Abstract
Many viruses have evolved mechanisms for evading the host immune system by synthesizing proteins that interfere with the normal immune response. The poxviruses are among the most accomplished at deceiving their hosts’ immune systems. The nucleotide sequence of the genome of the human cutaneous poxvirus, molluscum contagiosum virus (MCV) type 1, was recently reported to contain a region that resembles a human chemokine. We have cloned and expressed the chemokine-like genes from MCV type 1 and the closely related MCV type 2 to determine a potential role for these proteins in the viral life cycle. In monocyte chemotaxis assays, the viral proteins have no chemotactic activity but both viral proteins block the chemotactic response to the human chemokine, macrophage inflammatory protein (MIP)-1α. Like MIP-1α, both viral proteins also inhibit the growth of human hematopoietic progenitor cells, but the viral proteins are more potent in this activity than the human chemokine. These viral chemokines antagonize the chemotactic activity of human chemokines and have an inhibitory effect on human hematopoietic progenitor cells. We hypothesize that the inhibition of chemotaxis is an immune evasion function of these proteins during molluscum contagiosum virus infection. The significance of hematopoietic progenitor cell inhibition in viral pathogenesis is uncertain.
Poxviruses are notorious for their ability to evade the host’s immune system by both active and passive mechanisms (1). Since the eradication of smallpox, the only poxvirus that naturally infects humans is molluscum contagiosum virus (MCV). MCV causes benign proliferative lesions of the skin in normal and immunocompromised individuals. Persons with acquired immune deficiency syndrome (AIDS) sometimes get extensive MCV infections that can be disfiguring (2, 3). There are at least two types of MCV based on restriction endonuclease cleavage patterns of viral DNA (4–6). Some epidemiologic studies have been reported, but there is no consensus on anatomic or patient group distribution of the two MCV types. Although MCV cannot be propagated in tissue culture, we and others have shown recently that MCV can be grown in human tissue implanted into immunodeficient mice (7, 8).
Recently, much attention has been focused on chemokines, small proteins produced by lymphocytes and a variety of other cells. Chemokines were originally identified by their ability to attract inflammatory cells, but they have since been shown to have a variety of additional activities (for review, see ref. 9). In addition to chemotaxis, some chemokines have been shown to cause decreased growth of hematopoietic stem and early subsets of myeloid progenitor cells (10–13) and some have been shown to block entry of HIV into lymphocytes (14, 15). Roles for chemokines in a variety of allergic and autoimmune diseases have also been postulated, although direct evidence is limited (reviewed in ref. 16).
It has been assumed that MCV, like other poxviruses, has immune evasion functions. The recent report of the nucleotide sequence of the MCV genome (17) has permitted identification of potential candidate viral proteins that may be involved in escape from the host immune system. One such putative viral protein is designated MC148R. This open reading frame potentially encodes a 104-aa protein with significant homology to β chemokines such as macrophage inflammatory protein (MIP)-1β. The putative MC148R protein contains a 5-aa deletion at the functionally important amino terminus of the mature protein, relative to MIP-1β. Only three other viruses, human herpes virus 6 (18), human herpes virus 8 (19, 20), and murine cytomegalovirus (21) are known to encode chemokine analogs. These herpesvirus chemokine analogs do not contain deletions, although there is only limited information about their ability to function as chemokine agonists. The pathogenic role (if any) that these viral proteins play is unknown. Our goal in the experiments reported here was to characterize the MCV-encoded chemokine analogs at the functional level in an effort to understand more about their potential role in the viral life cycle. In addition, we wished to explore the possibility that the functional differences between these truncated viral proteins and the human analogs could be of more general interest in understanding chemokine structure and function.
MATERIALS AND METHODS
Extraction of DNA.
Specimens were collected by curettage from patients who were being treated for multiple molluscum lesions. The extracted lesion contents were minced with a scalpel blade and incubated overnight in a solution containing protease K (200 μg/ml) and 1% SDS. The mixture was then extracted with phenol and chloroform:isoamyl alcohol (24:1), and the DNA was precipitated with ethanol. Determination of MCV type was done by restriction endonuclease digestion of viral DNA as previously described (8).
Cloning.
PCR primers specific for the MC148R gene were synthesized on Applied Biosystems oligonucleotide synthesizer. The 5′ primer used was CCAAGAGGACGAGCTAA, and the 3′ primer was GTTCTCCTGCTCCATGACGA. PCRs were carried out in the Perkin–Elmer GeneAmp PCR System 9600 using 35 cycles of three temperatures of 94°C, 55°C, and 72°C for 1 min each. The product obtained was sequenced in bulk on the Applied Biosystems Prism sequencer to confirm the predicted nucleotide sequence. This PCR product was then cloned into the cloning vector, pCR3.1 (Invitrogen) using standard procedures specified by the manufacturer. Sequencing of the clone was carried out on the Applied Biosystems Prism sequencer to confirm that the cloned sequence was identical to the sequence of the bulk PCR product. The cloned insert was then excised by BamHI/SmaI double digestion and ligated into the baculovirus expression vector, pVL 1392 (PharMingen).
Protein Production and Purification.
The pVL vector with the MC148R gene was cotransfected with baculovirus DNA containing a lethal deletion into Sf9 insect cells according to the manufacturer’s directions. Resulting recombinant virus was amplified twice to high titer, and Sf9 cells were infected in spinner flasks using protein-free medium. Medium was then clarified by centrifugation at 10,000 × g and passed over a heparin–Sepharose column (Pharmacia). Heparin-binding proteins were eluted with a gradient of KCl. Only the initial fraction contained the protein of interest as judged by SDS/PAGE. High molecular weight contaminants were removed by centrifugation through a Centricon-30 column (Amicon), and the fraction passing through the membrane was collected and used in functional assays. Protein was quantified using the Bradford assay (Pierce).
Chemotaxis Assays.
Twenty milliliters of venous blood was collected from healthy volunteers in 10 cc heparinized tubes. Blood was mixed 1:1 with PBS, and 10 cc of Histopaque (Sigma) was underlayed. Tubes were centrifuged at 400 × g for 25 min. The band of mononuclear cells at the interface was collected and washed twice with PBS. Cells were resuspended in Dulbecco’s minimal essential medium (GIBCO/BRL) with antibiotics at a density of 106/ml. Bovine serum albumin was added to cells as described previously (22).
Chemotaxis was assayed using a modification of the method described by Boyden (23, 24). One hundred microliters of the mononuclear cell suspension was added to each Transwell insert (Costar). Dulbecco’s minimal essential medium with antibiotics, 0.2 mg/ml BSA, and the protein to be tested were placed in the bottom wells of a 24-well plate. All samples were done in triplicate, and at least two experiments for each protein were performed. Inserts were placed in the 24-well plate and incubated at 37°C in 5% CO2 for 90 min. At the end of the incubation, inserts were removed, the top of the insert was scraped with a rubber policeman to remove adherent cells, and the filters were stained with Wright–Giemsa stain. Cells adherent to the bottom of the filter were counted under three high-power fields, and cells that had migrated to the bottom of the 24-well plate were counted under three high-power fields as well. These two values were summed and expressed as total migrating cells (23). For presentation of results, the total migrating cells in each experiment were expressed as a percentage of the positive control for that experiment (10 ng of MIP-1α obtained from Collaborative Biomedical Products, Bedford, MA) and the results for different experiments were averaged.
Hematopoietic Progenitor Assays.
Hematopoietic colony formation assays were performed as described previously (11–13). Volunteer human bone marrow cells were collected from donors after obtaining informed consent. Low-density human marrow cells at 5 × 104/ml were plated in 1% methylcellulose in Iscove’s modified medium supplemented with 30% fetal calf serum, recombinant human (rHu) erythropoietin (1 unit/ml), rHu interleukin 3 (IL-3) (100 unit/ml), and rHu steel factor (50 ng/ml) for colony-forming unit granulocyte/macrophage, colony-forming unit granulocyte/erythrocyte/macrophage/megakaryocyte, or burst-forming unit erythrocyte analysis. These assays respectively allow quantitation of effects on early subsets of granulocyte-macrophage, multipotential, and erythroid progenitor cells. Various concentrations of partially purified MC148R protein (as described in the Results section) were compared with purified MIP-1α (Collaborative Biomedical Products). Cultures were incubated at 37°C in 5% CO2 and low (5%) oxygen tension for 14 days and then scored using an inverted microscope. Three plates were scored per data point per experiment and expressed as a percent of control for that experiment.
RESULTS
Amplification and Cloning of the MC148R Region from MCV Types 1 and 2.
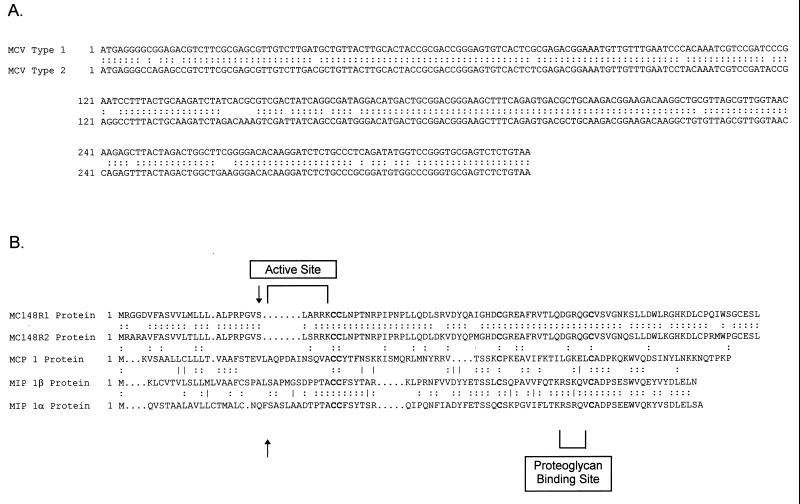
Based on the published sequence of MCV 1 DNA, primers flanking the MC148R region were designed and used with either MCV 1 or MCV 2 DNA that had been extracted from clinical specimens. The same primer pair was used for both MCV 1 and MCV 2. The resulting DNA fragment produced by PCR synthesis was ≈400 base pairs in size for both types. This fragment was then cloned into the pCR3.1 vector and amplified in One Shot cells (Invitrogen). As shown in Fig. 1, sequencing of these cloned PCR products showed that the MCV 1 sequence was identical to the previously published sequence (17). The MCV 2 sequence had 89% homology to the type 1 sequence. Amino acid comparisons showed 87% homology of the complete sequences and 86% when the putative leader sequence was removed. The four conserved cysteine residues characteristic of C—C chemokines appear to be conserved in both of the MCV 1 and MCV 2 proteins. Based on sequence comparisons with macrophage chemoattractant protein-1 (MCP-1), the first 24 amino acids compose a leader sequence and the next 5 amino acids constitute the active site. This contrasts with MCP-1, which has 10 amino acids at the amino-terminal active site. MCV 1 and MCV 2 have very similar amino acid compositions at the putative active site, except for a serine in MCV 2 instead of an alanine at position 26. Both proposed signal peptides share conserved regions near the putative cleavage sites.
Figure 1.
(A) Complete nucleotide sequences of MC148R1 and MC148R2 regions, showing sequence alignments. Note that the sequence of MC148R1 is identical to that previously reported (17). For the MC148R2 sequence, both the bulk PCR product and a clone made from the PCR product were sequenced and were identical. The MC148R2 sequence has been deposited in GenBank (accession number U96749). (B) Predicted protein sequences of MC148R1 and MC148R2, showing sequence alignments with MIP-1β and MCP-1. Putative active sites are highlighted, and the predicted signal peptide cleavage sites are indicated by vertical arrows.
Expression and Purification of the MC148R Proteins.
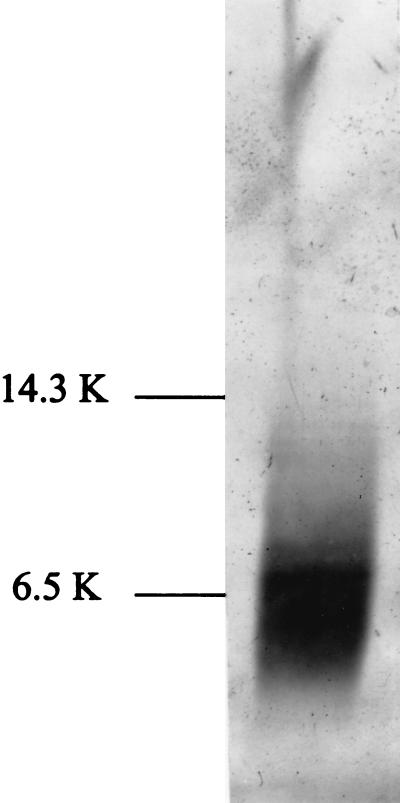
The PCR products containing the MC148R regions from each MCV type were excised from pCR3.1 and subcloned into the baculovirus expression vector, pVL1392. Sf9 insect cells were cotransfected with the pVL-MC1 or pVL-MC2 plasmid and baculovirus DNA containing a lethal deletion to make recombinant baculovirus. Sf9 cells infected with recombinant virus were then grown in protein-free medium. Because all known chemokines bind strongly to heparin, the clarified medium was then passed over a heparin–Sepharose column and eluted with a KCl salt gradient. The fraction that contained a discernible band of 8 kDa (the predicted molecular mass of the mature protein) on SDS/PAGE was then partially purified by removing high molecular mass contaminants. After partial purification, each preparation consisted of a prominent 8-kDa band on both Coomassie-stained and silver-stained gels. The only other band that was visible on the Coomassie-stained gel was a faint band at ≈4 kDa. A silver-stained gel of the partially purified protein is shown in Fig. 2. A prominent band at ≈8 kDa is seen in addition to a few minor bands. The protein concentration was measured, and the partially purified preparations of the two viral chemokines were used in functional assays. Medium from cells that had been transfected with the pVL 1392 vector alone was similarly processed and showed no band in the 8-kDa size range (data not shown). The preparation from cells transfected with vector alone was used as a control for the functional assays.
Figure 2.
SDS/polyacrylamide gel of recombinant baculovirus supernatants after partial purification, showing expression of the viral chemokine-like protein. The gel was stained using Silver Stain Plus (Bio-Rad). Lane 1 shows typical results for the partial purification of MC148R2. (Some silver-staining artifact is present at the upper portion of the gel.)
Chemotactic Activity of MC148R1 and MC148R2 Proteins.
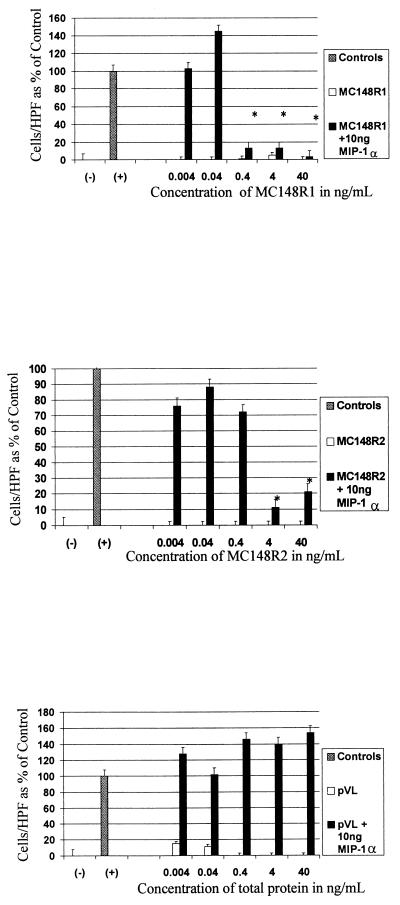
Because all chemokines are chemotactic by definition, the next series of experiments were designed to test whether the synthesized proteins were chemotactic for monocytes. The partially purified MC148R1 and MC148R2 proteins were assayed for chemotactic activity by placing the test protein in medium, which was separated from human peripheral blood mononuclear cells by a membrane. The number of cells migrating through the membrane in response to the protein were counted and expressed as a percentage of controls. As shown in Fig. 3, neither viral protein had significant chemotactic activity over a 10,000-fold concentration range compared with MIP-1α, the positive control. However, when either viral protein was present in the same well as MIP-1α, there was a marked inhibition of the expected chemotactic response. This effect was dependent on the concentration of the viral protein present. At very low concentrations of the viral protein, the number of migrating cells was statistically no different from the number migrating to MIP-1α alone. At higher concentrations of the viral protein, the number of migrating cells became no different than the number of cells migrating to the negative control. The recombinant viral proteins had no effect on cell viability. All experimental and control wells had 95–97% viability (measured by trypan blue exclusion) after incubation under assay conditions. The protein preparation from cells transfected with vector alone gave no chemotactic response and did not show any ability to block the chemotactic response of MIP-1α.
Figure 3.
Results of chemotaxis assays. Negative controls consisted of media alone, and positive controls consisted of media and 10 ng of MIP-1α. Concentrations of the viral proteins are for the partially purified protein preparations and include protein impurities in the total concentration. ∗, P < 0.05 when compared with positive controls; HPF, high power field. Results are expressed as percentages of the positive control above baseline and represent the mean of at least two independent experiments. (A) Results for MC148R1 shown as protein alone and in competition with MIP-1α. (B) Results for MC148R2. (C) Results for pVL vector control protein preparation.. Note that in each panel the viral protein (or vector control) alone bars are at or near the baseline.
Effect of MC148R1 and MC148R2 on Hematopoietic Progenitor Cell Growth.
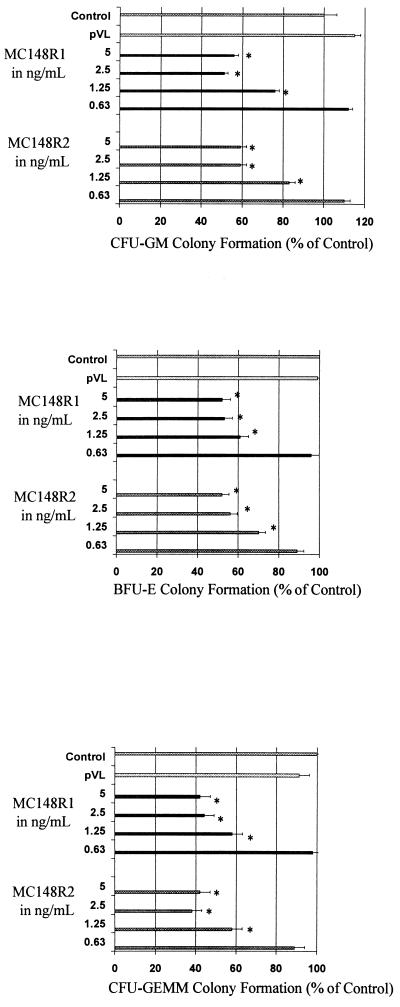
Chemokines have been shown to inhibit colony formation of hematopoietic progenitor cells (11–13). To determine whether the viral proteins shared this property, mononuclear cell fractions of bone marrow cells obtained from normal human donors were cultured in the presence of erythropoietin, interleukin 3, and steel factor (early-acting cytokines that allow detection of early, more immature subsets of myeloid progenitor cells (11–13)) in the presence or absence of the viral chemokine-like proteins. Both viral proteins inhibited colony formation of myeloid, erythroid, and multipotential progenitor cells by nearly 50% at concentrations as low as 1.25 to 2.5 ng/ml (Fig. 4). The plateau concentrations of MIP-1α that inhibit colony formation are 50–100 ng/ml, with less inhibition noted at 25 ng/ml and no inhibition at 1–10 ng/ml (11–13). Thus, the inhibitory concentrations of the viral proteins that show inhibition in this assay are lower than these concentrations of MIP-α. The degree of inhibition (between 40 and 60%) was equal to that seen in the same experiments using 100 ng/ml of MIP-1α (data not shown). Preparations from transfections using the pVL 1392 vector alone had no effect on cell growth or differentiation in this assay.
Figure 4.
Effects of the viral chemokine-like proteins on the proliferation of hematopoietic progenitor cells. (A) Effects on colony-forming unit granulocyte/macrophage colony formation. Control colonies consist of media alone. MC148R1 and MC148R2 were added to media and serially diluted. ∗, P < 0.01 when compared with control. (B) Effects on burst-forming unit erythrocyte colony formation. ∗, P < 0.01. (C) Effects on colony-forming unit granulocyte/erythrocyte/macrophage/megakaryocytecolony formation. ∗, P < 0.01. The results shown are the average of three separate experiments in which the control number of colony-forming unit granulocyte/macrophage, burst-forming unit erythrocyte, and colony-forming unit granulocyte/erythrocyte/macrophage/megakaryocyte colonies of individual experiments, respectively, ranged from 44 ± 7 to 69 ± 3, 24 ± 5 to 84 ± 7, and 16 ± 3 to 18 ± 3.
DISCUSSION
Like many other poxviruses, molluscum contagiosum probably uses a variety of methods to escape the immune system. We have demonstrated evidence of a novel mechanism for escape from immune system surveillance, namely, blocking the signal for migration of effector cells to the site of infection. We have also shown that MCV 2 contains a chemokine-like gene that is very similar to the MCV 1 counterpart at the DNA and protein level. Both viral chemokine-like proteins tested are quite potent in their ability to block chemotaxis by MIP-1α, a property necessary for a successful competitive inhibitor. Our experiments may underestimate the potency of the inhibitory effects because our viral chemokine preparations were only partially purified. Neither viral protein exhibits chemotactic properties even at high concentrations. Lesions in vivo are characterized by a lack of inflammatory cell infiltrates, consistent with a mechanism for immune system escape (25). Neither T cells nor natural killer cells are found at the base of molluscum lesions (26). Furthermore, lesions undergoing spontaneous resolution often show mononuclear cell infiltrates, confirming that these types of cells are critical in the immune response to molluscum contagiosum (27). Other studies have shown that mature molluscum lesions contain the C-X-C chemokines GROα and IL-8 within the molluscum body itself that are released on decomposition (28). It is possible that the immune response is blocked by chemokine antagonists early in infection, but later the physical barrier of the molluscum body prevents detection by the immune system. Further studies on the patterns of expression of these viral chemokines are needed to clarify this issue.
The deletion present at the amino terminus of MC148R with respect to MIP-1β and MCP-1 may explain the biologic behavior observed in these experiments. Previous reports on structure–function relationships of the C—C chemokines showed that for MCP-1, the first five amino acids constitute the “activation site,” and the next five constitute the “binding site” (29). The deletion of the first 5 amino acids in the viral proteins would then result in a protein that has only the binding site and would, thus, not be expected to activate the receptor. Alternatively, a proposed internal site, named the proteoglycan binding site, may explain the lack of chemotaxis. One mutational analysis of MIP-1α showed that placing neutral or acidic amino acids at this site resulted in the loss of chemotaxis while maintaining other chemokine functions (30). Curiously, both viral proteins contain neutral or acidic amino acids at this site. This change probably accounts for the observation that the viral chemokines elute from heparin–Sepharose at a lower salt concentration (0.25 M) than that required to elute other chemokines such as MIP-1α (0.6 M; ref. 31). The relative importance of this internal site versus the amino-terminal deletion of the viral proteins is not clear.
The inhibitory effect of chemokines on hematopoietic stem and progenitor cells has been studied intensely. A number of studies suggest a regulatory role for chemokines in hematopoiesis (reviewed in ref. 13). It is not known if these viral proteins reach the bone marrow during natural infection, so the effect on hematopoietic cells may not be relevant to molluscum contagiosum virus pathogenesis. However, the fact that the viral proteins do inhibit hematopoiesis suggests that they are able to activate at least some chemokine receptors and could have clinically relevant effects on other cell types. At present, it is not clear which portion of the chemokine protein is responsible for inhibiting hematopoiesis. Consistent with previous studies is the fact that modification of the proteoglycan binding site does not have an effect on inhibition of hematopoiesis (30). The inhibition of hematopoiesis would then be due to another active site that is retained in the viral proteins.
This study reports a viral chemokine that blocks the chemotactic activity of natural chemokines. This novel mechanism of immune escape underscores the importance of chemokines in natural infection. These viral chemokines also have been shown to block growth of hematopoietic progenitor cells. Further studies are needed to delineate the exact role of these viral proteins in vivo. The potential for use of these viral chemokines in clinical settings to block or reduce inflammatory reactions is uncertain. However, these viral proteins have properties that make them candidates for therapeutic applications.
Acknowledgments
We thank Scott Cooper for excellent technical assistance with the hematopoietic progenitor cell assays. This work was supported in part by Public Health Service Grants R01 HL56416 and R01 HL54037 and by a project in P01 HL53586 from the National Institutes of Health to H.E.B.
ABBREVIATIONS
- MCV
molluscum contagiosum virus
- MIP
macrophage inflammatory protein
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. U96749).
References
- 1.Pickup D J. Infect Agents Dis. 1994;3:116–127. [PubMed] [Google Scholar]
- 2.Smith K J, Skelton H G, Yeager J, Ledsky R, McCarthy W, Baxter D, Wagner K F. J Am Acad Dermatol. 1994;31:746–754. doi: 10.1016/s0190-9622(94)70236-5. [DOI] [PubMed] [Google Scholar]
- 3.Ray M C, Gately L E., III Infect Dis Clinics North Am. 1994;8:583–605. [PubMed] [Google Scholar]
- 4.Darai G, Reisner H, Scholz J, Schnitzler P, Lorbacher de Ruiz H. J Med Virol. 1986;18:29–39. doi: 10.1002/jmv.1890180105. [DOI] [PubMed] [Google Scholar]
- 5.Porter C D, Blake N W, Archard L C, Muhlemann M F, Rosedale N, Cream J J. Br J Dermatol. 1989;120:37–41. doi: 10.1111/j.1365-2133.1989.tb07763.x. [DOI] [PubMed] [Google Scholar]
- 6.Thompson C H, De Zwart-Steffe R T, Biggs I M. J Med Virol. 1990;32:1–9. doi: 10.1002/jmv.1890320102. [DOI] [PubMed] [Google Scholar]
- 7.Buller R M L, Burnett J, Chen W, Kreider J. Virology. 1995;213:655–659. doi: 10.1006/viro.1995.0037. [DOI] [PubMed] [Google Scholar]
- 8.Fife K H, Whitfeld M, Faust H, Goheen M P, Bryan J, Brown D R. Virology. 1996;226:95–101. doi: 10.1006/viro.1996.0631. [DOI] [PubMed] [Google Scholar]
- 9.Baggiolini M, DeWald B, Mossier B. Adv Immunol. 1994;55:97–179. [PubMed] [Google Scholar]
- 10.Graham G, Wright E, Hewick R, Wolpe S, Wilkie N, Donaldson D, Lorimore S, Pragnell I. Nature (London) 1990;344:442–444. doi: 10.1038/344442a0. [DOI] [PubMed] [Google Scholar]
- 11.Broxmeyer H E, Sherry B, Lu L, Cooper S, Oh K O, Tekamp-Olson P, Kwon B S, Cerami A. Blood. 1990;76:1110–1116. [PubMed] [Google Scholar]
- 12.Broxmeyer H E, Sherry B, Cooper S, Lu L, Maze R, Beckmann M P, Cerami A, Ralph P. J Immunol. 1993;150:3448–3458. [PubMed] [Google Scholar]
- 13.Broxmeyer H E, Cooper S, Hague N, Benninger L, Sarris A, Cornetta K, Vadhan-Raj S, Hendrie P, Mantel C. Ann Hematol. 1995;71:235–246. doi: 10.1007/BF01744373. [DOI] [PubMed] [Google Scholar]
- 14.Cocchi F, DeVico A L, Garzino-Demo A, Arya S K, Gallo R C, Lusso P. Science. 1995;270:1811–1815. doi: 10.1126/science.270.5243.1811. [DOI] [PubMed] [Google Scholar]
- 15.Oberlin E, Amara A, Bachelerie F, Bessia C, Virelizier J L, Arenzana-Seisdedos F, Schwartz O, Heard J M, Clark-Lewis I, Legler D F, Loetscher M, Baggiolini M, Mossier B. Nature (London) 1996;382:833–835. doi: 10.1038/382833a0. [DOI] [PubMed] [Google Scholar]
- 16.Mantovani A, Locati M, Allavena P, Sozzani S. Immunobiology. 1996;195:522–549. doi: 10.1016/S0171-2985(96)80020-9. [DOI] [PubMed] [Google Scholar]
- 17.Senkevich T G, Bugert J J, Sisler J R, Koonin E V, Darai G, Moss B. Science. 1996;273:813–816. doi: 10.1126/science.273.5276.813. [DOI] [PubMed] [Google Scholar]
- 18.Gompels U A, Nicholas J, Lawrence G, Jones M, Thompson B J, Martin M E D, Efstathiou S, Craxton M, Macaulay H A. Virology. 1995;209:29–51. doi: 10.1006/viro.1995.1228. [DOI] [PubMed] [Google Scholar]
- 19.Moore P S, Boshoff C, Weiss R A, Chang Y. Science. 1996;274:1739–1744. doi: 10.1126/science.274.5293.1739. [DOI] [PubMed] [Google Scholar]
- 20.Nicholas J, Ruvolo V, Burns W, Sandford G, Wan X, Ciufo D, Hendrickson S, Guo H, Hayward G, Reitz M. Nat Med. 1997;3:287–292. doi: 10.1038/nm0397-287. [DOI] [PubMed] [Google Scholar]
- 21.MacDonald M R, Li X Y, Virgin H W. J Virol. 1997;71:1671–1678. doi: 10.1128/jvi.71.2.1671-1678.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martinet Y, Martinet N, Vignaud J, Plenat F. J Immunol Methods. 1994;174:209–214. doi: 10.1016/0022-1759(94)90024-8. [DOI] [PubMed] [Google Scholar]
- 23.Keller H U, Borel J F, Wilkinson P C, Hess M W, Cottier H. J Immunol Methods. 1972;1:165–168. doi: 10.1016/0022-1759(72)90043-9. [DOI] [PubMed] [Google Scholar]
- 24.Boyden S V. J Exp Med. 1962;115:453–460. doi: 10.1084/jem.115.3.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Birthistle K, Carrington D. J Infect. 1997;34:21–28. doi: 10.1016/s0163-4453(97)80005-9. [DOI] [PubMed] [Google Scholar]
- 26.Viac J, Chardonnet Y. J Cutan Pathol. 1990;17:202–205. doi: 10.1111/j.1600-0560.1990.tb00085.x. [DOI] [PubMed] [Google Scholar]
- 27.Gottlieb S L, Myskowski P L. Int J Dermatol. 1994;33:453–461. doi: 10.1111/j.1365-4362.1994.tb02853.x. [DOI] [PubMed] [Google Scholar]
- 28.Takematsu H, Tagami H. Arch Dermatol Res. 1994;287:102–106. doi: 10.1007/BF00370727. [DOI] [PubMed] [Google Scholar]
- 29.Clark-Lewis I, Kim K, Rajarathnam K, Gong J, Dewald B, Mossier B, Baggiolini M, Sykes B. J Leukocyte Biol. 1995;57:703–711. doi: 10.1002/jlb.57.5.703. [DOI] [PubMed] [Google Scholar]
- 30.Graham G J, Wilkinson P C, Nibbs R J B, Lowe S, Kolset S O, Parker A, Freshney M G, Tsang M L S, Pragnell I B. EMBO J. 1996;15:6506–6515. [PMC free article] [PubMed] [Google Scholar]
- 31.Wolpe S D, Davatelis G E, Sherry B, Beutler B, Hesse D G, Nguyen H T, Moldawer L L, Nathan C F, Lowry S F, Cerami A. J Exp Med. 1988;167:570–581. doi: 10.1084/jem.167.2.570. [DOI] [PMC free article] [PubMed] [Google Scholar]