Abstract
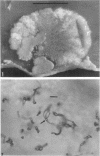
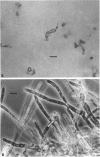
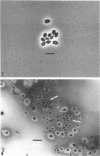
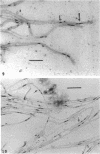
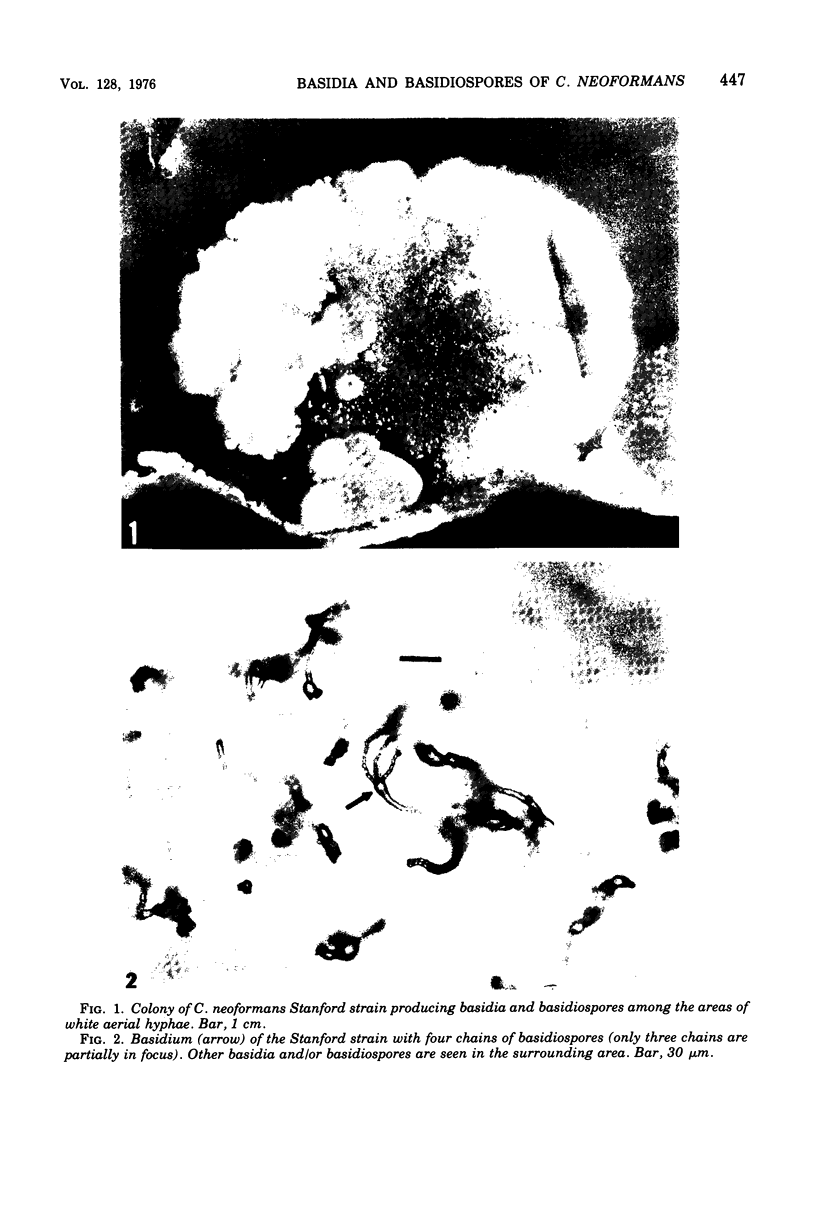
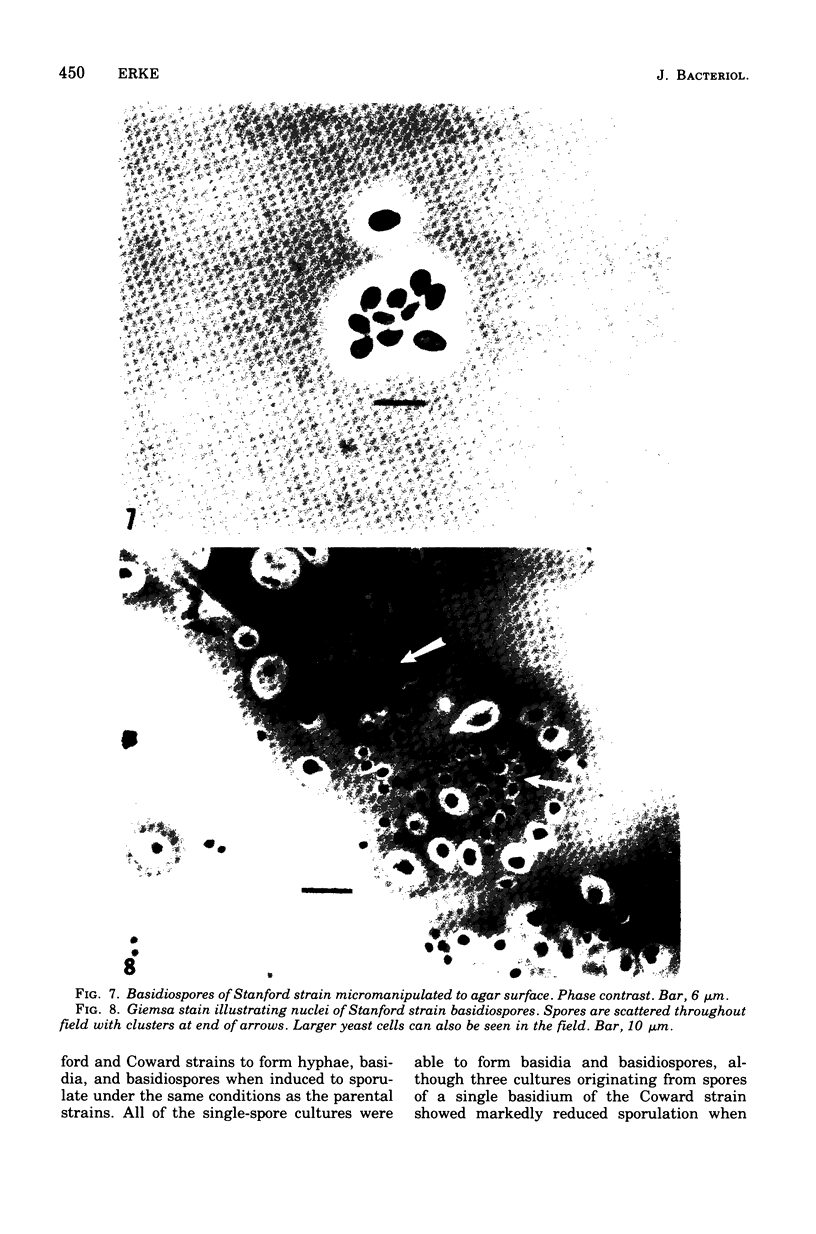
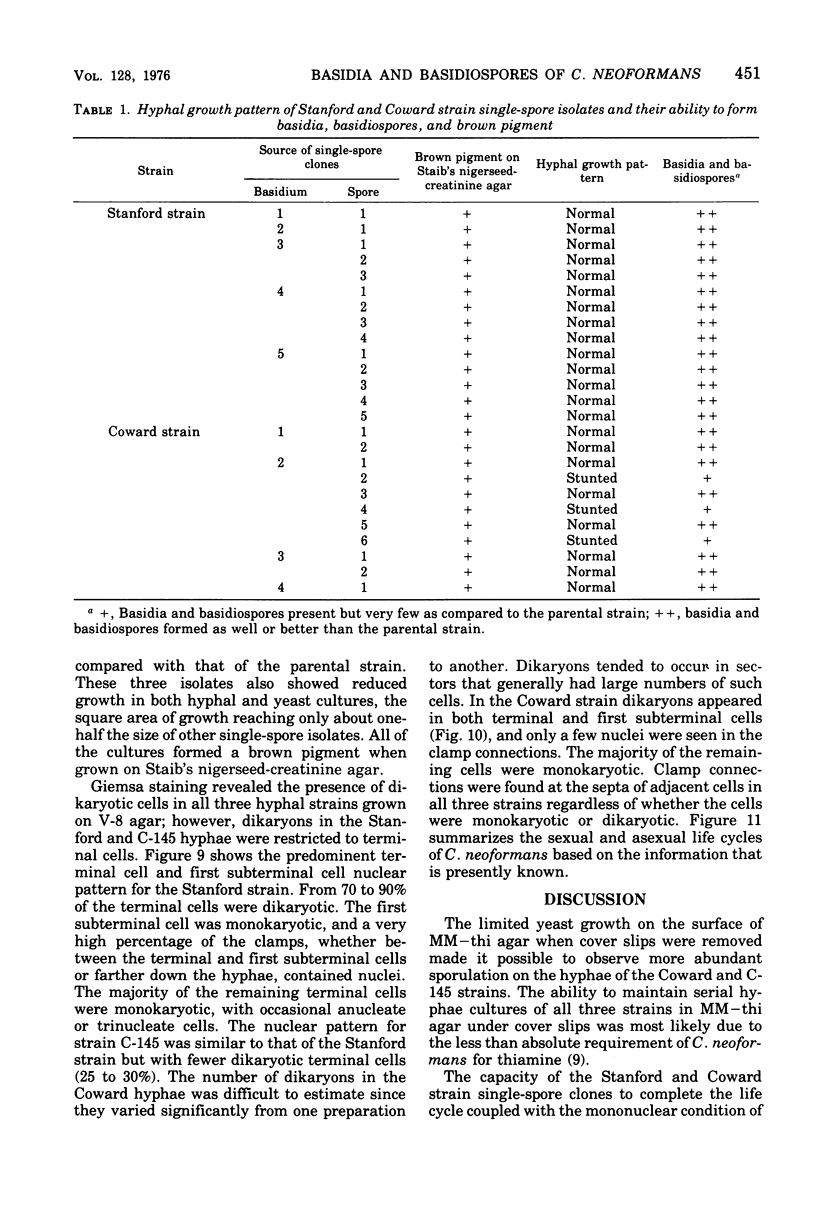
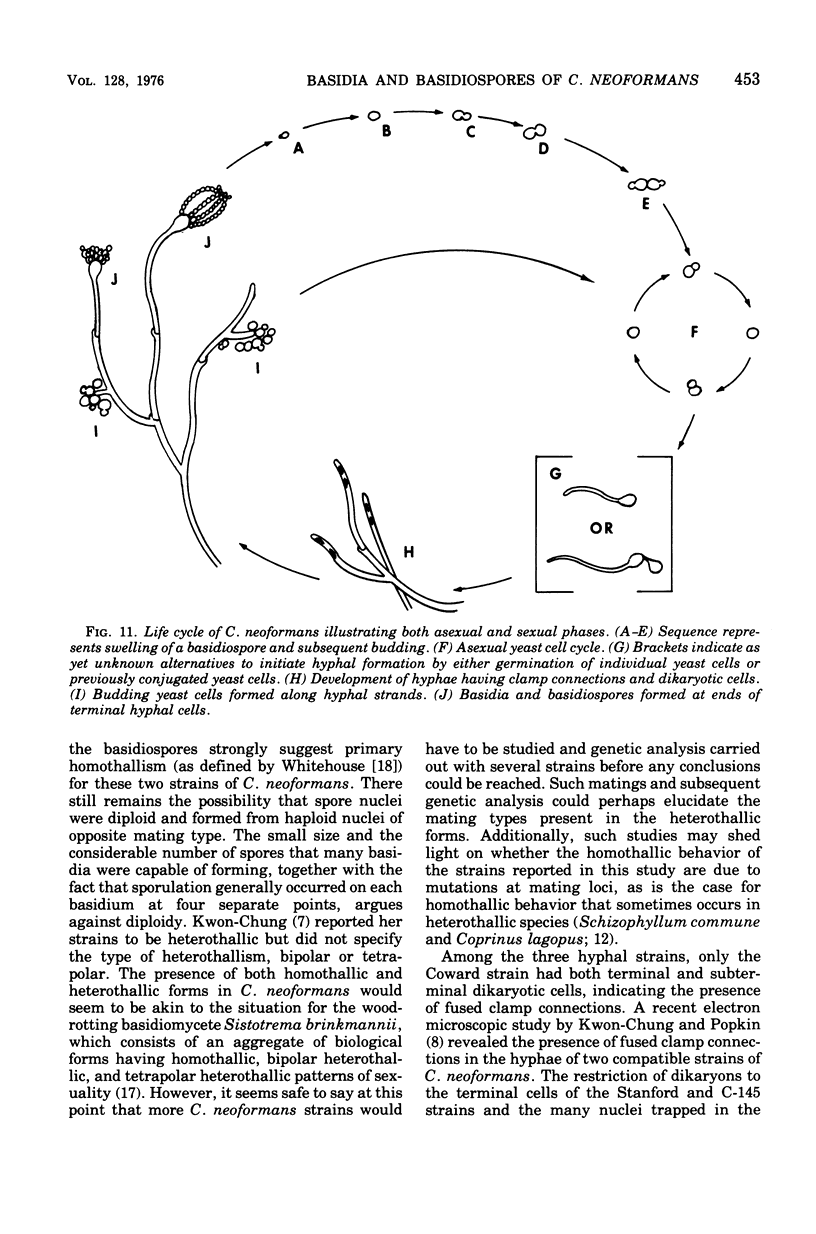
Three hypha-forming strains of Cryptococcus neoformans were induced to form basidia and basidiospores. Light microscopy showed that basidia formed at the ends of terminal hyphal cells and were able to produce from a few to many basidiospores. The morphology of the sexual structures indicated that these strains belonged to the recently described perfect state of C. neoformans, Filobasidiella neoformans. The average dimensions of the basidiospores were 1.9 mum in width by 2.7 mum in length. Giemsa staining revealed that dikaryotic cells were formed in all three strains. Only one strain had both terminal and subterminal dikaryons, indicating functional clamp connections, whereas the two remaining strains had dikaryons restricted to the terminal cells. Basidiospores of two strains were mononucleate, and yeast cell clones derived from single basidiospores of these two strains were able to complete the sexual life cycle, thus indicating their primary homothallic nature.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Cutler J. E., Erke K. H. Ultrastructural characteristics of Coccidioides immitis, a morphological variant of Cryptococcus neoformans and Podosypha ravenelii. J Bacteriol. 1971 Jan;105(1):438–444. doi: 10.1128/jb.105.1.438-444.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erke K. H., Schneidau J. D., Jr Relationship of some Cryptococcus neoformans hypha-forming strains to standard strains and to other species of yeasts as determined by deoxyribonucleic acid base ratios and homologies. Infect Immun. 1973 Jun;7(6):941–948. doi: 10.1128/iai.7.6.941-948.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farhi F., Bulmer G. S., Tacker J. R. Cryptococcus neoformans IV. The Not-So-Encapsulated Yeast. Infect Immun. 1970 Jun;1(6):526–531. doi: 10.1128/iai.1.6.526-531.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez F., Fu Y. S., Lurie H. Cryptococcosis histologically resembling histoplasmosis. A light and electron microscopical study. Arch Pathol. 1975 Jul;99(7):347–352. [PubMed] [Google Scholar]
- Kwon-Chung K. J. A new genus, filobasidiella, the perfect state of Cryptococcus neoformans. Mycologia. 1975 Nov-Dec;67(6):1197–1200. [PubMed] [Google Scholar]
- Kwon-Chung K. J., Popkin T. J. Ultrastructure of septal complex in Filobasidiella neoformans (Cryptococcus neoformans). J Bacteriol. 1976 Apr;126(1):524–528. doi: 10.1128/jb.126.1.524-528.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell K. E., Dahl B. A., Weeks R. J., Tosh F. E. Airborne Cryptococcus neoformans: particles from pigeon excreta compatible with alveolar deposition. J Infect Dis. 1972 Apr;125(4):412–415. doi: 10.1093/infdis/125.4.412. [DOI] [PubMed] [Google Scholar]
- STAIB F. MEMBRANFILTRATION UND NEGERSAAT (GUIZOTIA ABYSSINICA)-NAEHRBODEN FUER DEN CRYPTOCOCCUS NEOFORMANS-NACHWEIS (BRAUNFARBEFFEKT) Z Hyg Infektionskr. 1963 Oct 25;149:329–336. [PubMed] [Google Scholar]
- Shadomy H. J., Lurie H. I. Histopathological observations in experimental cryptococcosis caused by a hypha-producing strain of Cryptococcus neoformans (Coward strain) in mice. Sabouraudia. 1971 Mar;9(1):6–9. doi: 10.1080/00362177185190031. [DOI] [PubMed] [Google Scholar]
- Shadomy H. J., Utz J. P. Preliminary studies on a hyphaforming mutant of Cryptococcus neoformans. Mycologia. 1966 May-Jun;58(3):383–390. [PubMed] [Google Scholar]
- Ullrich R. C., Raper J. R. Primary Homothallism-relation to Heterothallism in the Regulation of Sexual Morphogenesis in Sistotrema. Genetics. 1975 Jun;80(2):311–321. doi: 10.1093/genetics/80.2.311. [DOI] [PMC free article] [PubMed] [Google Scholar]