Abstract
The lack of Na+/I− symporter (NIS) gene expression in some thyroid cancer patients has been a major hurdle that limits the efficacy of standard radioactive iodide therapy. The molecular mechanism that contributes to low NIS expression is not well understood. Activated NIS gene expression is stimulated by thyroid-stimulating hormone-mediated cAMP/protein kinase A signaling through a NIS upstream enhancer (NUE). The cAMP pathway is also stimulated by forskolin. In the current work, we studied the mechanism of transcriptional activation of NIS in normal thyroid cells and thyroid cancer cells. We identified the cAMP response element modulator (CREM) activator as a new component of the transcription complex that is important for NIS gene expression. The CREM complex is seen in the normal thyroid cells and BRAF (V600E) thyroid cancer cells (BHP 17–10) but is missing in rearranged in transformation/papillary thyroid carcinoma-1 rearrangement thyroid cancer cells (BHP 2–7). This complex is believed to be responsible for the loss of NUE activity and reduced NIS expression in the BHP 2–7 cell line. In BHP 2–7 cells, forskolin stimulated the thyroid-specific transcription factor Pax 8, but CREM activator mRNA did not increase, and this produced a small increase in NUE activity. Ectopic expression of CREM activator enhanced activity of the NUE, indicating that CREM is an essential regulator of NIS gene expression.
THYROID CANCER IS the most prevalent endocrine carcinoma (1). Papillary thyroid cancer makes up about 80% of all thyroid cancers (2) and is a differentiated carcinoma derived from follicular cells. Because radioiodide is concentrated in thyroid follicles, radioiodide-131 has been used for treatment of patients with differentiated thyroid carcinoma (3). Radioiodide is actively transported into normal and cancerous thyroid cells mediated by the sodium/iodide symporter (NIS) positioned on the basolateral membrane of thyroid follicular cells. NIS gene expression and iodide uptake are stimulated by thyroid-stimulating hormone (4,5,6). Endogenous or exogenous thyroid-stimulating hormone enhances the efficacy of radioiodide therapy. Thyroid-stimulating hormone and forskolin (FSK) both stimulate NIS gene expression in a cAMP-dependent manner, mediated by protein kinase A dependent and independent pathways (7,8,9). About 20% of differentiated thyroid carcinomas do not take up radioiodide, resulting in a poor prognosis (10). Generally, the loss of active iodide transport is the result of either faulty NIS gene expression or the loss of NIS protein translocation to the membrane.
Both rat NIS and human NIS (hNIS) promoters have been studied by several groups (11,12,13,14,15,16). They vary in functional regions, but both retain activity in different proximal and upstream regions. Taki et al. (8 found a strong thyroid-stimulating hormone and FSK-responsive far upstream enhancer between −9847 and −8968. Within this segment there is an essential Pax 8 site and cAMP response element (CRE)-like site along with an inactive thyroid transcription factor-1 site. The hNIS upstream enhancer (NUE) appears to be different from the rat NUE in promoter position and sequences. However, there is a region in the human promoter between −3927 and −3686 that is homologous to the rat promoter, but no activity has been found in this region (17).
Thyroid-specific and nonspecific transcription factors are required for thyroid-stimulating hormone and FSK-stimulated NUE activation of NIS gene expression. The thyroid-specific transcription factor Pax 8 is required for thyroid development and differentiated function. Pax 8 is essential for thyroid-stimulating hormone-stimulated NUE-activated hNIS gene expression (8). Pax 8 protein levels have increased with thyroid-stimulating hormone treatment up to 24 h in FRTL-5 cells [Fischer rat thyroid cells in low serum (5% calf serum)] (18). The downstream effects of thyroid-stimulating hormone stimulation are generally mediated by cAMP, which stimulates the small palindromic cis-acting element CRE (19,20,21,22). Both rat and human NUE contain degenerate CRE sites that require the traditional pathway for stimulation (8,15). The CRE element recruits transcription factors in the cAMP response element binding protein (CREB) family that include cAMP response element modulator (CREM) and activating transcription factor (ATF)-1 (23,24). CREM isoforms act as transcriptional activators by using glutamine-rich domains (Q1 and Q2) and serine phosphorylation (P-Box), and act as repressors that lack Q1 and Q2 domains (25).
Previously, Chun et al. (16) showed, by gel mobility supershift analysis, that CREB 1, ATF-2, c-Jun, and c-fos all bind to the CRE site in the rat NUE. Using a similar technique, Taki et al. (8) showed that CREB was involved in complexes bound to the CRE-like site in the human NUE. Furthermore, Taki et al. found that one CRE complex was thyroid-stimulating hormone induced, and was not shifted by antibodies to some CRE binding proteins. This complex was not observed in the papillary thyroid cancer cells (BHP 2–7) that have the rearranged in transformation/papillary thyroid carcinoma (RET-PTC)-1 rearrangement. The observation that factor S was not found with BHP 2–7 nuclear extract and that this cancer cell line does not show NUE activity implied an important role for the thyroid-stimulating hormone-induced complex in activated NUE stimulation of NIS gene expression.
In the present study, we aimed to identify the transcription factors involved in stimulating NIS gene expression through the NUE in functional thyroid cells (FRTL-5), and to understand the transcriptional defects that cause reduced endogenous NIS in mutually exclusive BHP 2–7 (RET-PTC1) and BHP 17–10 (BRAF) thyroid cancer cells.
Materials and Methods
Materials
Mouse CREM-1 (τ1αγ) polyhistidine-tagged fusion protein is a commercial product from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). All chemicals were purchased from Sigma Chemicals (St. Louis, MO) unless otherwise stated.
Cell culture
FRTL-5 rat thyroid cells, kindly provided by Dr. Leonard D. Kohn (Ohio University, Athens, OH), were grown in Coon’s modified Ham’s F-12 medium (Sigma Chemicals) supplemented with 5% calf serum and a six hormone mixture containing thyroid-stimulating hormone (1 mU/ml), insulin (10 μg/ml), cortisol (10 nm), transferrin (5 μg/ml), glycyl-l-histidyl-l-lysine acetate (10 ng/ml), and somatostatin (10 ng/ml) (26). In some experiments FRTL-5 cells were cultured for 6 d in medium without thyroid-stimulating hormone [five hormone mixture (5H)] after reaching approximately 80% confluency before use.
BHP 2–7 cells, derived from a human papillary thyroid cancer and recently shown by genetic analysis to be a subclone of the thyroid papillary carcinoma-1 cell line, kindly provided by Dr. Sissy Jhiang (Ohio State University, Columbus, OH) (27,28), and BHP 17–10 cells, recently shown by genetic analysis to be a subclone of the NPA 87 cell line, kindly provided by Dr. Guy Juillard (University of California Los Angeles, Los Angeles, CA), were both grown in RPMI 1640 medium (Sigma Chemicals) supplemented with 10% fetal bovine serum, 0.1 mm nonessential amino acids (Life Technologies, Inc., Gaithersburg, MD), and 1 mm sodium pyruvate (Life Technologies, Inc.).
Reporter plasmids and expression vectors
pGL3 basic vector (Promega Corp., Madison, WI) and pGL3 promoter vector with NIS proximal promoter (−812 to −268) and NUE enhancer region (−9847 to −8968) are described elsewhere (8). The CREM-τ2α expression vector pRc/CMV was kindly provided by Dr. G. N. Europe-Finner (University of Newcastle upon Tyne, Newcastle upon Tyne, UK) (29).
Transient expression analysis
Transient transfection and luciferase assay analysis were performed as described (8), with minor modifications. Briefly, FRTL-5 cells were grown in 12-well plates to approximately 80% confluency, and BHP 2–7 cells were grown to approximately 50–60% confluency (BHP 2–7 cells grow faster). One hundred fifty nanograms of firefly luciferase reporter construct were transiently transfected with 10 ng pRL-CMV (Renilla luciferase expression plasmid; Promega) to normalize the transfection efficiency. The Diethylaminoethyl (DEAE)-Dextran transfection system (Promega) was used for the transient transfection study of FRTL-5 and BHP 2–7 cells with minor modifications of the manufacturer’s protocol. Briefly, the cells were exposed to 250 μl of a plasmid-DEAE-Dextran-PBS mixture containing 10 μg DEAE-Dextran for 20 min at 37 C in a CO2 incubator, followed by the addition of 1 ml fresh medium, and continued incubation for 2.5 h. Eighteen to 21 h later, the medium was replaced with 1 ml fresh medium with or without FSK as indicated in different experiments, followed by another 24-h incubation. Firefly and Renilla luciferase activities were determined 48–72 h after the beginning of transfection with the Dual-Luciferase Reporter Assay System (Promega). Measurements were made using a TD-20/20 luminometer (Turner Designs, Sunnyvale, CA). Cotransfection experiments with 150 ng expression vectors were added to the DNA-DEAE-Dextran-PBS mixture and performed as described previously.
Nuclear extracts
Nuclear extracts from FRTL-5 and BHP 2–7 cells were prepared as described (30). Briefly, the cells were grown to 80% confluency and scraped free from the flask bottom. After a slow centrifugation, the pellet was washed with PBS (pH 7.4), then resuspended in 100 μl modified buffer A [10 mm HEPES (pH 7.9), 10 mm KCl, 1.5 mm MgCl2, 300 mm sucrose, 0.05% NP40, Roche protease inhibitor cocktail (Roche Diagnostics, Indianapolis, IN), 0.5 mm dithiothreitol (DTT), 0.1 μg/ml leupeptin, 1 μg/ml pepstatin-A, and 0.5 mm phenylmethylsulfonylfluoride]. Cells were gently homogenized, and the suspension was put on ice for 10 min, followed by centrifugation at 300 rpm, 3 min, 4 C. The cytoplasmic fractions were collected, and the pellet was resuspended in 50 μl modified buffer B [20 mm HEPES (pH 7.9), 0.2 mm EDTA (pH 8), 20% glycerol, 420 mm NaCl, 0.5 mm DTT, 0.1 μg/ml leupeptin, 1 μg/ml pepstatin-A, 0.5 mm phenylmethylsulfonylfluoride, and Roche protease inhibitor cocktail]. After 30 min on ice, the suspension was centrifuged at 14,000 rpm, 15 min, 4 C, and the nuclear extract was collected. Nuclear extract was used in EMSAs and Western blots after the quantification of protein using the Bradford reagent (Bio-Rad Laboratories, Inc., Hercules, CA).
Western blot analysis
Nuclear extracts (∼15 ng) were electrophoresed on a 10% sodium dodecyl sulfate-polyacrylamide gel and transferred to a nitrocellulose membrane (Hybond-ECL; Amersham Biosciences, Buckinghamshire, UK). The membrane was incubated in a 3% enhanced chemiluminescence blocking buffer at least 1 h, washed two times in Tris-buffered saline with Tween 20 (TBST). The membrane was incubated with mouse monoclonal ATF-1 (25C10G) (Santa Cruz Biotechnology) overnight. The membrane was washed two times in TBST and then incubated with horseradish peroxidase-conjugated antimouse IgG (Promega) for 45 min. Membrane was washed for 1 h in TBST. The proteins were illuminated with the enhanced chemiluminescence advance Western blotting detection kit (Amersham Biosciences), exposed to film, and analyzed using an imaging system (Alpha Innotech Corp., San Leandro, CA). Membranes were stripped [Pierce (Rockford, IL) stripping buffer] and reblotted with mouse monoclonal α-tubulin (Santa Cruz Biotechnology), rabbit monoclonal Pax 8 (31), rabbit CREB (Sigma Chemicals), or rabbit CREB PSer133 (Sigma Chemicals) antibody and treated as described previously.
EMSA
EMSAs were performed as previously described (14,30,32). Briefly, the 4-pmol top strand of synthesized single-strand oligonucleotides was labeled with γ-32P ATP (MP Biomedicals, Solon, OH) by T4 polynucleotide kinase and purified using the QIAquick Nucleotide Removal Kit (QIAGEN, Inc., Valencia, CA). The 60 μl mixture containing 4 pmol kinased oligo mixed with 60 μl containing 6 pmol complementary strand in 20 mm Tris-HCl (pH 7.4), 50 mm NaCl, was annealed by rapid heating to 95 C and slow cooling to room temperature in a PCR thermocycler (MJ Research, Inc., Waltham, MA). The resulting annealed probes were used in all EMSAs performed. FRTL-5, BHP 2–7 nuclear extracts, or CREM purified protein (Santa Cruz Biotechnology) was incubated with or without competitor oligonucleotides in the following buffer K [4.5% glycerol, 60 mm KCl, 0.05 μg polydeoxyinosinic-deoxycctidylic acid, 0.33% NP40, 250 mm HEPES NaOH (pH 7.8), 50 mm MgCl2, 5 mm EDTA (pH 8), and 5 mm DTT] for 20 min at room temperature. Labeled probe was added, and incubation was continued for an additional 20 min at room temperature. DNA-protein complexes were separated on 5.3% native polyacrylamide gels at 4 C. Gels were dried and exposed to film overnight in the −70 C freezer, and analyzed using an imaging system (Alpha Innotech), or placed in a cassette overnight and analyzed on the storm 840 phosphoimager (General Electric Co., Fairfield, CT). Supershift assays were performed using 2 μg antibody to CREB 1 (CHEMICON International, Inc., Temecula, CA), CREB 1 (Sigma Chemicals), CREB PSer133 (Sigma Chemicals), CREM (X-12), ATF-1 (25C10G), ATF-1 (C41–5.1) (Santa Cruz Biotechnology), or CREMτ (33) mixed with nuclear extracts and buffer K before labeled probe addition, as described previously.
RT-PCR analysis
Experiments were performed as described previously (34). Briefly, total RNAs were prepared from FRTL-5 and BHP 2–7 cells cultured with or without 10 μm FSK at various time points and prepared using the RNeasy kit (QIAGEN). RNA was reversed transcribed into cDNA with Oligo(dT)20 using the SuperScript III First-Strand Synthesis System (Invitrogen, Carlsbad, CA). Primer design was performed using “Primer3”; for human and rat primers see Table 1. The PCR mixture contained 0.2 pmol 5′ and 3′ primers, one 20th cDNA with buffers from the Taq DNA polymerase kit (QIAGEN) for 27, 30, or 34 cycles using a PTC-200 PCR machine (MJ Research). Resulting products were run on a 1% agarose gel and visualized using an imaging system (Alpha Innotech).
Table 1.
Primers used for PCR amplification
| Forward (5′–3′) | Reverse (5′–3′) | |
|---|---|---|
| Human primers | ||
| hATF-1 | cacaagagtaccacgtcagagacag | atatgagaaatgtgagctccctgaa |
| hCREB 1 | cgagaaccagcagagtggaga | gcatagatacctgggctaatgtgg |
| hCREM Q1 | gaagaggctccccagctgtaa | ccatggctgtggtgtctgaa |
| hCREM Q2 | tgatggtgttcagggactgc | ggctgcctgggacaaagaac |
| hGAPDH | gagtcaacggatttggtcgta | catgggtggaatcatattgga |
| hPax8A | tcactcacccttcgccataaa | aaaggcggagctagataaagagga |
| Rat primers | ||
| rATF-1 | atggaagattcccacaagagtaaca | gacaacgatgacacctgatgga |
| rCREB | gacaaccagcagagtggagatg | atggatacctgggctaatgtgg |
| rCREM Q1 | ggatcaggcactggaagagg | cgatggatgtggtgtctgaa |
| rCREM Q2 | cagggactccaggcattaacaa | caaagaactgctgtgtgccatc |
| rGAPDH | ccagcaaggatactgagagcaag | tgttatggggtctgggatgg |
| rPax 8 | cctgacaccttccaatacacctct | ctggggtttcctgctttatgg |
GAPDH, Glyceraldehyde-3-phosphate dehydrogenase.
Results
FSK stimulation increases the CRE binding transcription factors ATF-1 and CREM activator in FRTL-5 cells
It is unclear which transcription factors are required for FSK-stimulated NUE activity. To address this, we evaluated several candidate transcription factors that may bind the CRE’ site and Pax8 element for their potential to be FSK stimulated. The most likely candidates to bind and activate these elements are CREB family basic region leucine zipper (bZIP) molecules and the paired domain transcription factor Pax 8. ATF-1, CREB, and CREM activators are the most common CREB family activators that bind to CRE’ sites. We designed primers to evaluate CREB 1, CREM, ATF-1, and Pax 8 mRNA expression levels specifically in rat (FRTL-5) cells. CREM splicing is complex, thus we created primers that evaluated both activating regions (Q1 and Q2) (35).
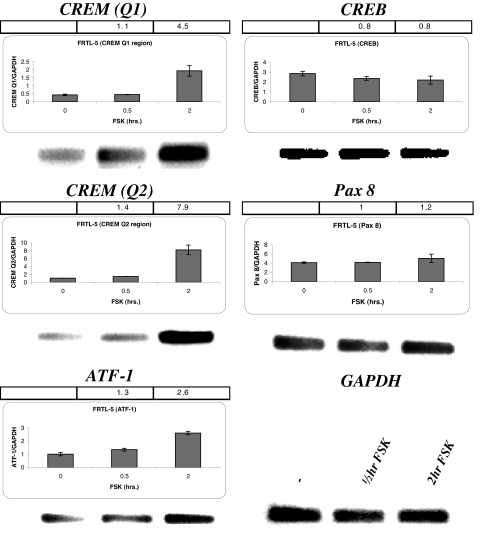
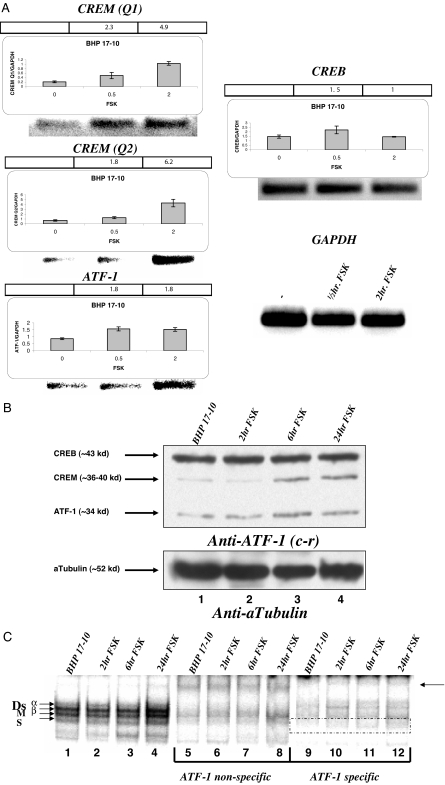
FRTL-5 cells were depleted of thyroid-stimulating hormone for 5–6 d to eliminate NIS gene expression, and then exposed to FSK to increase NIS mRNA levels. Presumably, this would raise transcription factors needed to stimulate NIS gene expression. FSK increased CREM activator (Q1 and Q2) mRNA expression 4.5 and 8-fold respectively (Fig. 1, top and middle left). ATF-1 mRNA expression also increased about 2.5-fold (Fig. 1, bottom left). In contrast, there was no detectable FSK stimulation of CREB and Pax 8 mRNA (Fig. 1, top and middle right) in FRTL-5 cells.
Figure 1.
PCR amplification effects of FSK on CREM activator, ATF-1, CREB, and Pax 8 mRNA expression in FRTL-5 cells. FRTL-5 cells were treated with 10 μm FSK for 0.5 and 2 h before collecting the mRNA and PCR amplification. Quantitative RT-PCR and PCR were performed for CREM Q1 (102 nt), CREM Q2 (106 nt), ATF-1 (82 nt), CREB (101 nt), and Pax8 (107 nt) denoted above each panel. The bar across the top of each panel is the fold difference of the FSK treated nuclear extract relative to the untreated nuclear extract. The y-axis of the bar graphs shows the amount of mRNA of the tested transcription factor relative to glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and the x-axis shows the time the cells were treated with FSK. Lower right, The gel below the graphs is the PCR products after 30 cycles of PCR amplification with glyceraldehyde-3-phosphate dehydrogenase included. Each experiment was done at least three times.
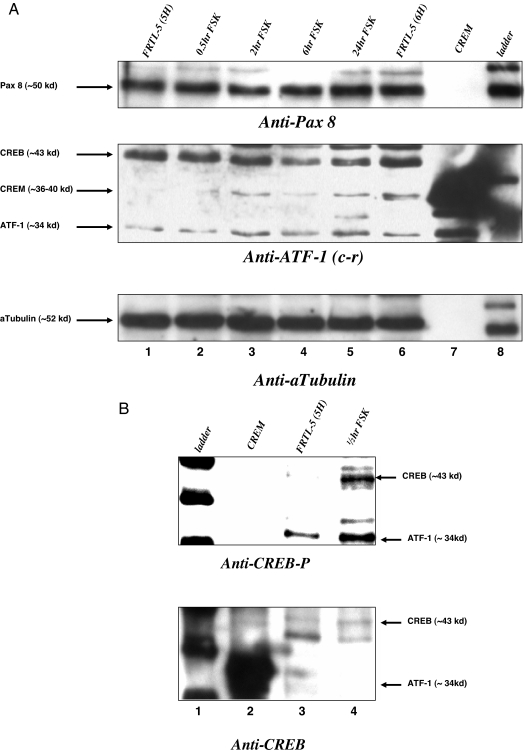
Nuclear extracts from FRTL-5 cells that were treated with FSK for 0.5, 2, 6, and 24 h were evaluated for candidate transcription factor protein expression. Figure 2A, top panel, shows that FRTL-5 cells grown without thyroid-stimulating hormone for 5 d (5H) have Pax 8 protein; treatment with FSK up to 24 h did not significantly increase Pax 8. Figure 2A, middle panel, is a blot with anti-ATF-1 (cross-reacting antibody) and shows that CREB protein does not change with FSK, whereas a CREM isoform and ATF-1 both increase with FSK between 2 and 24 h (lanes 3–5). There may be other CREM protein isoforms that we are unable to account for with our antibody. CREB protein is active when phosphorylated at serine 133 for up to 1-h FSK stimulation. Thus, we looked for FSK-stimulated modification of CREB using a phospho-CREB (CREB-P) antibody (Fig. 2B). Figure 2B, top, lane 3 vs. 4, shows that 0.5-h FSK stimulation increases CREB-P. Figure 2B, lane 2, contains commercial CREM protein that appears to be strongly detected with anti-CREB (bottom panel). In FRTL-5 cells, FSK stimulates CREM activator and ATF-1 mRNA and protein, as well as CREB phosphorylation.
Figure 2.
Western blot effects of FSK on CREM activator, ATF-1, CREB, CREB-P, and Pax 8 protein in FRTL-5 cells. A, FRTL-5 cells were treated with 10 μm FSK for 0.5, 2, 6, and 24 h before collecting the nuclear extract and performing Western blots with anti-Pax 8, anti-ATF-1 [cross-reacting (c-r) to CREB, CREM, and ATF-1], and anti-α-tubulin. Lanes 1–5 show increasing time of FSK treatment, lane 6 is nuclear extract from cells treated with thyroid-stimulating hormone, lane 7 is purified CREM protein, and lane 8 is a protein ladder. The same gel was stripped and reblotted. Bottom panel, The amounts of protein loaded in each lane are similar at about 10 μg, by evaluating α-tubulin. B, FRTL-5 cells were treated with 10 μm FSK for 30 min, and evaluated for CREB phosphorylation (top) and total CREB (bottom) using anti-CREB-P and anti-CREB. Lane 1 is the protein ladder, lane 2 is CREM purified protein, lane 3 is nuclear extract without FSK, and lane 4 is nuclear extract with FSK for 30 min.
Thyroid cancer cells (BHP 2–7) cannot increase ATF-1 and CREM activator, but FSK increases the Pax 8 transcription factor
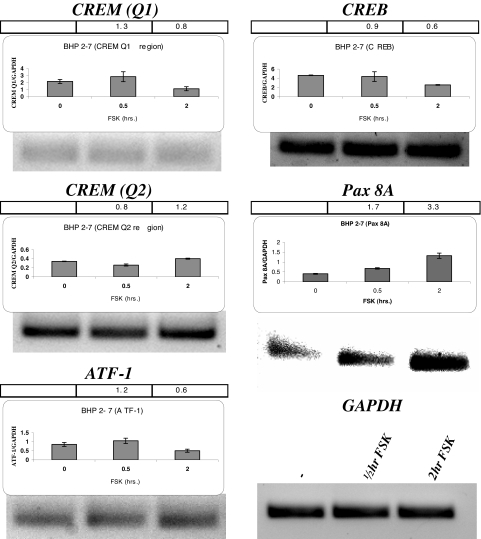
To understand the transcription factor differences between normal thyroid cells and thyroid cancer cells, we evaluated the transcription factors that are involved in BHP 2–7 cells. Figure 3 shows the mRNA expression of the NUE binding transcription factor candidates with FSK stimulation in BHP 2–7 cells. BHP 2–7 cells have a vastly different mRNA expression pattern from that of the FRTL-5 cells. CREM activator Q1 had low mRNA expression, even with four additional PCR amplification cycles, and Q1 and Q2 CREM activator regions did not change with FSK stimulation (Fig. 3, top left and middle left). In addition, ATF-1 required additional amplification cycles for a signal and was not affected by FSK (Fig. 3, bottom left). FSK did not stimulate CREB mRNA. The only FSK-stimulated mRNA effect in BHP 2–7 cells was a greater than 3-fold increase of Pax 8 mRNA (Fig. 3, middle right).
Figure 3.
PCR amplification effects of FSK on CREM activator, ATF-1, CREB, and Pax 8 mRNA expression in BHP 2–7 cells. BHP 2–7 cells were treated with 10 μm FSK for 0.5 and 2 h before collecting the mRNA and PCR amplification. Quantitative RT-PCR and PCR were performed for CREM Q1 (89 nt), CREM Q2 (128 nt), ATF-1 (71 nt), CREB (104 nt), and Pax8 (85 nt) denoted above each panel, and the representation is the same as described previously. The gel below the graphs is the PCR products after 30 cycles of PCR amplification with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) included (lower right), except CREM Q1 and ATF-1 are PCR products after 34 cycles, Each experiment was done at least three times.
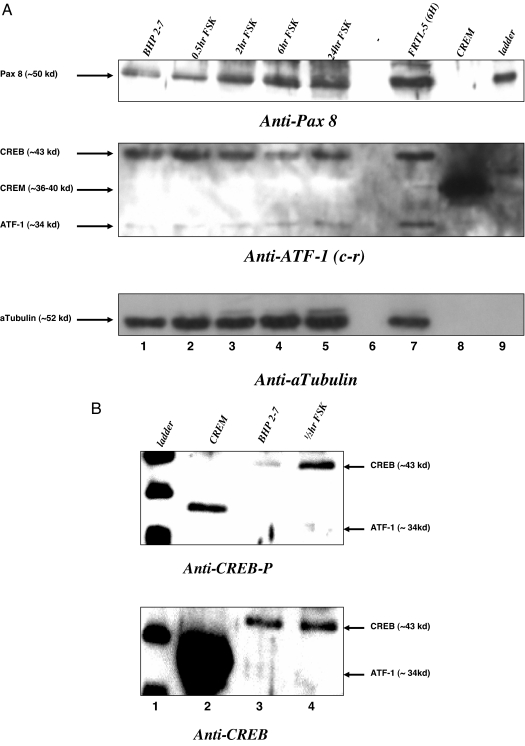
Nuclear extracts from BHP 2–7 cells that were treated with FSK for 0.5, 2, 6, and 24 h were evaluated for candidate transcription factor protein expression. Figure 4A, top panel, shows that FSK-treated BHP 2–7 nuclear extracts had increased Pax 8 protein, starting at 2 h and up to 24 h (Fig 4A, lanes 3–5). Figure 2A, middle panel, shows that the CREB protein did not change with FSK and that CREM was absent (Fig. 4A). ATF-1 showed a faint band that appears to be unchanged with FSK treatment (Fig. 4A). We evaluated CREB phosphorylation at 0.5-h FSK treatment in the BHP 2–7 nuclear extract (Fig. 4B) using CREB-P antibody. Figure 2B top, lane 3 vs. 4, shows that 0.5-h FSK stimulation increased CREB-P, and the total CREB remained unchanged (Fig. 4B, bottom, lane 3 vs. 4). In response to FSK treatment, BHP 2–7 cells are able to phosphorylate CREB protein and increase the amount of Pax 8 mRNA and protein but were defective in stimulating CREM activator and ATF-1.
Figure 4.
Western blot effects of FSK on CREM activator, ATF-1, CREB, CREB-P, and Pax 8 Protein in BHP 2–7 cells. A, BHP 2–7 cells were treated with 10 μm FSK for 0.5, 2, 6, and 24 h before collecting the nuclear extract and performing Western blots with anti-Pax 8, anti-ATF-1 [cross-reacting (c-r) to CREB, CREM, and ATF-1], and anti-α-tubulin. Lanes 1–5 show increasing time of FSK treatment, lane 6 is empty, lane 7 is nuclear extract from FRTL-5 cells treated with thyroid-stimulating hormone, lane 8 is purified CREM protein, and lane 9 is a protein ladder. The same gel was stripped and reblotted. Bottom panel, The amounts of protein loaded in each lane are similar at about 10 μg, by evaluating α-tubulin. B, BHP 2–7 cells were treated with 10 μm FSK for 30 min, and evaluated for CREB phosphorylation (top) and total CREB (bottom) using anti-CREB-P and anti-CREB. Lane 1 is the protein ladder, lane 2 is CREM purified protein, lane 3 is nuclear extract without FSK, and lane 4 is nuclear extract with FSK for 30 min.
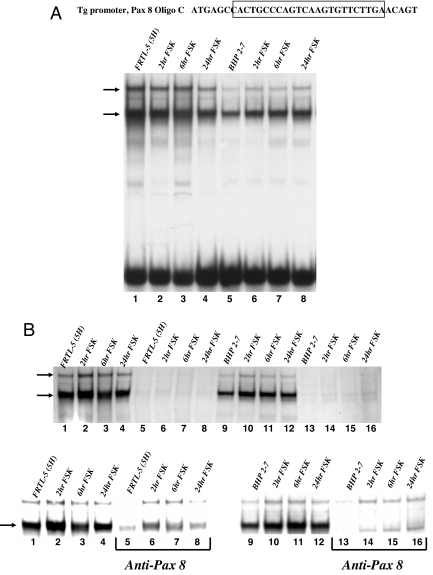
Characterization of complexes formed on Pax 8 and CRE’ sites to identify FSK-stimulated complex differences between FRTL-5 thyroid cells and thyroid cancer cells (BHP 2–7)
Pax 8 and CRE’ binding was assessed to compare the differences between FRTL-5 and BHP 2–7 complex formation, and to determine the transcription factors bound in these complexes. We examined nuclear extracts at various times of FSK treatment from FRTL-5 cells and BHP 2–7 cells using probes designed to optimize binding and complex specificity. Two major complexes form with the TG promoter Pax 8 binding site (Fig. 5A, arrows). Figures 2A and 4A show that both cells have Pax 8. Figure 5A (lanes 1–4) indicates that, with or without FSK, FRTL-5 cells have constant Pax 8 protein bound to the probe. BHP 2–7 cells show a gradual increase in the lower of the two complexes that is FSK dependent (Fig. 5A, top, lane 5 vs. 6–8). Both of these bands were specifically competed away with 100-fold excess cold probe (Fig. 5B, top). Pax 8 antibody diminished a much larger portion of the lower complex, suggesting that Pax 8 protein occupies this position. It appears that the FRTL-5 Pax 8 binding signals are stronger than the BHP 2–7 Pax 8 binding signals, (Fig. 5B, bottom panels). Thus, it is likely that anti-Pax 8 inhibition of the FRTL-5 complexes is not as complete as seen with the BHP 2–7 complexes. However, it is possible that an additional protein occupies the Pax 8 binding site in FRTL-5 nuclear extract that is absent in BHP 2–7 nuclear extract. In BHP 2–7 cells, the lower complex is enhanced with FSK, and thus an increase in Pax 8 protein results in an increase in Pax 8 bound to the probe (Fig. 5A).
Figure 5.
EMSA shows that FSK stimulates Pax 8 binding complexes in BHP 2–7 cells. A, Top is a representation of Pax 8 probe (designed from the TG promoter) used with the boxed region representing Pax 8 binding site. Lanes 1–4 are FRTL5 nuclear extract stimulated with FSK at 0, 2, 6, and 24 h. Lanes 5 and 6 are a repeat using BHP 2–7 nuclear extract. Arrows on the left represent the two major complexes bound to the Pax 8 probe. B, Top panel is a competition of the lanes above with 100-fold excess cold probe. FRTL-5 nuclear extract with FSK time increments in lanes 1–4; these were repeated with cold probe in lanes 5–8. The next eight lanes are repeats using BHP 2–7 nuclear extract. Arrows represent potential Pax 8 binding complexes. The bottom panel is organized as previously described except that FRTL-5 lanes 5–8 and BHP 2–7 lanes 9–16 are incubated with anti-Pax 8. Arrow represents Pax 8 binding complex.
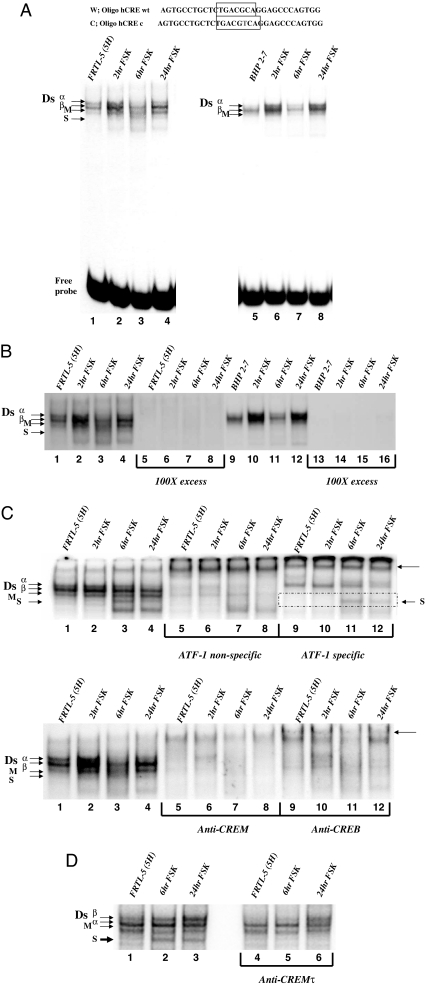
To assess possible differences in CRE’ complexes between the cell lines, we repeated the gel shifts using a CRE consensus probe. Figure 6A shows four previously described complexes (Dsα, Dsβ, M, and S) with FRTL-5 nuclear extract and three complexes (Dsα, Dsβ, and M) with BHP 2–7 nuclear extract (8). It is noticeable that FSK stimulation for 2–24 h greatly enhances the S complex in FRTL-5 (Fig. 6A, lane 1 vs. 2–4). All of these bands have specificity for the CRE probe (Fig. 6B). The S complex is enhanced with FSK stimulation and is absent in the BHP 2–7 lanes. This FSK-stimulated complex appears only in FRTL-5 cells, suggesting that the FSK-stimulated transcription factors, ATF-1 and CREM activator, may be components of the factor S complex.
Figure 6.
EMSA shows that FSK stimulated a CREM complex (factor S) on a CRE’ probe in FRTL-5 cells. A, Top shows the NUE CRE’ site compared with a CRE consensus probe, the CRE protein binding region is boxed. Left gel shows FRTL-5 nuclear extracts bound to the CRE consensus probe treated with FSK for 0, 2, 6, and 24 h (lanes 1–4). Complexes are designated on the left of the gel in order of migration, Ds a, Ds b, M and S, and free probe. The same orientation is used for BHP 2–7 nuclear extracts that lack the S complex. B, A competition of the lanes seen previously with 100-fold excess cold probe. Complexes are in the order as described previously with FRTL-5 lanes 5–8 repeated with cold probe and lanes 13–16 repeated with cold probe. C, FRTL-5 nuclear extracts are supershifted with various antibodies. Top panel, Lanes 5–8 are shifted with an ATF-1 cross-reacting antibody, and lanes 9–12 are shifted with an ATF-1 specific antibody (reacts with CREB, CREM, and ATF-1). The arrow on the right of the gel indicates supershift. The bottom panel is a repeat with lanes 5–8 incubated with anti-CREM and lanes 9–12 incubated with anti-CREB. D, FRTL-5 nuclear extracts are dissociated with CREMτ antibody. Lanes 1–3 are FRTL-5 extract without FSK, 6-h FSK treated, and 24-h FSK treated. Lanes 4–6 are in the same order with CREMτ antibody. c, Consensus; wt, wild type.
To identify the transcription factors associating with the FSK-stimulated complex, we tested antibodies that could inhibit and/or supershift the complex. We evaluated ATF-1 specific and cross-reacting antibodies to determine if ATF-1 is part of the factor S complex. Supershifts with the ATF-1 specific antibody suggest the Dsα complex (Fig. 6C, top, lanes 9–12) and S complex (Fig. 6C, top, lanes 11 and 12, boxed region) are still intact, whereas most complexes are sensitive to the ATF-1 cross-reacting antibody (Fig. 6C, top, lanes 5–8). It appears that the M complex is mildly resistant to the ATF-1 cross-reacting antibody. We further tested two other CREB family cross-reacting antibodies, to CREB and CREM, to verify that the factor S complex consists of CREB family proteins. Both cross-reacting antibodies disrupted most of the complexes, including the factor S complex (Fig. 6C, bottom, lanes 5–12). A similar pattern of supershifts was seen with the BHP 2–7 nuclear extracts. ATF-1 specific antibody did not alter the Dsα complex, but all other complexes were disrupted with ATF-1, CREB, or CREM cross-reacting antibodies (data not shown). Three CREB family cross-reacting antibodies, but not ATF-1 specific antibody, shifted the factor S complex. CREM, but not CREB, and the factor S complex are stimulated by FSK treatment, suggesting that CREM and the factor S complex share features that link their appearance in FRTL-5 cells.
To determine if the FSK-stimulated S complex consists of CREM activator, we performed gel shifts using CREMτ antibody, specific to all of the CREM isoforms (33). Using FRTL-5 nuclear extracts from 5H cells, 6-h FSK, and 24-h FSK, we evaluated complex dissociation with the CREMτ antibody (Fig. 6D). The 6- and 24-h FSK extracts have the FSK-stimulated S complex that is dissociated with the CREMτ antibody (Fig. 6D, compare lanes 2 and 3 with lanes 5 and 6). CREMτ specific antibody strongly dissociated the FSK-stimulated S complex, strengthening the argument that CREM is, at least, part of this important complex needed for full activation of the NUE and NIS expression.
BRAF (V600E) thyroid cancer cell line retains CREM activator expression and FSK-stimulated S complex
A different thyroid papillary cancer cell line that has a BRAF (V600E) mutation, BHP 17–10, was studied for the presence of CREM activator and FSK-stimulated S complex. Figure 7A shows the mRNA expression of the NUE binding transcription factor candidates with FSK stimulation in BHP 17–10 cells. BHP 17–10 cells have a very similar mRNA expression pattern to FRTL-5 cells, except for the Pax 8 expression. Pax 8 was absent in the BHP 17–10 BRAF cells (no signal seen). However, CREM Q1 and Q2 activator mRNA are both stimulated by FSK (Fig. 7A, top left and middle left). In addition, ATF-1 was FSK stimulated (Fig. 7A, bottom left), and FSK did not stimulate CREB mRNA (Fig. 7A, right).
Figure 7.
BHP 17–10 cells have FSK-stimulated CREM activator and S complex but are missing the Pax 8 transcription factor. A, BHP 17–10 cells were treated with 10 μm FSK for 0.5 and 2 h before collecting the mRNA and PCR amplification. Quantitative RT-PCR and PCR were performed for CREM Q1 (89 nt), CREM Q2 (128 nt), ATF-1 (71 nt), and CREB (104 nt) denoted above each panel, and the representation is the same as described previously. The gel below the graphs is the PCR products after 30 cycles of PCR amplification with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) included (lower right). Each experiment was done at least three times. B, BHP 17–10 cells were treated with 10 μm FSK for 2, 6, and 24 h before collecting the nuclear extract and performing Western blots with anti-ATF-1 [cross-reacting (c-r) to CREB, CREM, and ATF-1], and anti-α-tubulin. Lanes 1–4 show increasing time of FSK treatment. The same gel was stripped and reblotted. The bottom panel shows the amounts of protein loaded in each lane are similar at about 10 μg, by evaluating α-tubulin. C, BHP 17–10 nuclear extracts bound to the CRE consensus probe treated with FSK for 0, 0.5, 2, 6, and 24 h (lanes 1–5). Complexes are designated on the left of the gel in order of migration, Ds a, Ds b, M, and S. Lanes 6–10 are shifted with anti-CREM cross-reacting antibody. The arrow on the right of the gel indicates supershift.
Nuclear extracts from BHP 17–10 cells that were treated with FSK for, 2, 6, and 24 h were evaluated for candidate transcription factor protein expression. Figure 7B, top panel, shows that FSK-treated BHP 17–10 nuclear extracts had increased CREM and ATF-1 protein at 6 and 24 h (lanes 3–4), and there was no visible change of CREB protein. Pax 8 protein did not show any signal (data not shown).
Because BHP 17–10 did not have Pax 8 mRNA and protein, only the CRE’ probe was evaluated in gel shift assays. Figure 7C shows that the BRAF cells have all four major complexes that are seen in normal thyroid cells (Dsα, Dsβ, M, and S). FSK treatment appears to enhance the S band slightly more than the other bands, but Dsα was weaker in all lanes relative to FRTL-5 shifts (Fig. 7C, lanes 1–4). The BHP 17–10 nuclear extracts were tested with ATF-1 cross-reacting and specific antibodies. ATF-1 cross-reacting antibody supershifted all complexes; however, the M complex appears to be more resistant to this antibody (Fig. 7C, lanes 5–8). ATF-1 specific antibody demonstrates a similar pattern of supershifts to FRTL-5 (Fig. 6C); Dsα and S complexes are more resistant than the other complexes (Fig. 7C, lanes 9–12). Thus, CREM activator and FSK-stimulated S complex are both present in the BRAF cells, as in the FRTL-5 thyroid cells.
FSK partially stimulates NUE activity in BHP 2–7 thyroid cancer cells
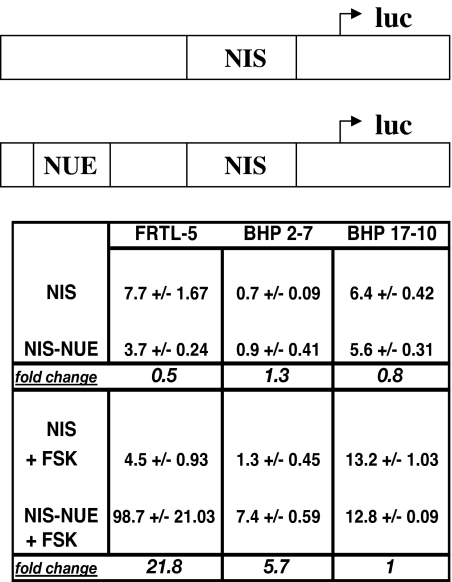
RET-PTC 1 thyroid cancer cells have not been shown to have NUE activity because they lack thyroid-stimulating hormone receptor (28). However, by using FSK we are able to bypass the need for thyroid-stimulating hormone receptor in thyroid cancer cells. Thus, we analyzed the NUE activity in FRTL-5 cells, BHP 2–7 cells, and BHP 17–10 cells with and without FSK. Prior work showed that FRTL-5 cells, depleted of thyroid-stimulating hormone, did not have NUE activity. However, adding back thyroid-stimulating hormone or FSK greatly enhanced luciferase expression through the NUE (8). We transiently cotransfected plasmids (pGL3 firefly luciferase) with the NIS proximal promoter alone or with the NUE attached (Fig. 8, top) along with a control plasmid (pRL-CMV Renilla luciferase) into FRTL-5 cells, BHP 2–7 cells, and BHP 17–10 cells, and evaluated the firefly luciferase normalized to the Renilla luciferase. FRTL-5 cells, when cotransfected with the NUE and with the addition of FSK, increased expression over 20-fold (Fig. 8, left column), as previously reported (8). BHP 2–7 cells showed more than a 5-fold increase in NUE activity in the presence of FSK (Fig. 8, middle column). BHP 2–7 cells had a much lower basal level of transcription than FRTL-5 cells (Fig. 8, 0.7 vs. 7.7) and an overall lower activation (Fig. 8, 7.4 vs. 98.7). BHP 17–10 cells maintained a high level of basal expression but did not respond to FSK (Fig. 8, right column). This is likely because BHP 17–10 cells lack Pax 8 that is needed for NUE activation. BHP 2–7 cells respond to FSK treatment and show NUE activity, whereas BHP 17–10 did not.
Figure 8.
The NUE is FSK stimulated in BHP 2–7 cells. Top, Plasmid constructs showing the proximal promoter NIS and enhancer NUE upstream of firefly luciferase reporter gene. FRTL-5 cells were cultured for 6 d in 5H medium, transfected with each luciferase reporter construct, and cultured an additional 18–21 h in 5H medium, treated with or without 10 μm FSK for 36–48 h. BHP 2–7 cells and BHP 17–10 cells were cultured in RPMI medium with or without 10 μm FSK. Results are expressed as a percentage of firefly luciferase to Renilla luciferase. A pRL-CMV vector was cotransfected to normalize for transfection efficiency. Values are the means of two to three separate experiments. Top of chart indicates the cells used, and along side the chart indicates the plasmid used. Upper section of chart is without FSK, and lower section is with FSK.
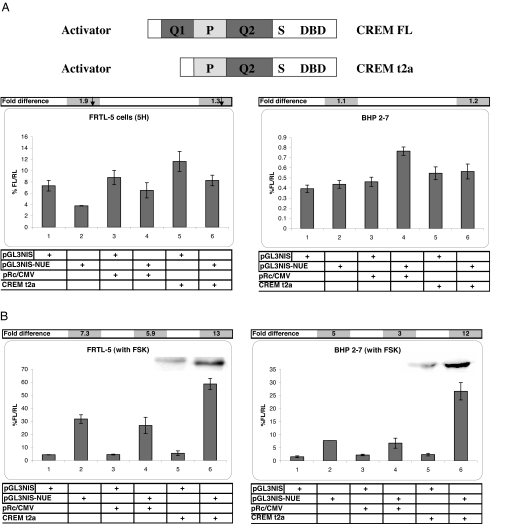
CREM activator enhances NUE activity in FSK-stimulated thyroid cancer cells
FSK induced CREM activator in normal thyroid cells, but not in thyroid cancer cells. It appears that the FSK-stimulated complex found on the CRE’ site consists of CREM activator. Thus, we cotransfected CREM τ2α (activator, Fig. 9A, top) into both FRTL-5 cells and BHP 2–7 cells to evaluate functionality of CREM activator on the NUE. We did not find activity with CREM activator in FRTL-5 cells or BHP 2–7 cells without FSK stimulation (Fig. 9A, bottom), even though CREM was expressed (data not shown). In FRTL-5 cells this may be due to low ATF-1 expression and/or CREB/CREM modification. In BHP 2–7 cells, this is likely due to low Pax 8 levels. In contrast, when both cell lines were cotransfected with CREM τ2α and FSK-stimulated, the NUE activity was enhanced (Fig. 9B, both panels, lanes 5 vs. 6). The FRTL-5 cells and BHP 2–7 cells had NUE activity close to 2-fold higher with CREM τ2α than without it. The insets of Fig. 9B show CREM τ2α protein overexpressed at approximately 39 kDa in both cell lines, as previously reported (29). This indicates that CREM activator is functional on the NUE during FSK treatment. To control for NUE activity, we tested a CREB expression vector in both cell lines with FSK, and there was a very slight increase in NUE activity (data not shown). The lack of a major increase in NUE activity is likely because both cell lines are saturated with endogenous CREB.
Figure 9.
CREM τ2α activator has activity on the NUE in both FRTL-5 cells and BHP 2–7 cells. A, Top, representation of CREM τ2α activator expression vector construct compared with CREM τ (full length) activator. Bottom left panel shows FRTL-5 cells without and with NUE in lanes 1 and 2, without NUE cotransfected with empty vector and CREM τ2α in lanes 3 and 4, and with NUE cotransfected with empty vector and CREM τ2α in lanes 5 and 6. Above the bottom right panel is the same orientation with BHP 2–7 cells. The bar across the top of each panel is the fold difference with and without the NUE in the absence and presence of expression vector. FRTL-5 (5H) and unstimulated BHP 2–7 cells with either an empty expression vector or CREM τ2α activator. Percentage of firefly luciferase to Renilla luciferase and sd values of FRTL-5 cells (lanes 1–6): 7.34 ± 0.94, 3.79 ± 0.05, 8.80 ± 1.26, 6.52 ± 1.35, 11.63 ± 1.76, and 8.27 ± 0.94. Percentage of firefly luciferase to Renilla luciferase and sd values of BHP 2–7 cells (lanes 1–6): 0.39 ± 0.04, 0.44 ± 0.04, 0.46 ± 0.05, 0.76 ± 0.04, 0.55 ± 0.06, and 0.56 ± 0.07. B, Panels have the same orientation as described previously, except both cell lines were treated with 10 μm FSK. Insets are Western blots of nuclear extracts that were cotransfected with expression vectors and visualized with CREM antibody. Insets are placed above the bars that represent the same conditions used for both sets of data. FSK-stimulated FRTL-5 cells and FSK-stimulated BHP 2–7 cells with empty expression vector or CREM τ2α activator. Percentage of firefly luciferase to Renilla luciferase and sd values of FRTL-5 (left panel lanes 1–6): 4.38 ± 0.13, 31.77 ± 3.31, 4.52 ± 0.36, 26.86 ± 5.94, 5.52 ± 1.79, and 58.60 ± 4.24. Percentage of firefly luciferase to Renilla luciferase and sd values for BHP 2–7 (right panel lanes 1–6): 1.54 ± 0.34, 7.74 ± 0.01, 2.22 ± 0.29, 6.75 ± 1.91, 2.36 ± 0.48, and 26.61 ± 3.31. Basic pGL3 vector was used in all experiments with a value between 0 and 0.09 activity.
Discussion
We characterized Pax 8 and CRE’ element binding transcription factors that are stimulated by FSK in FRTL-5, BHP 2–7, and BHP 17–10 cell lines, and correlated the data with NUE activity leading to NIS gene expression. The main observation of the current data shows that CREM activator is FSK induced, and contributes to the binding and activity of the NUE needed for NIS expression. In addition, the data elucidate the differences in transcription factors that are stimulated and modified by FSK in all three cell lines. FSK stimulates CREM activator and ATF-1 mRNA and protein in FRTL-5 and BHP 17–10 cells; in contrast, FSK stimulates Pax 8 protein in BHP 2–7 cells. FSK-stimulated FRTL-5 and BHP 17–10 nuclear extracts have an additional CRE’ binding complex (factor S) that is absent in BHP 2–7 cells. The finding that FSK stimulates CREM activator together with the gel supershift data suggests that factor S consists of CREM activator. The FSK-stimulated factor S complex appears to provide strong NUE activity for NIS gene expression when Pax 8 is present. However, in BHP 2–7 cells, FSK mildly stimulates the NUE without the factor S complex by increasing the Pax 8 protein to DNA complex. Furthermore, we demonstrate that CREM activator can increase NUE activity in both FRTL-5 and BHP 2–7 cell lines. The results give insight into the importance of the factor S complex in NUE activation.
FSK-stimulated complex is required for strong NUE activity
The data show that FSK-stimulated FRTL-5 cells, BHP 2–7 cells, and BHP 17–10 cells express different patterns of transcription factors that influence NUE activation. FRTL-5 and BHP 17–10 nuclear extracts show a FSK-stimulated increase in a complex (factor S) that was defined previously (8). The factor S complex was supershifted with cross-reacting antibodies ATF-1, CREM and CREB, but not with ATF-1 specific antibody (Fig. 6C, lanes 11 and 12 boxed region and Fig. 7C, lanes 10–12 boxed region). More specifically, the CREMτ antibody dissociated the S complex (Fig. 6D). The data indicate that the FSK-stimulated factor S complex consists of CREM activator. FSK stimulation and the factor S complex are required for NUE activation of NIS gene expression. However, NUE activation may include other unknown FSK-stimulated activators in FRTL-5 cells.
In contrast, BHP 2–7 nuclear extracts showed FSK-stimulated increases in Pax 8 complex in EMSA. FSK stimulates Pax 8 protein and Pax 8 EMSA complexes, strongly suggesting that BHP 2–7 cells use Pax 8 to gain minimum NUE activity. Similarly, De Vita et al. (36) reported that a RET/PTC1 rearrangement cell line has reduced levels of Pax 8 protein. The lack of factor S complex probably prevents BHP 2–7 cells from gaining full NUE activation and, therefore, NIS gene expression. It is possible that NIS endogenous expression in BHP 2–7 cells is suppressed by transactive repressor(s) acting on the NIS promoter region (37). FSK-treated BHP 17–10 cells were unable to gain NUE activation, but this was expected because they lack Pax 8.
CRE sites can potentially bind to a variety of bZIP molecules. CRE sites are preferentially bound by homodimers and heterodimers of CREB family proteins, and bear similarity to activator protein (AP)-1 sites that are strongly bound by dimers through the FOS/JUN subfamily of bZIP molecules. The CREB family of bZIP molecules has cross talked and heterodimerized with the FOS/JUN bZIP molecules (24,38,39). Chun et al. (16) demonstrated that cJun and c-fos bZIP molecules bind the CRE site of the rat NUE by supershift analysis, even though these proteins were not implicated in binding to the CRE’ site in the human NUE (8). It is possible that the M complex, which is relatively stable in the presence of CREB family cross-reacting antibodies, consists of AP-1 binding protein or unknown proteins. Thus, we cannot exclude the possibility that AP-1 binding proteins or other unknown proteins contribute to the stimulation of the NUE.
NUE has activity in thyroid cancer cells, and CREM τ2α activator enhances NUE activity in both FRTL-5 cells and BHP 2–7 cells
It has been well documented that FSK stimulates NUE activity in FRTL-5 cells. However, there was no available evidence of FSK-stimulated NUE activity in BHP 2–7 cells. We provide evidence that the NUE is active in BHP 2–7 cells (Fig. 8), but the FSK-stimulated activity is significantly lower than that in FRTL-5 cells. We evaluated the effects of adding a CREM activator (CREM τ2α) in both cell lines with and without FSK. Only with FSK treatment did both cell lines respond positively to CREM overexpression by doubling expression from the NUE. This signifies the productive enhancement of NUE activity through CREM activator interaction. It may be that FSK-stimulated factors are needed to interact with CREM activator for optimal NUE activity. Alternatively, FSK stimulation of CREM phosphorylation may be required for activation of the NUE. It is presumed that CREM is phosphorylated at serine 117 by protein kinase A, similar to CREB phosphorylation. However, it is generally believed that CREM may be able to bypass phosphorylation and still interact with CREB binding protein (CBP) to activate gene expression (25). In either case, CREM activator clearly enhances NUE activity in BHP 2–7 cells, allowing for potential NIS gene expression to occur.
Implications for activating NIS gene expression through an active NUE in cancer cells
BHP 2–7 was reported to have a RET-PTC 1 rearrangement (40,41), and lacked NIS and thyroid-stimulating hormone receptor mRNA analyzed by Northern blot (28,30). Thyroid-stimulating hormone receptor is absent in the majority of human thyroid cancer cell lines, but its expression is often maintained in cancer tissue. We report here that basal level transcription of the NIS proximal promoter in BHP 2–7 cells appears to be significantly lower than in FRTL-5 cells. FSK stimulation of Pax 8 increases NUE activity in BHP 2–7 cells up to the basal level of FRTL-5 cells, but this is not adequate for NIS gene expression. BHP 2–7 cells do not transport iodide but can do so if a NIS expression vector is transiently introduced into the cell (30). Thus, BHP 2–7 cells are capable of translocating an active NIS protein to the membrane, FSK-stimulated NUE activity, coupled with increased CRE’ binding transcription factors, may enhance NIS gene expression to a level consistent with providing BHP 2–7 cells with enough active NIS protein to transport iodide. By understanding the mechanisms responsible for NIS gene expression, we may be able to induce NIS gene expression and iodide uptake in various cancer cells, thus allowing for effective radioiodide therapy.
Acknowledgments
We thank Drs. Gregory Brent and Takahiko Kogai for kindly providing the human and rat glyceraldehyde-3-phosphate dehydrogenase and Na+/I− symporter primers, and for enlightening discussions. We also thank Dr. G. N. Europe-Finner for kindly providing the cAMP response element modulator expression vector, Dr. Peter Kopp for kindly providing the cAMP response element binding protein expression vector, Dr. Paolo Sassone-Corsi for kindly providing the cAMP response element modulator τ-antibody, and Dr. Katsumi Taki for helpful discussions.
Footnotes
This work was supported by Merit Award Research Funds from the Department of Veterans Affairs (to J.M.H.).
Disclosure Statement: The authors have nothing to disclose.
First Published Online January 17, 2008
Abbreviations: AP, Activator protein; ATF, activating transcription factor; bZIP, basic region leucine zipper; CRE, cAMP response element; CREB, cAMP response element binding protein; CREB-P, phospho-cAMP response element binding protein; CREM, cAMP response element modulator; DEAE, diethylaminoethyl; DTT, dithiothreitol; 5H, five hormone mixture; FSK, forskolin; NIS, sodium/iodide symporter; NUE, sodium/iodide symporter upstream enhancer; RET-PTC, rearranged in transformation/papillary thyroid carcinoma; TBST, Tris-buffered saline with Tween 20.
References
- Klein M, Aubert V, Weryha G, Leclere J 1996 [Classification and epidemiology of thyroid tumors]. Rev Prat 46:2288–2295 (French) [PubMed] [Google Scholar]
- Hundahl SA, Cady B, Cunningham MP, Mazzaferri E, McKee RF, Rosai J, Shah JP, Fremgen AM, Stewart AK, Holzer S 2000 Initial results from a prospective cohort study of 5583 cases of thyroid carcinoma treated in the united states during 1996. U.S. and German Thyroid Cancer Study Group. An American College of Surgeons Commission on Cancer Patient Care Evaluation study. Cancer 89:202–217 [DOI] [PubMed] [Google Scholar]
- Shen DH, Kloos RT, Mazzaferri EL, Jhian SM 2001 Sodium iodide symporter in health and disease. Thyroid 11:415–425 [DOI] [PubMed] [Google Scholar]
- Kogai T, Curcio F, Hyman S, Cornford EM, Brent GA, Hershman JM 2000 Induction of follicle formation in long-term cultured normal human thyroid cells treated with thyrotropin stimulates iodide uptake but not sodium/iodide symporter messenger RNA and protein expression. J Endocrinol 167:125–135 [DOI] [PubMed] [Google Scholar]
- Saito T, Endo T, Kawaguchi A, Ikeda M, Nakazato M, Kogai T, Onaya T 1997 Increased expression of the Na+/I- symporter in cultured human thyroid cells exposed to thyrotropin and in Graves’ thyroid tissue. J Clin Endocrinol Metab 82:3331–3336 [DOI] [PubMed] [Google Scholar]
- Kogai T, Endo T, Saito T, Miyazaki A, Kawaguchi A, Onaya T 1997 Regulation by thyroid-stimulating hormone of sodium/iodide symporter gene expression and protein levels in FRTL-5 cells. Endocrinology 138:2227–2232 [DOI] [PubMed] [Google Scholar]
- Weiss SJ, Philp NJ, Ambesi-Impiombato FS, Grollman EF 1984 Thyrotropin-stimulated iodide transport mediated by adenosine 3′,5′-monophosphate and dependent on protein synthesis. Endocrinology 114:1099–1107 [DOI] [PubMed] [Google Scholar]
- Taki K, Kogai T, Kanamoto Y, Hershman JM, Brent GA 2002 A thyroid-specific far-upstream enhancer in the human sodium/iodide symporter gene requires Pax-8 binding and cyclic adenosine 3′,5′-monophosphate response element-like sequence binding proteins for full activity and is differentially regulated in normal and thyroid cancer cells. Mol Endocrinol 16:2266–2282 [DOI] [PubMed] [Google Scholar]
- Venkateswaran A, Marsee DK, Green SH, Jhiang SM 2004 Forskolin, 8-Br-3′,5′-cyclic adenosine 5′-monophosphate, and catalytic protein kinase A expression in the nucleus increase radioiodide uptake and sodium/iodide symporter protein levels in RET/PTC1-expressing cells. J Clin Endocrinol Metab 89:6168–6172 [DOI] [PubMed] [Google Scholar]
- Dulgeroff AJ, Hershman JM 1994 Medical therapy for differentiated thyroid carcinoma. Endocr Rev 15:500–515 [DOI] [PubMed] [Google Scholar]
- Ryu KY, Tong Q, Jhiang SM 1998 Promoter characterization of the human Na+/I- symporter. J Clin Endocrinol Metab 83:3247–3251 [DOI] [PubMed] [Google Scholar]
- Tong Q, Ryu KY, Jhiang SM 1997 Promoter characterization of the rat Na+/I- symporter gene. Biochem Biophys Res Commun 239:34–41 [DOI] [PubMed] [Google Scholar]
- Endo T, Kaneshige M, Nakazato M, Ohmori M, Harii N, Onaya T 1997 Thyroid transcription factor-1 activates the promoter activity of rat thyroid Na+/I- symporter gene. Mol Endocrinol 11:1747–1755 [DOI] [PubMed] [Google Scholar]
- Ohmori M, Endo T, Harii N, Onaya T 1998 A novel thyroid transcription factor is essential for thyrotropin-induced up-regulation of Na+/I- symporter gene expression. Mol Endocrinol 12:727–736 [DOI] [PubMed] [Google Scholar]
- Ohno M, Zannini M, Levy O, Carrasco N, di Lauro R 1999 The paired-domain transcription factor Pax8 binds to the upstream enhancer of the rat sodium/iodide symporter gene and participates in both thyroid-specific and cyclic-AMP-dependent transcription. Mol Cell Biol 19:2051–2060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun JT, Di Dato V, D’Andrea B, Zannini M, Di Lauro R 2004 The CRE-like element inside the 5′-upstream region of the rat sodium/iodide symporter gene interacts with diverse classes of b-Zip molecules that regulate transcriptional activities through strong synergy with Pax-8. Mol Endocrinol 18:2817–2829 [DOI] [PubMed] [Google Scholar]
- Jhiang SM 2000 Regulation of sodium/iodide symporter. Rev Endocr Metab Disord 1:205–215 [DOI] [PubMed] [Google Scholar]
- Fabbro D, Pellizzari L, Mercuri F, Tell G, Damante G 1998 Pax-8 protein levels regulate thyroglobulin gene expression. J Mol Endocrinol 21:347–354 [DOI] [PubMed] [Google Scholar]
- Lalli E, Sassone-Corsi P 1994 Signal transduction and gene regulation: the nuclear response to cAMP. J Biol Chem 269:17359–17362 [PubMed] [Google Scholar]
- Rosenberg D, Groussin L, Jullian E, Perlemoine K, Bertagna X, Bertherat J 2002 Role of the PKA-regulated transcription factor CREB in development and tumorigenesis of endocrine tissues. Ann NY Acad Sci 968:65–74 [DOI] [PubMed] [Google Scholar]
- Yamamoto KK, Gonzalez GA, Biggs 3rd WH, Montminy MR 1988 Phosphorylation-induced binding and transcriptional efficacy of nuclear factor CREB. Nature 334:494–498 [DOI] [PubMed] [Google Scholar]
- Gonzalez GA, Montminy MR 1989 Cyclic AMP stimulates somatostatin gene transcription by phosphorylation of CREB at serine 133. Cell 59:675–680 [DOI] [PubMed] [Google Scholar]
- Meyer TE, Habener JF 1992 Cyclic AMP response element binding protein CREB and modulator protein CREM are products of distinct genes. Nucleic Acids Res 20:6106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hai T, Curran T 1991 Cross-family dimerization of transcription factors Fos/Jun and ATF/CREB alters DNA binding specificity. Proc Natl Acad Sci USA 88:3720–3724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Cesare D, Fimia GM, Sassone-Corsi P 1999 Signaling routes to CREM and CREB: plasticity in transcriptional activation. Trends Biochem Sci 24:281–285 [DOI] [PubMed] [Google Scholar]
- Ambesi-Impiombato FS, Parks LA, Coon HG 1980 Culture of hormone-dependent functional epithelial cells from rat thyroids. Proc Natl Acad Sci USA 77:3455–3459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta K, Pang XP, Berg L, Hershman JM 1996 Antitumor actions of cytokines on new human papillary thyroid carcinoma cell lines. J Clin Endocrinol Metab 81:2607–2612 [DOI] [PubMed] [Google Scholar]
- Ohta K, Pang XP, Berg L, Hershman JM 1997 Growth inhibition of new human thyroid carcinoma cell lines by activation of adenylate cyclase through the β-adrenergic receptor. J Clin Endocrinol Metab 82:2633–2638 [DOI] [PubMed] [Google Scholar]
- Tyson-Capper AJ, Bailey J, Krainer AR, Robson SC, Europe-Finner GN 2005 The switch in alternative splicing of cyclic AMP-response element modulator protein CREMτ2α (activator) to CREMα (repressor) in human myometrial cells is mediated by SRp40. J Biol Chem 280:34521–34529 [DOI] [PubMed] [Google Scholar]
- Kogai T, Hershman JM, Motomura K, Endo T, Onaya T, Brent GA 2001 Differential regulation of the human sodium/iodide symporter gene promoter in papillary thyroid carcinoma cell lines and normal thyroid cells. Endocrinology 142:3369–3379 [DOI] [PubMed] [Google Scholar]
- Ohmori M, Harii N, Endo T, Onaya T 1999 Tumor necrosis factor-α regulation of thyroid transcription factor-1 and Pax-8 in rat thyroid FRTL-5 cells. Endocrinology 140:4651–4658 [DOI] [PubMed] [Google Scholar]
- Fenton MS, Gralla JD 2001 Function of the bacterial TATAAT-10 element as single-stranded DNA during RNA polymerase isomerization. Proc Natl Acad Sci USA 98:9020–9025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmas V, Laoide BM, Masquilier D, de Groot RP, Foulkes NS, Sassone-Corsi P 1992 Alternative usage of initiation codons in mRNA encoding the cAMP-responsive-element modulator generates regulators with opposite functions. Proc Natl Acad Sci USA 89:4226–4230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogai T, Kanamoto Y, Li AI, Che LH, Ohashi E, Taki K, Chandraratna RA, Saito T, Brent GA 2005 Differential regulation of sodium/iodide symporter gene expression by nuclear receptor ligands in MCF-7 breast cancer cells. Endocrinology 146:3059–3069 [DOI] [PubMed] [Google Scholar]
- Uyttersprot N, Miot F 1997 Dog CREM transcription factors: cloning, tissue distribution, and identification of new isoforms. Biochem Biophys Res Commun 237:74–78 [DOI] [PubMed] [Google Scholar]
- De Vita G, Zannini M, Cirafici AM, Melillo RM, Di Lauro R, Fusco A, Santoro M 1998 Expression of the RET/PTC1 oncogene impairs the activity of TTF-1 and Pax-8 thyroid transcription factors. Cell Growth Differ 9:97–103 [PubMed] [Google Scholar]
- Li W, Venkataraman GM, Ain KB 2007 Protein synthesis inhibitors, in synergy with 5-azacytidine, restore sodium/iodide symporter gene expression in human thyroid adenoma cell line, KAK-1, suggesting trans-active transcriptional repressor. J Clin Endocrinol Metab 92:1080–1087 [DOI] [PubMed] [Google Scholar]
- Hai TW, Liu F, Coukos WJ, Green MR 1989 Transcription factor ATF cDNA clones: an extensive family of leucine zipper proteins able to selectively form DNA-binding heterodimers. Genes Dev [Erratum (1990) 4:682] 3:2083–2090 [DOI] [PubMed] [Google Scholar]
- Masquilier D, Sassone-Corsi P 1992 Transcriptional cross-talk: nuclear factors CREM and CREB bind to AP-1 sites and inhibit activation by Jun. J Biol Chem 267:22460–22466 [PubMed] [Google Scholar]
- Vitagliano D, Carlomagno F, Motti ML, Viglietto G, Nikiforov YE, Nikiforova MN, Hershman JM, Ryan AJ, Fusco A, Melillo RM, Santoro M 2004 Regulation of p27Kip1 protein levels contributes to mitogenic effects of the RET/PTC kinase in thyroid carcinoma cells. Cancer Res 64:3823–3829 [DOI] [PubMed] [Google Scholar]
- Xu J, Moatamed F, Caldwell JS, Walker JR, Kraiem Z, Taki K, Brent GA, Hershman JM 2003 Enhanced expression of nicotinamide n-methyltransferase in human papillary thyroid carcinoma cells. J Clin Endocrinol Metab 88:4990–4996 [DOI] [PubMed] [Google Scholar]