Interfaces between the detection, signaling, and repair of DNA damage. Science 297: 547–551 [DOI] [PubMed] [Google Scholar]
- Sanchez Y, Desany BA, Jones WJ, Liu Q, Wang B, Elledge SJ (1996) Regulation of RAD53 by the ATM-like kinases MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science 271: 357–360 [DOI] [PubMed] [Google Scholar]
- Sanders SL, Portoso M, Mata J, Bahler J, Allshire RC, Kouzarides T (2004) Methylation of histone H4 lysine 20 controls recruitment of Crb2 to sites of DNA damage. Cell 119: 603–614 [DOI] [PubMed] [Google Scholar]
- Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek Baer R, Lukas J, Jackson SP (2007) Human CtIP promotes DNA end resection. Nature 450: 509–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shroff R, Arbel-Eden A, Pilch D, Ira G, Bonner WM, Petrini JH, Haber JE, Lichten M (2004) Distribution and dynamics of chromatin modification induced by a defined DNA double-strand break. Curr Biol 14: 1703–1711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara N, Ira G, Haber JE (2000) DNA length dependence of the single-strand annealing pathway and the role of Saccharomyces cerevisiae RAD59 in double-strand break repair. Mol Cell Biol 20: 5300–5309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney FD, Yang F, Chi A, Shabanowitz J, Hunt DF, Durocher D (2005) Saccharomyces cerevisiae Rad9 acts as a Mec1 adaptor to allow Rad53 activation. Curr Biol 15: 1364–1375 [DOI] [PubMed] [Google Scholar]
- Toh GW, O'Shaughnessy AM, Jimeno S, Dobbie IM, Grenon M, Maffini S, O'Rorke A, Lowndes NF (2006) Histone H2A phosphorylation and H3 methylation are required for a novel Rad9 DSB repair function following checkpoint activation. DNA Repair (Amst) 5: 693–703 [DOI] [PubMed] [Google Scholar]
- Ubersax JA, Woodbury EL, Quang PN, Paraz M, Blethrow JD, Shah K, Shokat KM, Morgan DO (2003) Targets of the cyclin-dependent kinase CDK1. Nature 425: 859–864 [DOI] [PubMed] [Google Scholar]
- Usui T, Ogawa H, Petrini JH (2001) A DNA damage response pathway controlled by Tel1 and the Mre11 complex. Mol Cell 7: 1255–1266 [DOI] [PubMed] [Google Scholar]
- van Attikum H, Fritsch O, Hohn B, Gasser SM (2004) Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell 119: 777–788 [DOI] [PubMed] [Google Scholar]
- van Leeuwen F, Gafken PR, Gottschling DE (2002) Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell 109: 745–756 [DOI] [PubMed] [Google Scholar]
- Vaze MB, Pellicioli A, Lee SE, Ira G, Liberi G, Arbel-Eden A, Foiani M, Haber JE (2002) Recovery from checkpoint-mediated arrest after repair of a double-strand break requires Srs2 helicase. Mol Cell 10: 373–385 [DOI] [PubMed] [Google Scholar]
- Vodenicharov MD, Wellinger RJ (2006) DNA degradation at unprotected telomeres in yeast is regulated by the CDK1 (Cdc28/Clb) cell-cycle kinase. Mol Cell 24: 127–137 [DOI] [PubMed] [Google Scholar]
- Wakayama T, Kondo T, Ando S, Matsumoto K, Sugimoto K (2001) Pie1, a protein interacting with Mec1, controls cell growth and checkpoint responses in Saccharomyces cerevisiae. Mol Cell Biol 21: 755–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert TA, Kiser GL, Hartwell LH (1994) Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes Dev 8: 652–665 [DOI] [PubMed] [Google Scholar]
- White CI, Haber JE (1990) Intermediates of recombination during mating type switching in Saccharomyces cerevisiae. EMBO J 9: 663–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocki R, Javaheri A, Allard S, Sha F, Cote J, Kron SJ (2005) Role of Dot1-dependent histone H3 methylation in G1 and S phase DNA damage checkpoint functions of Rad9. Mol Cell Biol 25: 8430–8443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L, Elledge SJ (2003) Sensing DNA damage through ATRIP recognition of RPA–ssDNA complexes. Science 300: 1542–1548 [DOI] [PubMed] [Google Scholar]
- Zou L, Liu D, Elledge SJ (2003) Replication protein A-mediated recruitment and activation of Rad17 complexes. Proc Natl Acad Sci USA 100: 13827–13832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubko MK, Guillard S, Lydall D (2004) Exo1 and Rad24 differentially regulate generation of ssDNA at telomeres of Saccharomyces cerevisiae cdc13-1 mutants. Genetics 168: 103–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubko MK, Maringele L, Foster SS, Lydall D (2006) Detecting repair intermediates in vivo: effects of DNA damage response genes on single-stranded DNA accumulation at uncapped telomeres in budding yeast. Methods Enzymol 409: 285–300 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures