Abstract
The complex prokaryote, Myxococcus xanthus, undergoes a program of multicellular development when starved for nutrients, culminating in sporulation. M. xanthus makes MglA, a 22-kDa, soluble protein that is required for both multicellular development and gliding motility. MglA is similar in sequence to the Saccharomyces cerevisiae SAR1 protein, a member of the Ras/Rab/Rho superfamily of small eukaryotic GTPases. The SAR1 gene, when integrated into the M. xanthus genome, complements the sporulation defect of a ΔmglA strain. A forward, second-site mutation on the M. xanthus chromosome, rpm, in combination with SAR1, restores fruiting body morphogenesis and gliding motility to a ΔmglA strain. The result that the rpm mutation suppresses the substitution of SAR1 for mglA suggests that Sar1p interacts with other M. xanthus proteins to control the motility-dependent aggregation of cells during development.
Keywords: Myxococcus xanthus, gliding, fruiting body morphogenesis, Sar1p, MglA
Myxococcus xanthus, a Gram-negative soil bacterium, undergoes multicellular development. When starved for nutrients, groups of >105 wild-type M. xanthus cells aggregate and form fruiting bodies containing heat-resistant spores (1). During the starvation-induced differentiation of M. xanthus, like that of the eukaryotic slime mold Dictyostelium discoideum, the interaction of multiple cell types (2) is required to produce cells with different terminally differentiated fates. A subset of developing M. xanthus cells differentiates into spherical spores that are resistant not only to heat, but also to ultraviolet light and desiccation. Other cells remain rod-shaped and form the base and sheath of the fruiting body (3).
Gliding motility plays a critical role in M. xanthus development, because development depends on a critical density of cells achieved by motility-dependent aggregation (4, 5). The genetic basis of gliding motility is complex and involves at least two independent mechanisms, adventurous and social (6, 7). Adventurous gliding enables individual cells to move dependent on interactions of cells with solid surfaces, whereas social gliding enables groups of cells to move dependent on cell-cell interactions (8). Although both mechanisms are required for the normal developmental program, a subset of mutants defective in social gliding can produce heat-resistant spores, and a subset of mutants defective in adventurous gliding can complete the morphogenesis of the fruiting structure (6, 7).
Mutations in only one known gene, mglA, affect both adventurous and social gliding mechanisms (6, 9). MglA is a 22-kDa cytoplasmic protein that is transcribed with MglB, an 18-kDa protein that is predicted to interact with and stabilize MglA (10). In the absence of MglB, M. xanthus makes normal levels of mglA transcript, but reduced levels of MglA protein. Although mglB mutants show reduced colony spreading on growth medium, they exhibit both adventurous and social gliding.
When starved for nutrients, mglA− strains fail to aggregate or bear fruit and produce less than 0.01% the wild-type complement of spores (11). The defect in sporulation is due, in part, to an inability to mediate the C-signal, an extracellular morphogen essential for sporogenesis during multicellular development (12). In contrast, mglB− strains aggregate into small, flat mounds and produce a nearly wild-type complement of heat-resistant spores (10). These results suggest that gliding and sporulation require different amounts of MglA protein, or that MglB is dispensable for sporulation.
The critical need for MglA in motility, sporulation, and morphogenesis suggests that MglA plays a central role in signal transduction in M. xanthus. MglA shares identity with the small molecular mass eukaryotic GTPases (13) such as Ras, Rab, Rho, Arf, and Sar1p (14). Subgroups of GTPases play critical roles in organelle function, cytoskeletal organization, and the regulation of vesicular trafficking of proteins through the exocytic and endocytic pathways (15). The protooncogene, Ras, controls cellular growth by modulating a mitogen-activated protein kinase signal transduction cascade (16–18). Sar1p has a different, essential function. It is required for protein transport from the endoplasmic reticulum to the Golgi in yeast (19) and mammals (20). Sar1p is regulated by the 70,000 Da integral membrane glycoprotein Sec12p, which is proposed to catalyze guanine-nucleotide exchange and recruit Sar1p to a vesicle formation site on the endoplasmic reticulum membrane (21), and by Sec23p, which activates the GTPase activity of Sar1p (22).
To explore the similarity between MglA and its eukaryotic homologs, Ha-RAS and SAR1 were introduced into a strain carrying a deletion of mgl. The yeast GTPase Sar1p is able to complement the sporulation defect of an mgl mutant. An allele of SAR1 with an altered GTP consensus fails to complement the sporulation defect. Complementation of the motility defect requires a second-site mutation in combination with SAR1, suggesting that Sar1p interacts with an M. xanthus protein to control gliding. These results establish the requirement for a GTPase in motility and development of M. xanthus.
MATERIALS AND METHODS
Growth of Bacterial Strains.
M. xanthus strains DK1622 (motile wild-type), DK3619 (motile, Ω1901 TetR), DK3662 (nonmotile, Ω1901 TetRmglA8), DK5208 (csgA::Ω205 TetR), and DK6204 (nonmotile, ΔmglA) used in this study have been described previously (13, 23, 24). M. xanthus strains were grown in CTPM medium (1% casitone/10 mM Tris/4 mM MgSO4/1 mM potassium phosphate, at a final pH of 7.5) at 250 rpm. Magnesium was omitted when tetracycline was added. Solid medium contained 1.5% agar. Phage Mx4 ts-18 ts-27 htf-1 hrm-1 (25) was used to mediate generalized transduction between strains of M. xanthus. Escherichia coli strain JM107 (26) was used as the host for construction and maintenance of plasmid DNA. Where applicable, media were supplemented with the antibiotics kanamycin sulfate (40 μg/ml), ampicillin (100 μg/ml), and/or tetracycline (12.5 μg/ml).
Construction of Plasmids.
Plasmids pPLH477 and pPLH490 were constructed to express SAR1 and Ha-RAS from the mgl promoter. Plasmid pMYY3–1 containing the cDNA clone for SAR1 was obtained from Akihiko Nakano (University of Tokyo, Japan). Plasmid pBR322-c-Ha-RAS was from Fuyuhiko Tamanoi (University of California, Los Angeles). The 0.67-kb SAR1 gene was removed by restriction with NcoI and EcoRI, purified, and ligated with a 4.4-kb EcoRI–NcoI fragment from pPLH325 (13) to produce pPLH477. Ha-RAS was amplified by the PCR using oligonucleotide 5′-AAGCTTCCAATGGCCGAATAC-3′ for the 5′ end of Ha-RAS, which introduces an NcoI site, and 5′-GAATTCGTCGACCTATCA3′ for the 3′ end of Ha-RAS, which introduces a SalI site. The 580-bp Ha-RAS PCR product was gel purified, digested with NcoI and SalI, and ligated with a 4.4-kb SalI–NcoI fragment from pPLH325 to produce pPLH490. The 4.4-kb fragment from plasmid pPLH325 used to construct pPLH477 and pPLH490 contains the ColE1 origin of replication (which does not function in M. xanthus), a kanamycin-resistance determinant, and the mgl promoter, but lacks all of mglB and mglA.
To determine the requirement for GTPase activity of SAR1 in sporulation of M. xanthus, lysine 36, a residue in the G1 domain that is essential for GTPase function in Ras (lysine 16) (27) was changed to threonine by site-directed mutagenesis using the PCR overlap method (28). Plasmid pPLH477 was used as template with oligonucleotides 5′-CAATGTGGTGGTACCGGCATTATCCAA-3′ (SAR1 K36T reverse) and 5′-ATCACAAGCTTAAGAGTCAGGCCCC-3′ (outside 1) to produce product A and 5′-TTGGATAATGCCGGTACCACCACATTG-3′ (SAR1 K36T forward) and 5′-ATAACAATTTCACACAGGAAACA-3′ (outside 2) to produce product B. PCR products A and B were purified, annealed, and used as template with the two outside oligonucleotides to amplify SAR1K36T and the mgl regulatory region. The 940-bp product was purified, digested with HindIII and SalI, and cloned into pBGS18 to produce pAGS101. The presence of a new KpnI site in pAGS101 confirmed that the SAR1 gene carried the K36T mutation.
Plasmids pPLH477, pPLH490, and pAGS101 were introduced into the chromosome of M. xanthus DK6204 (ΔmglA; ref. 13), by selection for homologous recombination between the mgl regulatory region on the plasmid and the homologous region on the chromosome to produce MxH1048, MxH1091, and MxH1116, respectively. Southern analysis (29) was used to confirm that recombinants carry a single, integrated copy of SAR1 (or the Ha-RAS) gene. Chromosomal DNA was prepared as described by Yee and Inouye (30), digested with EcoRI, HindIII, and AscI, separated in 0.7% agarose, and transferred to Nytran. Membranes were probed with 32P-labeled DNA corresponding to mglA from pPLH325, SAR1 from pMYY3–1, and the Ha-RAS PCR product (data not shown).
Immunoblot Analysis.
Anti-Sar1p polyclonal antibody was obtained from Randy Schekman (University of California, Berkeley). Soluble extracts of M. xanthus cells were prepared from strains grown in CTPM broth, lysed by passage through a French pressure cell at 20,000 psi, and centrifuged at 200,000 × g. Aliquots containing about 15 μg of total protein were separated on 12.5% tricine gels (31), transferred to nitrocellulose, and probed with rabbit anti-Sar1p antibody, diluted 1:2000 in Tris-buffered saline, at pH 7.6 with 0.1% Tween-20 (32). Antigen-antibody reactions were visualized with ECL (Amersham) according to manufacturer’s instructions.
Genetic and Molecular Analysis of Strains.
M. xanthus strain MxH1056 is a motile derivative of MxH1048 obtained after UV mutagenesis. Cells of MxH1048 were washed with TPM buffer (which is identical to CTPM medium, except that casitone is omitted) and exposed to ultraviolet light in a Stratalinker (Stratagene) for 2 min. Aliquots of cells were then spotted on CTPM agar and incubated at 33°C. MxH1056 was identified as a flare that moved away from the initial spot of growth. To determine if motility of M. xanthus MxH1056 was due to a mutation in SAR1 or in the mgl promoter, genetic linkage between the KanR determinant that integrated with SAR1 and the motility phenotype was measured. Mx4 phage lysate of MxH1056 was transduced into the nonmotile parent, DK6204, and KanR transductants were selected. Transductants (145) were transferred to CTPM Kan agar, and motility of all 145 was indistinguishable from the parent, MxH1048 (Δmgl SAR1). Chromosomal DNA from MxH1056 was digested with EcoRI and ligated to recover SAR1 and the mgl promoter for sequencing. KanR plasmids were recovered in E. coli and sequenced on a LiCor automated sequencer using commercial M13 forward and reverse primers. The sequence of SAR1 from MxH1056 and the original plasmid pPLH477 is identical to the published sequence of SAR1 (19). These results show that rpm is not linked genetically with SAR1 and that the gain of motility in MxH1056 is not due to a mutation in SAR1.
To determine if SAR1 and rpm both were required for motility, experiments were designed to replace the integrated copy of SAR1 with other mgl alleles or to disrupt the SAR1 gene. Strains DK3619, DK3662, and MxH1095 carry TetR insertions linked with mglA+, mglA8, and ΔmglA, respectively, and were used as donors for introducing mgl alleles by Mx4 transduction into SAR1 backgrounds. MxH1095 was constructed by transduction (DK3619 × DK6204) with myxophage Mx4; its genotype was confirmed by Southern analysis (not shown). KanR recipient strains were MxH1048 (ΔmglA SAR1) and MxH1056 (ΔmglA SAR1 rpm). Transductants were selected on CTP agar containing tetracycline. The TetR marker is about 1–2 kb from the mgl operon, and TetR transductants obtain the linked mgl allele at a frequency >80% (24). Individual TetR transductants then were transferred to plates with tetracycline and kanamycin to determine the number of transductants that carried both markers. Colonies were examined visually for motility.
Upon infection of MxH1056 with phage lysates prepared from the three Ω1901 TetR donor strains, only the mglA8 donor strain gave rise to KanS TetR replacements. To confirm that the replacements carried the mglA8 allele, chromosomal DNA was prepared from eight independent TetR KanS recombinants for restriction fragment length polymorphism analysis. The mglA gene was amplified with oligonucleotides 5′-GCAAAAGCTTAACCACCCTACTTGAGCT-3′ and 5′-ATCACAAGCCTAAGAGTCAGGCCCC-3′ using PCR, and PCR products were purified and restricted with FokI, to confirm the loss of a FokI site caused by the mglA8 mutation. PCR and Southern analysis were used to show the absence of SAR1 in the KanS TetR strains.
Analysis of Motility and Development.
The gliding phenotypes of wild-type and mutant cells were assayed visually, microscopically, and by measuring the spreading areas of 1.5 × 107 cells on 0.3% and 1.5% casitone agar media as described by Shi and Zusman (33). Fruiting body morphogenesis and sporogenesis were compared for mutant ΔmglA SAR1 cells and otherwise isogenic mglA+ and ΔmglA strains. To initiate development, cells were grown in CTPM medium to 5 × 108 cells/ml, concentrated by centrifugation at 12,000 xg for 10 min, suspended in TPM buffer to 5 × 109 cells/ml, spotted (20 μl) on TPM starvation plates and incubated at 33°C. Morphogenesis was monitored microscopically at 6-hr intervals for several days. Events such as rippling, formation of translucent mounds, and progression to dark mounds serve as temporal markers for development, and were monitored visually for 70 hr with a Nikon SMZ-U stereomicroscope. After incubation at 33°C for 5 days, plates were incubated at 50°C for 2 hr to kill undifferentiated cells, and samples were scraped into TPM buffer for spore assays. Samples were sonicated for 5 s at 10% power using a Microson cell disruptor (Heat Systems/Ultrasonics) to uniformly disperse spores, and serial dilutions in TPM buffer were plated on CTPM agar. Colonies formed from germinating spores were counted after incubation at 33°C for 3–4 days.
RESULTS
Eukaryotic GTPases Are Expressed and Stably Maintained in M. xanthus.
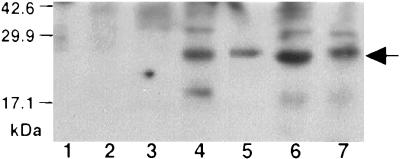
The predicted amino acid sequence of MglA protein shows that it is related to other members of the Ras family of GTPases (13). MglA shares 24% identity and 47% similarity with Sar1p, a member of the Rab subfamily of small GTPases (34) (Fig. 1), and 15% identity (28% similarity) with Ha-Ras (data not shown). To determine if Ha-Ras or Sar1p could complement the defects of a strain with a deletion of mglA, the SAR1 and Ha-RAS genes were introduced in single copy in place of the mglA gene on the M. xanthus genome and expressed under control of the mgl promoter. Immunoblots prepared from extracts of strains expressing SAR1 or Ha-RAS were probed with anti-Sar1p antibody. A 21-kDa protein, indistinguishable from Sar1p in yeast, was detected in M. xanthus stains expressing an integrated copy of SAR1 (Fig. 2), or Ha-RAS (data not shown).
Figure 1.
MglA shares 24% identity with Sar1p. The complete amino acid sequence for MglA and Sar1p is shown. The primary sequence of the 22-kDa MglA contains 195 amino acids, and the 21-kDa Sar1p contains 190 amino acids. Residues that comprise the conserved GTP consensus ae underlined in bold. | indicates identity; ∗ indicates similarity. # indicates the site of the mutation in mglA8. The method of Altschul et al. (44) was used to align the sequences.
Figure 2.
The 22-kDa yeast protein Sar1p is produced in M. xanthus. Immunoblot of extracts from S. cerevisiae and M. xanthus strains was probed with a 1:2,000 dilution of rabbit anti-Sar1p antibody. Lane 1, Bio-Rad Kaleidoscope prestained standards; carbonic anhydrase (42,600); soybean trypsin inhibitor (29,900) and lysozyme (17,100). Lane 2, M. xanthus DK1622 (wild type). Lane 3, M. xanthus DK6204 (ΔmglA). Lane 4, M. xanthus MxH1048 (ΔmglA SAR1). Lane 5, S. cerevisiae. Lane 6, M. xanthus MxH1056 (ΔmglA SAR1 rpm). Lane 7, M. xanthus MxH1116 (ΔmglA SAR1 lys36thr).
Sar1p Can Substitute for One Function of MglA, the Production of Heat-Resistant Spores.
As shown in Table 1, heat-resistant spores are formed by the ΔmglA SAR1 strain, MxH1048, with an efficiency close to that of the wild-type (mglA+) strain and 104-fold higher than that observed for the parental ΔmglA strain. Hence, the SAR1 gene complements the inability of the ΔmglA strain to produce heat-resistant spores. The spores produced by MxH1048 are slightly ovoid, whereas the spores produced by the wild type are spherical. The significance of the spore shape is unknown, although variations in spore shape have been observed with other mutants (35). Although Ha-RAS encodes a GTPase similar in size to MglA, it is not able to complement the sporulation defect of the ΔmglA mutant.
Table 1.
SAR1 rescues the sporulation defect of a ΔmglA strain
| Strain | Genotype | Phenotype*
|
Viable spores, ml | Viable spores†, wild type | |
|---|---|---|---|---|---|
| Motility | Fruiting | ||||
| DK1622 | Wild type | + | + | 5 × 108 | 100.00% |
| DK6204 | ΔmglA | − | − | 5 × 104 | 0.01% |
| MxH1048 | ΔmglA SAR1 | − | − | 4.6 × 108 | 92.00% |
| MxH1116 | Δmgl SAR1 K36T | − | − | 9 × 104 | 0.02% |
| MxH1091 | ΔmglA Ha-RAS | − | − | 2 × 105 | 0.04% |
Assays for motility and morphogenesis are described in Materials and Methods.
Approximately 5-10% of wild-type cells differentiate into heat-resistant spores by day 5.
To determine if the GTPase activity of SAR1 was required to complement the sporulation defect of the Δmgl mutant, a mutant allele of SAR1 was constructed and introduced into the M. xanthus ΔmglA strain. Lysine 36 of SAR1 is in the G1 domain of the GTP consensus and is predicted to make contact with main-chain oxygens of the phosphate groups of GTP (36). Replacement of lysine 36 with threonine did not affect expression or stability of Sar1p in M. xanthus (Fig. 2), but abolished the ability of Sar1p to rescue the sporulation defect of ΔmglA. These data show that the GTPase activity of Sar1p is required for complementation of the sporulation defect.
The SAR1 gene does not complement the defects in motility or fruiting body morphogenesis caused by a ΔmglA mutation even though ΔmglA SAR1 cells can differentiate from rod-shaped vegetative cells into refractile spores. This result shows that gliding motility and the differentiation of heat-resistant spores involve different functions of MglA. Sar1p can complement one, but not both, of these functions.
Sar1p Interacts with Rpm to Complement Motility.
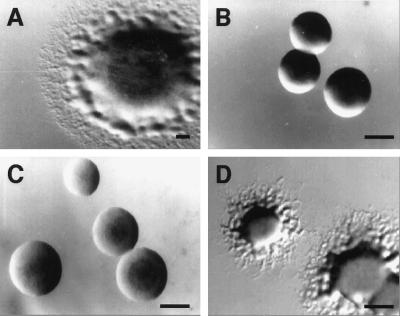
Small GTPases like Sar1p, function by interacting with, and modifying the activities of, other proteins including GTPase-activating proteins, guanine-nucleotide exchange factors, and guanine-nucleotide release factors (15). Sar1p complements the sporulation defect caused by a deletion of the mglA gene, but it does not complement the motility defect of this mutant. The simplest hypothesis to account for this observation is that Sar1p interacts with a protein that normally interacts with MglA to control sporulation, but cannot interact with a protein that normally interacts with MglA to control motility. To test this hypothesis, a mutation that restores interactions between Sar1p and the target of MglA interaction required for gliding motility was selected. The nonmotile Δmgl SAR1 strain, MxH1048, was mutagenized, and a motile derivative was isolated. The spreading ratio for this mutant, designated MxH1056, on 0.3 and 1.5% agar is similar to that of the wild-type strain (ratio = 1.8), which suggests that both adventurous and social modes of gliding have been restored. Microscopic examination of cells at the edges of a colony edges supports this claim. However, because the spreading rate for the motile derivative is only 20% that of the wild-type strain on surfaces of both agar concentrations, MxH1056 is designated Δmgl SAR1 rpm because it restores partial gliding motility (Fig. 3).
Figure 3.
A second-site mutation restores motility to ΔmglA SAR1. Morphology of M. xanthus colonies growing on CTPM 1.5% agar plates. (A) M. xanthus DK1622. (B) ΔmglA (DK6204). (C) ΔmglA SAR1 (MxH1048). (D) ΔmglA SAR1 rpm (MxH1056). Photographs were taken with a Nikon SMZ-U stereomicroscope. (Bar = 3 mm.)
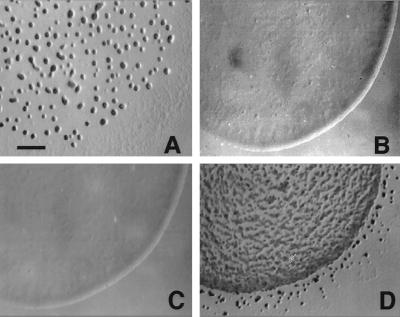
Cells of MxH1056 (Δmgl SAR1 rpm) were starved for nutrients to induce fruiting body formation and sporulation. In contrast with cells of the ΔmglA SAR1 parent, cells of the rpm derivative aggregate to form numerous mounds (Fig. 4D), which are smaller and less erect than wild-type mounds (Fig. 4A). The mounds produced by the rpm derivative mature to a brown color at the same time as the wild-type cells (30 hr), coincident with their production of spherical, refractile, heat-resistant myxospores.
Figure 4.
The motile ΔmglA SAR1 rpm strain forms fruiting aggregates. Morphogenesis of fruiting bodies on TPM starvation medium. (A) M. xanthus DK1622. (B) ΔmglA (DK6204). (C) ΔmglA SAR1 (MxH1048). (D) ΔmglA SAR1 rpm (MxH1056). The parent, ΔmglA SAR1, fails to aggregate (C), although heat-resistant spores are produced. Cells with the ΔmglA mutation (B) fail to aggregate and fail to differentiate into spores. Photographs were taken 48 hr after the onset of development with a Nikon SMZ-U stereomicroscope. (Bar = 50 mm.)
Mutations in M. xanthus that bypass the requirement of MglA for motility have yet to be found. Therefore, the rpm mutation is predicted to be in SAR1 or in a gene encoding a protein that interacts with Sar1p to control motility. To begin to map the rpm mutation, the integrated pmglSAR1 kanamycin-resistant (KanR) plasmid was transduced from the Δmgl SAR1 rpm mutant into mglA+ and ΔmglA genetic backgrounds, and KanR transductants were selected and scored for motility. Transduction of pmglSAR1 into both motile (mglA+) and nonmotile (ΔmglA) genetic backgrounds yielded only nonmotile recombinants (88/88 and 145/145, respectively). Thus, the rpm mutation that confers the gain of motility is not due to a mutation in the integrated pmglSAR1 gene, because it is not linked genetically with the KanR marker adjacent to SAR1.
A series of reciprocal crosses was performed to test whether SAR1 and rpm are required in combination for motility, or if rpm bypasses the requirement for Sar1p (and MglA). If SAR1 and rpm are both required for motility, then replacement of SAR1 with a mutant allele of mgl should yield nonmotile recombinants. In these crosses, myxophage Mx4 was grown on donor cells carrying a TetR Ω1901 insertion linked (>80% cotransduction) with mglA (24). TetR strains that carry Ω1901 linked with mglA+, ΔmglA, and mglA8 were used as donors in these crosses; the KanR recipients were the motile Δmgl SAR1 rpm (MxH1056) strain and its nonmotile parent, Δmgl SAR1 (MxH1048).
As shown in Table 2, replacement of SAR1 with mglA8 abolishes gliding motility and shows that Sar1p is required for gliding. Surprisingly, replacement of the SAR1 gene with mglA+ or ΔmglA occurred only in the parental Δmgl SAR1 strain, and did not occur in the rpm genetic background. Although TetR transductants were obtained with Δmgl SAR1 rpm in crosses using mglA+ and ΔmglA strains as donors, they retain the integrated SAR1 KanR plasmid. These data suggest that either the rpm mutation is synthetic-lethal in combination with the ΔmglA or mglA+ alleles, or that the Δmgl SAR1 rpm strain is a poor recipient for generalized transduction. To exclude the latter possibility, a transducing lysate was grown on M. xanthus strain DK5208 (provided by L. Shimkets, University of Georgia, Athens) which carries the TetR csgA::Ω205 insertion and is not linked with mglA. The numbers of TetR KanR recombinants arising from crosses with Δmgl SAR1 and Δmgl SAR1 rpm recipients were similar, 108 and 79, respectively, indicating that the mutant Δmgl SAR1 rpm strain is a good recipient for generalized transduction.
Table 2.
Disruption of SAR1 in the ΔmglA SAR1 rpm strain abolishes gliding motility
| TetR donor | KanR recipient | TetR transductants*
|
||||
|---|---|---|---|---|---|---|
| Total TetR | KanR
|
KanS
|
||||
| Motile | Nonmotile | Motile | Nonmotile | |||
| Ω1901 mglA+ (motile) | ΔmglA SAR1 | 113 | 0 | 10 | 103 | 0 |
| ΔmglA SAR1 rpm | 63 | 63† | 0 | 0 | 0 | |
| Ω1901 ΔmglA (nonmotile) | ΔmglA SAR1 | 78 | 0 | 11 | 0 | 67 |
| ΔmglA SAR1 rpm | 49 | 49† | 0 | 0 | 0 | |
| Ω1901 mglA8 (nonmotile) | ΔmglA SAR1 | 89 | 0 | 9 | 0 | 80 |
| ΔmglA SAR1 rpm | 44 | 12† | 0 | 0 | 32‡ | |
Transductants initially were selected on plates with tetracycline and subsequently were scored for resistance to kanamycin as described in Materials and Methods.
Motile transductants were identical to the ΔmglA SAR1 rpm recipient and did not carry the donor mglA allele.
DNA from eight transductants was assayed for the presence of the mglA8 and SAR1 genes. In all eight, the mglA8 allele had replaced the integrated copy of SAR1.
The mglA8 strain DK3662 carries a missense mutation predicted to result in a conservative amino acid substitution (Leu-93-Phe) in MglA protein (11). Western immunoblot and ELISA assays using anti-MglA antibody show that DK3662 produces a normal level of the inactive, missense MglA8 protein (unpublished results). The Leu-93-Phe mutation is predicted to make MglA8 more like Sar1p, which may account for the ability of MglA8 to substitute for Sar1p in the rpm background. Nevertheless, recombinants with the genotype Ω1901 (TetR) mglA8 rpm are not motile and fail to produce heat-resistant spores during starvation-induced development. Therefore, the rpm mutation does not suppress the mglA8 defect. When the pmglSAR1 plasmid, pPLH477, is reintroduced and integrated into this host, both motility and sporulation are restored.
DISCUSSION
Multiple types of small GTPases have been found in eukaryotic organisms where they are required for a wide range of cellular functions, including signal transduction, cytoskeletal assembly, and transport. Nearly all members of this expanding family of proteins are essential. MglA is not required for growth, but represents a new member of the GTPase family required for motility and sporulation. GTP-binding proteins have been identified in prokaryotic organisms, including EF-Tu (37), Era (38, 39), FtzZ (40), and Obg (41), but they are larger in size and possess extra domains not found in the Ras-like eukaryotic proteins. To date, the M. xanthus mglA gene encodes the only prokaryotic analog of the lower molecular mass (20–25 kDa) GTPases.
Members of the Ras superfamily have been divided into subgroups based on identity and function. The prototype, Ha-Ras, controls a kinase casade regulating gene expression and cell morphology. Sar1p, a distant relative of Ha-Ras with only 20–25% identity overall with the Ras and Rab families, lacks the C-terminal cysteine for prenylation and the N-terminal glycine for myristylation, and is essential for vesicular transport. MglA is related to each of these proteins, with 15% and 24% identity with Ras and Sar1p, respectively, and has characteristics of each of these subfamilies. A critical glycine residue in the G1 domain of Ras (G12) is conserved in MglA (G21), whereas Sar1p has an aspartate residue in this position (D32). In contrast, both Sar1p and MglA contain a threonine in the G1 domain, in place of the conserved serine residue in the Ras subfamily.
To confirm the requirement of a small GTPase in sporulation and motility of M. xanthus, and to define the subfamily to which MglA belongs, Ha-Ras and Sar1p were substituted for MglA in M. xanthus. The strains expressing Ras and Sar1p produced colonies that resembled the ΔmglA parent on agar and grew at the same rate as the wild-type strain. Hence, neither Ras nor Sar1p complement the motility defect of an mglA mutant. However, when cells expressing Sar1p were induced to develop by starvation, the cells differentiated into refractile, heat-resistant spores. Expression of Sar1p having a defective GTPase, or the more distantly related Ha-Ras, failed to complement the sporulation defect. Hence, complementation of the sporulation defect of a Δmgl mutant requires a functional GTPase.
The ability of Sar1p, but not Ras, to complement may reflect the closer similarity between MglA and Sar1p than between MglA and Ha-Ras. What particular amino acid residues in the sequences of small GTPases determine their specificity of function, by facilitating interactions with other proteins? Clearly, these residues include Leu-93 in MglA, which is near the active (GTP-binding) site of these effectors. The ability of mglA8 to replace SAR1 in the rpm strain may be due to the nature of the mglA8 mutation, which changes leucine-93 to phenylalanine, making MglA8 protein more like Sar1p.
SAR1 in combination with rpm can restore fruiting body morphogenesis and gliding motility to a ΔmglA host. The interaction of the rpm mutation with the mglA locus is allele-specific because the rpm mutation, which is not linked with the mglA locus, is lethal in combination with the ΔmglA and mglA+ alleles, but not the SAR1 or mglA8 alleles. The genetic specificity of this interaction most likely reflects the specificity of interactions between the small GTPase, MglA, and its GTPase-activating and guanine nucleotide exchange partners, because the activity of Sar1p, like Ras, Rab, and other small GTPases, is modulated through its interactions with other effectors. Sar1p function requires the guanine nucleotide exchange factor Sec12p (42) and the GTPase-activating protein Sec23p (43). The ability of Sar1p to function in M. xanthus suggests that functional homologs of these proteins and others may exist in this prokaryote. Work is underway to identify the gene affected by the rpm mutation.
The finding that M. xanthus encodes a small, nonessential GTPase, a defect in which may be complemented by an essential, eukaryotic GTPase, facilitates the genetic analysis of small GTPase structure and function. This demonstrates that a eukaryotic GTPase can complement the defects of a prokaryotic GTPase. The majority of small eukaryotic GTPases are involved in essential cellular functions. Thus, the complex prokaryote M. xanthus provides a system in which mutations that inactivate Sar1p or other GTPases will have a conditional phenotype that is not a conditional-lethal phenotype. Because the small GTPase, MglA, is required for both motility and development, the powerful genetic selections for motility (and for development) may be used to explore the interactions of MglA or its homolog, Sar1p, with other proteins in novel ways.
Acknowledgments
I thank Angela Stassinopoulos for excellent technical assistance, Phil Youderian for helpful discussions, Akihiko Nakano, Fuyuhiko Tamanoi, and Randy Schekman for materials, and Dale Kaiser, Scott Minnich, Akihiko Nakano, Larry Shimkets, and Phil Youderian for critical reading of the manuscript. This work was supported by the National Science Foundation (MCB-9206996) and the National Institutes of Health (GM50962).
ABBREVIATIONS
- CTPM
1% casitone/10 mM Tris/4 mM MgSO4/1 mM potassium phosphate, at a final pH of 7.5
- TPM
CTPM with the casitone omitted
References
- 1.Dworkin M. Microbiol Rev. 1996;60:70–102. doi: 10.1128/mr.60.1.70-102.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Laue B E, Gill R E. J Bacteriol. 1995;177:4089–4096. doi: 10.1128/jb.177.14.4089-4096.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O’Connor K A, Zusman D R. J Bacteriol. 1991;173:3318–3333. doi: 10.1128/jb.173.11.3318-3333.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim S K, Kaiser D. Genes Dev. 1990;4:896–905. doi: 10.1101/gad.4.6.896. [DOI] [PubMed] [Google Scholar]
- 5.Sager B M. Genes Dev. 1996;10:2237–2250. doi: 10.1101/gad.10.18.2237. [DOI] [PubMed] [Google Scholar]
- 6.Hodgkin J, Kaiser D. Mol Gen Genet. 1979;171:177–191. [Google Scholar]
- 7.MacNeil S D, Calara F, Hartzell P L. Mol Microbiol. 1994;14:61–71. doi: 10.1111/j.1365-2958.1994.tb01267.x. [DOI] [PubMed] [Google Scholar]
- 8.Hartzell P L, Youderian P. Arch Microbiol. 1995;164:309–323. doi: 10.1007/BF02529977. [DOI] [PubMed] [Google Scholar]
- 9.Stephens K, Kaiser D. Mol Gen Genet. 1987;207:256–266. [Google Scholar]
- 10.Hartzell P, Kaiser D. J Bacteriol. 1991;173:7625–7635. doi: 10.1128/jb.173.23.7625-7635.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stephens K, Hartzell P, Kaiser D. J Bacteriol. 1989;171:819–830. doi: 10.1128/jb.171.2.819-830.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kroos L, Hartzell P, Stephens K, Kaiser D. Genes Dev. 1988;117:252–266. doi: 10.1101/gad.2.12a.1677. [DOI] [PubMed] [Google Scholar]
- 13.Hartzell P, Kaiser D. J Bacteriol. 1991;173:7615–7624. doi: 10.1128/jb.173.23.7615-7624.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Balch W E. Trends Biochem Sci. 1990;15:473–477. doi: 10.1016/0968-0004(90)90301-q. [DOI] [PubMed] [Google Scholar]
- 15.Macara I G, Lounsbury K M, Richards S A, McKiernan C, Bar-Sago D. FASEB J. 1996;10:625–630. doi: 10.1096/fasebj.10.5.8621061. [DOI] [PubMed] [Google Scholar]
- 16.Hall A. Science. 1994;264:1416–1414. [Google Scholar]
- 17.Leevers S J, Paterson H F, Marshall C J. Nature (London) 1994;369:411–414. doi: 10.1038/369411a0. [DOI] [PubMed] [Google Scholar]
- 18.Stokoe D, Macdonald S G, Cadwallader K, Symons M, Hancock J F. Science. 1994;264:1463–1467. doi: 10.1126/science.7811320. [DOI] [PubMed] [Google Scholar]
- 19.Nakano A, Muramatsu M. J Cell Biol. 1989;109:2677–2691. doi: 10.1083/jcb.109.6.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shen K A, Hammond C M, Moore H-P. FEBS Lett. 1993;335:380–385. doi: 10.1016/0014-5793(93)80423-r. [DOI] [PubMed] [Google Scholar]
- 21.Barlowe C, Schekman R. Nature (London) 1993;365:347–349. doi: 10.1038/365347a0. [DOI] [PubMed] [Google Scholar]
- 22.Yoshihisa T, Barlowe C, Schekman R. Science. 1993;259:1466–1468. doi: 10.1126/science.8451644. [DOI] [PubMed] [Google Scholar]
- 23.Kaiser D. Proc Natl Acad Sci USA. 1979;76:5952–5956. doi: 10.1073/pnas.76.11.5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sodergren E, Kaiser D. J Mol Biol. 1983;167:295–310. doi: 10.1016/s0022-2836(83)80337-4. [DOI] [PubMed] [Google Scholar]
- 25.Campos J M, Geisselsoder J, Zusman D R. J Mol Biol. 1978;119:167–178. doi: 10.1016/0022-2836(78)90431-x. [DOI] [PubMed] [Google Scholar]
- 26.Yanisch-Perron C, Viera J, Messing J. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 27.Sigal I S, Gibbs J B, D’Alonzo J S, Temeles G L, Wolanski B S, Socher S H, Scolnick E M. Proc Natl Acad Sci USA. 1986;83:952–956. doi: 10.1073/pnas.83.4.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook J, Fitsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 30.Yee T, Inouye M. J Bacteriol. 1981;145:1257–1265. doi: 10.1128/jb.145.3.1257-1265.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schagger H, VonJagow G. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 32.Harlow E, Lane D. Antibodies: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. [Google Scholar]
- 33.Shi W, Zusman D R. Proc Natl Acad Sci USA. 1993;90:3378–3382. doi: 10.1073/pnas.90.8.3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bourne H R, Sanders D A, McCormick F. Nature (London) 1991;349:117–127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- 35.MacNeil S D, Mouzeyan A, Hartzell P L. Mol Microbiol. 1994;14:785–796. doi: 10.1111/j.1365-2958.1994.tb01315.x. [DOI] [PubMed] [Google Scholar]
- 36.Pai E F, Kabsch W, Krengel U, Holmes K C, John J, Wittinghofer A. Nature (London) 1989;341:209–214. doi: 10.1038/341209a0. [DOI] [PubMed] [Google Scholar]
- 37.Jurnak F. Science. 1985;230:32–36. doi: 10.1126/science.3898365. [DOI] [PubMed] [Google Scholar]
- 38.Ahnn J, March P E, Takiff H E, Inouye M. Proc Natl Acad Sci USA. 1986;83:8849–8853. doi: 10.1073/pnas.83.23.8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gollop N, March P E. J Bacteriol. 1991;173:2265–2270. doi: 10.1128/jb.173.7.2265-2270.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Boer P, Crossley R, Rothfield L. Nature (London) 1992;359:254–256. doi: 10.1038/359254a0. [DOI] [PubMed] [Google Scholar]
- 41.Trach K, Hoch J A. J Bacteriol. 1989;171:1362–1371. doi: 10.1128/jb.171.3.1362-1371.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barlowe C, d’Enfert C, Schekman R. J Biol Chem. 1993;268:873–879. [PubMed] [Google Scholar]
- 43.Hicke L, Schekman R. EMBO J. 1989;8:1677–1684. doi: 10.1002/j.1460-2075.1989.tb03559.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Altschul S F, Gish W, Miller W, Myers E W, Lipman D. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]