Abstract
A visual pigment-like protein, referred to as peropsin, has been identified by large-scale sequencing of cDNAs derived from human ocular tissues. The corresponding mRNA was found only in the eye, where it is localized to the retinal pigment epithelium (RPE). Peropsin immunoreactivity, visualized by light and electron microscopy, localizes the protein to the apical face of the RPE, and most prominently to the microvilli that surround the photoreceptor outer segments. These observations suggest that peropsin may play a role in RPE physiology either by detecting light directly or by monitoring the concentration of retinoids or other photoreceptor-derived compounds.
Visual pigments are retinal-binding chromoproteins that constitute a subfamily of G protein-coupled receptors. The most extensively studied visual pigments are those located in retinal photoreceptors. These proteins detect light by monitoring the 11-cis to all-trans photoisomerization of a covalently bound retinal chromophore. Within this group, pairwise comparisons among vertebrate family members typically show greater than 40% amino acid identity, whereas pairwise comparisons between vertebrate and invertebrate visual pigments show approximately 20% amino acid identity. All visual pigments described to date have the following features: (i) a lysine in the middle of the seventh putative transmembrane segment, corresponding to Lys-296 in bovine rhodopsin, which is the site of covalent binding of the chromophore via a retinylidene Schiff base; (ii) a pair of cysteines, corresponding to Cys-110 and Cys-187 in bovine rhodopsin, which are presumed to form a disulfide bond connecting the first and second extracellular loops; (iii) the sequence (glu/asp)-arg-tyr, or a close match to this sequence, at the beginning of the second cytosolic loop; and (iv) one or more serine or threonine residues in the cytosolic carboxyl terminus, which in bovine rhodopsin are the sites of light-dependent phosphorylation by rhodopsin kinase (1).
Molecular cloning studies have revealed the sequences of three members of the visual pigment family that are either present in nonretinal tissues or that have been hypothesized to play a role other than phototransduction. Pinopsin is a pineal-specific visual pigment identified in chickens that is likely to mediate pineal photosensitivity; its sequence closely resembles those of vertebrate rod and cone pigments (2, 3). Retinochrome is a retinal chromoprotein present in the inner segments of cephalopod photoreceptors that photoconverts all-trans retinal to 11-cis retinal (4, 5). Retinal pigment epithelium (RPE) G protein-coupled receptor (RGR) is a vertebrate protein present in the intracellular membranes of both the RPE and Muller cells in the retina (6, 7). RGR purified from bovine RPE binds to all-trans but not 11-cis retinal and absorbs both visible and ultraviolet light (8, 9). The sequences of retinochrome and RGR opsin form a distinct and highly divergent branch within the visual pigment family (6, 10). Whether retinochrome and RGR act as signal-transducing light receptors, participate in the visual cycle as retinal isomerases, or function in both capacities, is not known.
In the vertebrate eye, the RPE lies adjacent to the photoreceptor cells and performs a number of functions critical for the viability and activity of the retina (11). The principal pathway by which the retina takes up nutrients and other small molecules from serum is from the choroidal circulation via transepithelial transport across the RPE. The RPE also engulfs, digests, and recycles approximately 10% of the mass of each photoreceptor outer segment per day (12), mediates the isomerization of retinal from all-trans to 11-cis (13), and stores retinoid precursors for visual pigment regeneration (14). Outer segment phagocytosis occurs on a diurnal schedule, with the peak of rod outer segment disc shedding occurring at dawn and the peak of cone outer segment disc shedding occurring at dusk (15, 16). In conjunction with the retina, the RPE contributes to the generation of a standing electrical potential across the eye and to the c-wave of the electroretinogram, a voltage response to illumination that occurs on a time scale of several seconds (17). The isolated RPE shows an electrical response to illumination only at intensities that are well above the physiological range; this response appears to be mediated by melanin (17).
In this paper we report the identification of a visual pigment-like protein that is present in the RPE, where it is localized to the microvilli that surround the photoreceptor outer segments. This protein may play a role in RPE physiology either by detecting light directly or by monitoring the concentration of retinoids or other photoreceptor-derived compounds.
MATERIALS AND METHODS
cDNA Cloning.
Oligo(dT)-primed cDNA libraries from adult human retinas (19) and P0–P7 mouse eyes (A. Lanahan, H.S., and J.N., unpublished work) were screened by DNA hybridization under standard conditions (20). The complete coding-region sequences of human and mouse peropsin were each obtained from two independent cDNA clones.
Interspecific Mouse Backcross Mapping.
Interspecific backcross progeny were generated by mating (C57BL/6J × Mus spretus) F1 females and C57BL/6J males as described (21). A total of 205 N2 mice were used to map the Rrh locus as described (22). A 0.7 kb probe containing part of the second exon of the mouse peropsin gene detected a fragment of 5.0 kb in PvuII-digested C57BL/6J DNA and a fragment of 2.9 kb was detected in PvuII-digested M. spretus DNA. The presence or absence of the 2.9-kb PvuII M. spretus-specific fragment was followed in backcross mice. A description of the probes and restriction fragment length polymorphisms for the loci linked to Rrh, including Egf and Nfkb1, has been reported previously (23). Recombination distances were calculated using map manager, version 2.6.5. Gene order was determined by minimizing the number of recombination events required to explain the allele distribution patterns.
RNase Protection and Northern Blot Hybridization.
Ten micrograms of total RNA from each mouse tissue or 10 μg of yeast tRNA was used for the RNase protection assay. For RNA blotting, 10 μg of bovine RNA was resolved by agarose gel electrophoresis under denaturing conditions, blotted, and hybridized with a human peropsin coding region probe (20).
In Situ Hybridization.
In situ hybridization was performed with digoxigenin-labeled riboprobes essentially as described (24) using 20 μm sections cut from unfixed albino rat eyes frozen in OCT.
Production of Antibodies and Immunoblotting.
Rabbits were immunized with a synthetic peptide corresponding to the carboxyl-terminal 20 residues of mouse peropsin conjugated via its amino terminus to BSA (25). For affinity purification, a fusion protein was produced in Escherichia coli in which the same 20 amino acids were displayed at the carboxyl terminus of the maltose binding protein. The maltose binding protein–peropsin fusion was purified by amylose affinity chromatography, coupled to an Affigel matrix (Bio-Rad), and used to affinity purify anti-peropsin antibodies. Protein extracts from mouse eye (following removal of the lens) or brain were prepared by homogenizing the tissue in SDS sample buffer, and insoluble material was removed by microcentrifugation.
Immunohistochemistry.
For plastic embedding, CD1 mice were perfused with PBS/4% paraformaldehyde for light microscopy, or PBS/1% glutaraldehyde/4% paraformaldehyde for electron microscopy. The anterior chamber and lens were removed, and eye cups were further incubated in the same fixative for 3 hr at 4°C and embedded in Unicryl resin (BBI, Cardiff, U.K.). For light microscopy, 1 μm sections were stained, as described below for the staining of frozen sections, with affinity-purified rabbit anti-peropsin and then incubated with goat anti-rabbit antibodies conjugated to 1 nm gold particles followed by silver enhancement. For post-embedding immuno-electron microscopy, 0.1 μm sections were blocked in 10 mM Tris (pH 7.5), 150 mM NaCl, 0.5% Tween 20 containing 5% normal goat serum, stained with affinity-purified rabbit anti-peropsin in the same solution overnight at 4°C, and then stained with goat anti-rabbit antibodies conjugated to 18 nm gold particles in 10 mM Tris (pH 7.5)/150 mM NaCl for 1 hr at room temperature. Following five washes in 10 mM Tris (pH 7.5)/150 mM NaCl and five washes in water, the grids were stained with 0.2% uranyl acetate in 0.1 M maleate (pH 6.2) for 15 min at room temperature. Some sections were also counterstained with lead acetate.
For immunostaining of frozen sections, eye cups from CD1 mice were fixed at 4°C in PBS/4% paraformaldehyde for 3 hr, cryoprotected in 30% sucrose overnight, and embedded in OCT. Ten μm frozen sections were preincubated for 1 hr at room temperature in PBS containing 5% normal goat serum/0.3% Triton X-100 and then incubated in the same buffer overnight at 4°C in affinity-purified primary antibody. Biotinylated goat anti-rabbit secondary antibody and Texas Red-conjugated streptavidin (Vector Laboratories) or avidin-horseradish peroxidase (ExtrAvidin peroxidase; Sigma) were used to visualize immunostaining.
RESULTS
Identification of a Novel Visual Pigment Homologue.
To efficiently identify novel genes expressed in ocular tissues, we have determined partial sequences from cDNA clones derived from these tissues (J. P. Macke, P. Smallwood, A. Rattner, J. Williams, and J.N., unpublished). In an initial group of 5,000 partial sequences derived from human retina cDNAs, one clone showed significant homology to visual pigments but was clearly distinct from known visual pigments. As described below, subsequent experiments revealed that the corresponding gene is expressed in the RPE but not in the retina. The presence of these sequences in the human retina cDNA library presumably reflects contamination by RPE of the human retina sample used to prepare the library. To indicate its RPE localization, this protein has been named peropsin, the first three letters of which are a rearrangement of RPE.
The amino acid sequences of human and mouse peropsin were deduced from full-length cDNA clones. The deduced amino acid sequences show all the hallmarks of typical G protein-coupled receptors, including seven segments of hydrophobic amino acids that presumably correspond to membrane-spanning α-helices, a (glu/asp)-arg-tyr triplet at the beginning of the second cytosolicloop, and two (mouse) or five (human) serine and threonine residues in the carboxyl-terminal tail that are potential sites for regulation by receptor kinases (Fig. 1). The human and mouse orthologues share 80% amino acid identity, with the regions of greatest divergence occurring in the amino- and the carboxyl-terminal tails. A potential site for asn-linked glycosylation is present within the first 10 residues of both human and mouse peropsin. In the discussion that follows, we will refer to amino acid sequence comparisons with human peropsin, but similar results were obtained with mouse peropsin.
Figure 1.
Amino acid sequences of human and mouse peropsin aligned with human rhodopsin, octopus rhodopsin, bovine RGR opsin, and squid retinochrome. The positions of the retinylidene Schiff base (Lys-296 in rhodopsin) and its counterion (Glu-113 in rhodopsin) are indicated by arrowheads. The region encompassing the seventh putative transmembrane segment of squid retinochrome has been manually aligned to bring Lys-275, the site of the retinylidene Schiff base (26), into register with Lys-296 in human rhodopsin. h, human; m, mouse; o, octopus; b, bovine; s, squid.
When the human peropsin amino acid sequence is compared with all GenBank protein sequences (May 1997 release) using the blast algorithm (27), the top 124 matches are with vertebrate and invertebrate visual pigments (Fig. 2). A large variety of hormone, neurotransmitter, and neuropeptide G protein-coupled receptors comprise the next most homologous group of sequences, of which the most homologous is a putative tachykinin receptor. Significantly, peropsin contains a lysine at the position corresponding to Lys-296 in bovine rhodopsin, the site of covalent attachment of 11-cis retinal in all visual pigments. To our knowledge, no G protein-coupled receptors other than members of the visual pigment family have a lysine at this position. The glutamate that serves in all vertebrate rod and cone pigments as the retinylidene Schiff base counterion (corresponding to Glu-113 in bovine rhodopsin) is occupied by tyrosine in peropsin, as it is in many invertebrate visual pigments.
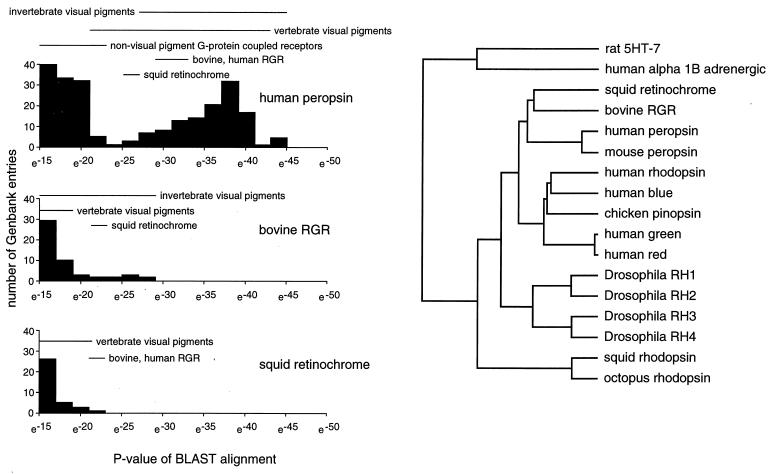
Figure 2.
(Left) Quantitative analysis of homologies between protein sequences in GenBank (May 1997 release) and human peropsin, bovine RGR, and squid retinochrome as determined by the blast algorithm (27). The P value is the probability of finding a comparable match by chance in a random sequence database of equivalent size. Horizontal bars indicate the range of P values for the indicated classes of sequences. Redundant database entries have not been eliminated for this analysis. (Right) Dendrogram of visual pigments and visual pigment-related proteins calculated using geneworks software with gap penalties set at 5 for each gap and 25 for lengthening a gap. In the dendrogram, the length of each line is proportional to the degree of amino acid sequence divergence. The rat 5HT-7 and human alpha 1B adrenergic receptors are two of the five nonvisual pigment G protein-coupled receptors in the GenBank database that are most homologous to peropsin.
The peropsin sequence also shows significant homology to bovine RGR and squid retinochrome that rank, respectively, 99th and 121st in a blast search of the GenBank protein database (May 1997 release). The blast alignment indicates that the similarity between peropsin and the majority of visual pigments is greater than that between peropsin and RGR or retinochrome (Fig. 2). This analysis also indicates that peropsin is overall more similar to visual pigments than are RGR or retinochrome. We note, however, that when sequence homologies among visual pigment family members are calculated using an algorithm (geneworks), which penalizes gaps less than the blast algorithm, peropsin falls within the RGR/retinochrome branch of the visual pigment family (Fig. 2).
Whole genome Southern blot hybridization at 42°C in the presence of 27% formamide/5× SSC/10% dextran sulfate using a probe containing the human peropsin coding and 5′ untranslated regions shows one or several discrete hybridizing fragments in DNA from human, cow, mouse, chicken, Xenopus, catfish, and zebrafish (data not shown), suggesting that peropsin orthologues are present within many vertebrates. Under these conditions, the known visual pigment genes would not be expected to hybridize to the peropsin probe.
Localization of Peropsin mRNA and Protein.
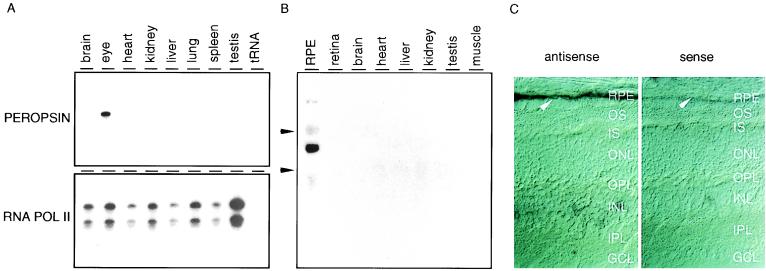
In the adult mouse, RNase protection with samples from brain, eye, heart, kidney, liver, lung, spleen, and testis shows peropsin transcripts only in the eye, and in situ hybridization to the adult rat eye localizes peropsin transcripts to the RPE (Fig. 3). The absence of peropsin from the retina and nonocular tissues, and its presence in the RPE, was also seen by Northern blot hybridization to total bovine RNA (Fig. 3).
Figure 3.
RNase protection, Northern blot analysis, and in situ hybridization localize peropsin transcripts to the RPE. (A) Total RNA from the indicated adult mouse tissues was used for RNase protection with a mouse peropsin probe. Peropsin transcripts are detected only in the eye. A control reaction with an RNA polymerase II probe is shown at bottom. (B) Northern blot of total RNA from the indicated bovine tissues was hybridized to a probe encompassing the human peropsin coding region. Peropsin transcripts are detected only in the RPE. Arrowheads indicate the mobilities of the ribosomal RNAs. (C) In situ hybridization using a digoxigenin-labeled mouse peropsin probe localizes peropsin transcripts in the adult rat to the RPE (arrowhead). (Left) Antisense probe. (Right) Sense probe. Albino rats were used for this experiment so that RPE melanin would not obscure the signal. At long substrate incubation times, a faint hybridization signal is seen in the inner nuclear layer; the origin of this signal is not known. Arrowhead, nucleus of RPE cell. OS, outer segments; ONL, outer nuclear layer; OPL, outer plexiform layer; INL, inner nuclear layer; IPL, inner plexiform layer; GCL, ganglion cell layer.
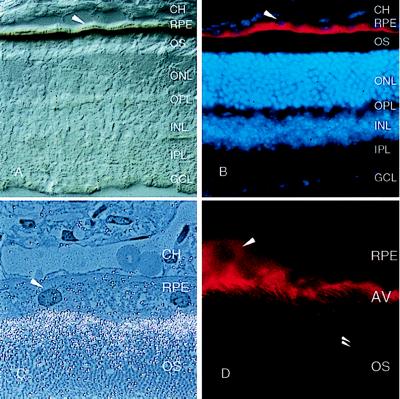
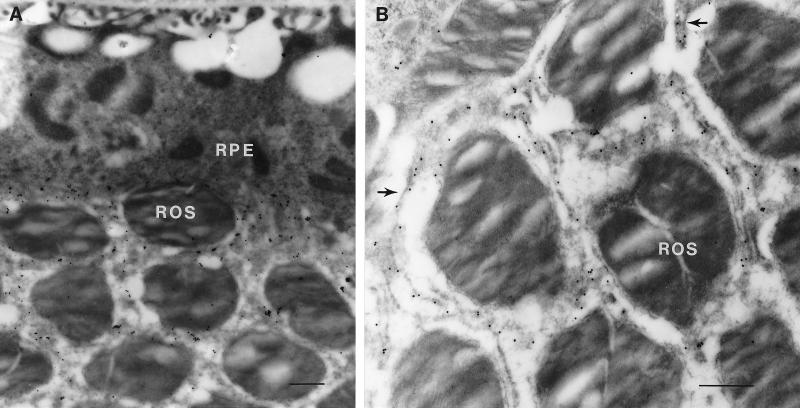
To localize the peropsin protein, antibodies were raised against a synthetic peptide corresponding to the carboxyl-terminal 20 residues of mouse peropsin. Immunoblots of total protein from mouse brain or eye (minus the lens) shows that the affinity-purified anti-peropsin recognizes a single eye-specific protein with an apparent molecular mass of approximately 38 kDa, close to the predicted molecular mass of 37 kDa for unglycosylated mouse peropsin (Fig. 4). Immunofluorescent and immunoperoxidase staining with affinity-purified anti-peropsin antibodies shows immunoreactivity exclusively on the apical face of the RPE, in the region containing the distal portion of rod photoreceptor outer segments (Fig. 5). When examined by confocal microscopy, the immunofluorescent anti-peropsin signal is seen within numerous microvilli projecting from the apical surface of the RPE. Occasional larger immunostained structures are observed projecting beyond the array of microvilli. Immunogold staining of 1 μm plastic sections also indicates a microvillar localization of peropsin, and shows that the immunostained region encompasses the distal one-third of the outer segment zone. To further assess the subcellular distribution of peropsin, ultra-thin sections were examined by postembedding immunoelectron microscopy (Fig. 6). This analysis confirms that peropsin is localized to the apical plasma membrane of the RPE, including the microvilli that occupy the space between rod outer segments, and that it is absent from rod outer segments.
Figure 4.
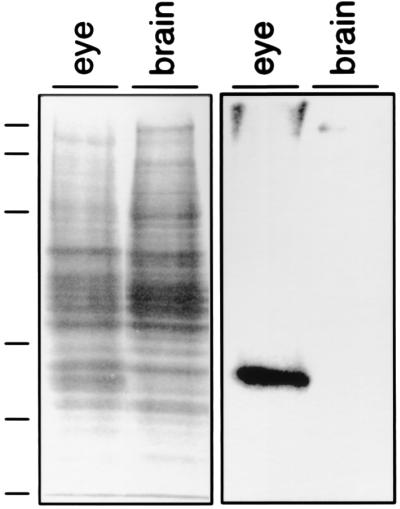
Affinity-purified anti-peropsin antibodies recognize a single protein with an apparent molecular mass of approximately 38 kDa. Fifteen micrograms of total protein from mouse brain or mouse eye from which the lens was removed were loaded on each lane of a 10% polyacrylamide/SDS gel and either stained with Coomassie blue (Left) or immunoblotted using affinity-purified rabbit antibody prepared against a synthetic 20 amino acid peptide corresponding to the carboxyl-terminus of mouse peropsin (Right). The molecular masses of protein size standards, indicated at the left, are, from top to bottom, 192, 127, 73, 43, 32, and 17 kDa.
Figure 5.
Histochemical localization of peropsin in the adult mouse eye. Albino (CD1) mice were used in all experiments so that RPE melanin would not obscure the signal. (A) Immunoperoxidase and (B) immunofluorescent staining with affinity-purified anti-peropsin antibodies localizes peropsin to the apical face of the RPE. (B) Nuclei are visualized with 4′,6-diamidino-2-phenylindole. (C) Immunogold staining with anti-peropsin antibodies of a 1 μm section of unicryl-embedded tissue localizes peropsin to the apical microvilli of the RPE. Gold particles were silver-enhanced and the section was counterstained with methylene blue. Photoreceptor outer segments are visible in the lower half of the micrograph. (D) Confocal image of anti-peropsin immunostaining of the RPE. Peropsin is localized to the dense network of microvilli that protrude from the apical face of the RPE; occasional larger structures (double arrowhead) protrude from the RPE and contain peropsin immunoreactivity. Single arrowhead, nucleus of RPE cell. CH, choroid; OS, outer segments; ONL, outer nuclear layer; OPL, outer plexiform layer; INL, inner nuclear layer; IPL, inner plexiform layer; GCL, ganglion cell layer; AV, apical microvilli.
Figure 6.
Immunoelectron microscopic localization of peropsin in the mouse eye. Immunogold labeling with anti-peropsin antibodies is localized to the apical face of the RPE and to the microvilli that surround the rod outer segments (ROS). (A) At low magnification, the full thickness of the RPE is seen. Numerous large phagosomes are present within the RPE, and the infoldings at the basal face of the RPE are seen immediately adjacent to Bruch’s membrane at the top of the photomicrograph. (B) At higher magnification, individual immunolabeled microvilli (arrows) are seen between unlabeled rod outer segments. (Bars = 0.5 μm.)
Chromosomal Location of the Mouse Peropsin Gene.
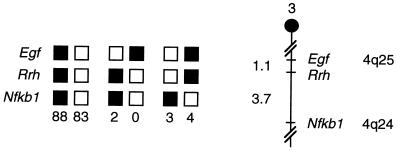
The peropsin gene will be referred to as Rrh, an abbreviation for “RPE-derived rhodopsin homologue.” The mouse chromosomal location of Rrh was determined by interspecific backcross analysis using progeny derived from matings of [(C57BL/6J × M. spretus) F1 × C57BL/6J] mice. The mapping panel has been typed for over 2,400 loci that are well distributed along all the autosomes and the X chromosome (21). Rrh is located in the distal region of mouse chromosome 3, linked to Egf and Nfkb1. Although 180 mice were analyzed for every marker and are shown in the segregation analysis (Fig. 7), up to 187 mice were typed for some pairs of markers. Each locus was analyzed in pairwise combinations for each recombination frequency using the additional data. The ratios of the total number of mice exhibiting recombinant chromosomes to the total number of mice analyzed for each pair of loci and the most likely gene order are: centromere - Egf - 2/185 - Rrh - 7/187 - Nfkb1. The recombination frequencies (expressed as genetic distances in centimorgans ± SEM) are: Egf - 1.1 ± 0.8 - Rrh - 3.7 ± 1.4 - Nfkb1. The distal region of mouse chromosome 3 shares a region of homology with human chromosome 4q (summarized in Fig. 7). The placement of Rrh in this interval in the mouse suggests that the human homologue will map to 4q.
Figure 7.
Rrh maps in the distal region of mouse chromosome 3. Rrh was placed on mouse chromosome 3 by interspecific backcross analysis. Segregation patterns of Rrh and flanking genes in 180 backcross animals that were typed for all loci are shown at the top. For individual pairs of loci, more than 180 animals were typed (see text). Each column represents the chromosome identified in the backcross progeny that was inherited from the (C57BL/6J × M. spretus) F1 parent. Solid boxes represent the presence of a C57BL/6J allele and open boxes represent the presence of a M. spretus allele. The number of offspring inheriting each type of chromosome is listed at the bottom of each column. A partial chromosome 3 linkage map showing the location of Rrh in relation to linked genes is shown at right. Recombination distances between loci, in centimorgans, are shown to the left of the chromosome and the positions of loci in human chromosomes, where known, are shown to the right. References for the human map positions of loci cited in this study can be obtained from the Genome Data Base, a computerized database of human linkage information maintained by The William H. Welch Medical Library of The Johns Hopkins University (Baltimore).
DISCUSSION
This paper describes the discovery and localization of peropsin, a novel visual pigment-like G protein-coupled receptor. Peropsin is localized to the apical microvilli of the RPE, where it is in close proximity to photoreceptor outer segments. Electron microscopic studies in many vertebrate retinas have demonstrated an intimate association between RPE microvilli and photoreceptor outer segments. This association is presumed to be important for adhesion of the retina to the RPE, transport of small molecules between the RPE and the photoreceptors, and orderly phagocytosis of photoreceptor outer segments by the RPE. The localization of peropsin to the microvilli suggests that this close association may also play a role in regulating RPE physiology via cell-surface G protein-coupled receptors. In contrast, RGR opsin is localized in the RPE to internal rather than surface membranes.
Based on its homology to the visual pigments, it seems reasonable to speculate that peropsin binds to a retinoid ligand. The presence of a lysine at a position homologous to Lys-296 in bovine rhodopsin further suggests that, like the visual pigments, peropsin may form a complex with an isomer of retinal via a Schiff base with the amino group of lysine. If retinal is the ligand, peropsin could have either of two distinct modes of action. First, peropsin could be a light receptor that signals in response to photoisomerization of the bound chromophore. In this context, the location of peropsin in the apical microvilli would permit efficient light absorption even in the presence of high concentrations of RPE melanin, because the vast majority of melanosomes are sequestered within the RPE cell bodies. Second, peropsin could be activated (or inactivated) by binding to one or more retinal isomers in a reaction that does not depend on illumination. In this model, peropsin might act as a sensor of retinal that is released by photobleaching of visual pigments in the adjacent outer segments or it might monitor retinal metabolism in the RPE. Although we favor retinal as the ligand, at present there is no experimental data to support this hypothesis. In preliminary experiments, we have attempted to reconstitute recombinant peropsin produced in transfected 293 cells or in baculovirus-infected Sf9 cells by incubation with a variety of retinal isomers. Using protocols that were developed for mammalian visual pigment reconstitution in vitro (28), we have not observed a photolabile absorption band in the visible region of the spectrum. However, it is possible that retinal may only bind to peropsin at a particular stage in its biosynthesis, as suggested for Drosophila visual pigments (29), that binding requires cofactors that are present in the RPE but absent in the tissue culture expression system, or that retinal binding produces a photostable or UV-absorbing pigment.
In an alternate model of peropsin action, peropsin might recognize a nonretinoid ligand, but share sequence similarity with visual pigments based on its evolutionary origin as a visual pigment gene that was recruited for RPE-specific expression. The possibility that the RPE and retina express divergent copies of genes that were recently derived from a common ancestor is consistent with the close embryological relationship between these two tissues. Both tissues are derived from the neuroepithelium of the optic vesicle, and in many species the RPE retains the ability to transdifferentiate into neural retina (30). In this model, the peropsin binding pocket would have diverged to generate specificity for a nonretinoid ligand that would presumably be present in the subretinal space.
All of the models described above envision a role for peropsin in regulating RPE function or activity. It will be of interest to determine which intracellular signal transduction pathway mediates peropsin action and how the transduced signal modulates RPE physiology. Whether regulation is based directly on light absorption or is via detection of a molecule that is released into the subretinal space, these findings further reinforce the idea that the RPE and retina function together as a coordinated unit.
Acknowledgments
We thank C. Riley, C. Davenport, J. Ptak, and M. Kazienko for oligonucleotide and peptide synthesis; J. Macke for determining the initial partial cDNA sequence of human peropsin; J. Li and M. Delannoy for assistance with confocal microscopy and cutting plastic sections; J. Chang, S. Zhang, P. Campochiaro, and D. Zack for the gift of the bovine tissue RNA blot; D. B. Householder for excellent technical assistance; and A. Rattner, P. Sherman, P. Tong, D. Valle, and K.-W. Yau for helpful comments on the manuscript. This research was supported by the Ruth and Milton Steinbach Fund, the Howard Hughes Medical Institute, and the National Cancer Institute, Department of Health and Human Services, under contract with Advanced BioScience Laboratories.
ABBREVIATIONS
- RPE
retinal pigment epithelium
- RGR
retinal G protein-coupled receptor
Footnotes
References
- 1.Khorana H G. J Biol Chem. 1992;267:1–4. [PubMed] [Google Scholar]
- 2.Okano T, Yoshizawa T, Fukada Y. Nature (London) 1994;372:94–97. doi: 10.1038/372094a0. [DOI] [PubMed] [Google Scholar]
- 3.Max M, McKinnon P J, Seidenman K J, Barrett R K, Applebury M L, Takahashi J S, Margolskee R F. Science. 1995;267:1502–1506. doi: 10.1126/science.7878470. [DOI] [PubMed] [Google Scholar]
- 4.Hara T, Hara R. Nature (London) 1967;214:573–575. doi: 10.1038/214573a0. [DOI] [PubMed] [Google Scholar]
- 5.Hara T, Hara R. Nature (London) 1968;219:450–454. doi: 10.1038/219450a0. [DOI] [PubMed] [Google Scholar]
- 6.Jiang M, Pandey S, Fong H K W. Invest Ophthalmol Visual Sci. 1993;34:3669–3678. [PubMed] [Google Scholar]
- 7.Pandey S, Blanks J C, Spee C, Jiang M, Fong H K W. Exp Eye Res. 1994;58:605–614. doi: 10.1006/exer.1994.1055. [DOI] [PubMed] [Google Scholar]
- 8.Shen D, Jiang M, Hao W, Tao L, Salazar M, Fong H K W. Biochemistry. 1994;33:13117–13125. doi: 10.1021/bi00248a022. [DOI] [PubMed] [Google Scholar]
- 9.Hao W, Fong H K W. Biochemistry. 1996;35:6251–6256. doi: 10.1021/bi952420k. [DOI] [PubMed] [Google Scholar]
- 10.Hara-Nishimura I, Matsumoto T, Mori H, Nishimura M, Hara R, Hara T. FEBS Lett. 1990;271:106–110. doi: 10.1016/0014-5793(90)80383-t. [DOI] [PubMed] [Google Scholar]
- 11.Zinn K M, Marmor M F. The Retinal Pigment Epithelium. Cambridge, MA: Harvard Univ. Press; 1979. pp. 226–244. [Google Scholar]
- 12.Young R W, Bok D. J Cell Biol. 1969;42:392–403. doi: 10.1083/jcb.42.2.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rando R R. Angew Chem. 1990;29:461–480. [Google Scholar]
- 14.Dowling J E. Nature (London) 1960;188:114–118. doi: 10.1038/188114a0. [DOI] [PubMed] [Google Scholar]
- 15.LaVail M M. Science. 1976;194:1071–1074. doi: 10.1126/science.982063. [DOI] [PubMed] [Google Scholar]
- 16.Young R W. Invest Ophthalmol. 1978;17:105–116. [PubMed] [Google Scholar]
- 17.Marmor M F, Lurie M. In: The Retinal Pigment Epithelium. Zinn K M, Marmor M F, editors. Cambridge, MA: Harvard Univ. Press; 1979. pp. 226–244. [Google Scholar]
- 18.Wang Y, Macke J P, Abella B S, Andreasson K, Worley P, Gilbert D J, Copeland N G, Jenkins N A, Nathans J. J Biol Chem. 1996;271:4468–4476. doi: 10.1074/jbc.271.8.4468. [DOI] [PubMed] [Google Scholar]
- 19.Nathans J, Thomas D, Hogness D S. Science. 1986;232:193–202. doi: 10.1126/science.2937147. [DOI] [PubMed] [Google Scholar]
- 20.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 21.Copeland N G, Jenkins N A. Trends Genet. 1991;7:113–118. doi: 10.1016/0168-9525(91)90455-y. [DOI] [PubMed] [Google Scholar]
- 22.Jenkins N A, Copeland N G, Taylor B A, Lee B K. J Virol. 1982;43:26–36. doi: 10.1128/jvi.43.1.26-36.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hamel C P, Jenkins N A, Gilbert D J, Copeland N G, Redmond T M. Genomics. 1994;20:509–512. doi: 10.1006/geno.1994.1212. [DOI] [PubMed] [Google Scholar]
- 24.Schaeren-Wiemers N, Gerfin-Moser A. Histochemistry. 1993;100:431–440. doi: 10.1007/BF00267823. [DOI] [PubMed] [Google Scholar]
- 25.Sung C-H, Makino C, Baylor D, Nathans J. J Neurosci. 1994;14:5818–5833. doi: 10.1523/JNEUROSCI.14-10-05818.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hara-Nishimura I, Kondo M, Nishimura M, Hara R, Hara T. FEBS Lett. 1993;335:94–98. doi: 10.1016/0014-5793(93)80447-3. [DOI] [PubMed] [Google Scholar]
- 27.Altschul S F, Gish W, Miller W, Myers E W, Lippman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 28.Merbs S, Nathans J. Photochem Photobiol. 1992;56:869–881. doi: 10.1111/j.1751-1097.1992.tb09708.x. [DOI] [PubMed] [Google Scholar]
- 29.Ozaki K, Nagatani H, Ozaki M, Tokunaga F. Neuron. 1993;10:1113–1119. doi: 10.1016/0896-6273(93)90059-z. [DOI] [PubMed] [Google Scholar]
- 30.Coulombre J, Coulombre A. Dev Biol. 1965;12:79–92. doi: 10.1016/0012-1606(65)90022-9. [DOI] [PubMed] [Google Scholar]