Abstract
Trichodiene synthase from Fusarium sporotrichioides contains two metal ion-binding motifs required for the cyclization of farnesyl diphosphate: the “aspartate-rich” motif D100DXX(D/E) that coordinates to Mg2+A and Mg2+C, and the “NSE/DTE” motif N225DXXSXXXE that chelates Mg2+B (boldface indicates metal ion ligands). Here, we report steady-state kinetic parameters, product array analyses, and X-ray crystal structures of trichodiene synthase mutants in which the fungal NSE motif is progressively converted into a plant-like DDXXTXXXE motif, resulting in a degradation in both steady-state kinetic parameters and product specificity. Each catalytically active mutant generates a different distribution of sesquiterpene products, and three newly detected sesquiterpenes are identified. In addition, the kinetic and structural properties of the Y295F mutant of trichodiene synthase were found to be similar to those of the wild-type enzyme, thereby ruling out a proposed role for Y295 in catalysis.
Keywords: enzyme kinetics, protein crystallography, terpenoid cyclase, farnesyl diphosphate
Sesquiterpene synthases catalyze the metal ion-dependent cyclization of the universal acyclic substrate, farnesyl diphosphate (FPP), to form one of more than 300 known monocyclic, bicyclic, or tricyclic hydrocarbon or alcohol products of widely varied structure and stereochemistry [1–5]. These cyclic products represent branch points in terpenoid biosynthesis. Further modifications of cyclic sesquiterpenes and diterpenes involving downstream enzymes such as the cytochrome P450s of Artemisia annua and Taxus brevifolia, or the Fe(II)-α-ketoglutarate-dependent hydroxylase PtlH of Streptomyces avermitilis, confer highly useful properties on these terpenoid templates by forming products with anticancer, antimalarial, and antibiotic properties [6–8].
The typical FPP cyclization cascade is initiated by the ionization of the allylic diphosphate moiety, triggered by a complex network of hydrogen bond and metal ion coordination interactions. Three catalytically obligatory Mg2+ (or Mn2+) ions are required for catalysis [5, 9, 10]. These metal ions are coordinated by two conserved signature sequence motifs that characterize all known terpene cyclases: an aspartate-rich motif, which usually appears as DDXX(D,E); and the so-called NSE/DTE motif, which usually appears as (N,D)DXX(S,T)XXX(E,D) (boldface type indicates typical metal ion ligands). Comparison of 54 plant, fungal, and microbial cyclases reveals that the second metal ion-binding motif is conserved as NDXXSXXXE in most fungal and microbial enzymes, while it occurs as DDXXTXXXE in most plant cyclases [11, current studies].
Trichodiene synthase from Fusarium sporotrichioides is an 89 kDa homodimer that catalyzes the cyclization of FPP to form trichodiene as the major product [10, 13] (Fig. 1). This cyclase contains the aspartate-rich motif D100DSKD (D100 coordinates to Mg2+A and Mg2+C; D104 does not coordinate to metal ions in this cyclase) and the NSE/DTE motif N225S229E233 (these residues chelate Mg2+B) [14]. Site-directed mutagenesis of the aspartate-rich motif has established that substitution of glutamate for D100 or D101 decreases the catalytic efficiency (kcat/KM) of the enzyme 22-fold or 6-fold, respectively, in comparison with the wild-type enzyme, and decreases the proportion of trichodiene formed with a concomitant increase in the generation of several additional sesquiterpene coproducts [15]. X-ray crystal structures of D100E trichodiene synthase and its complex with Mg2+3-PPi reveal that the diphosphate-triggered active site closure observed in the wild-type enzyme [14] is highly attenuated in this mutant, perhaps as a consequence of incomplete metal ion binding (Mg2+C is not observed) [16]. Compromised metal ion binding is thus associated with a decrease in both the rate and the specificity of catalysis.
Fig. 1.
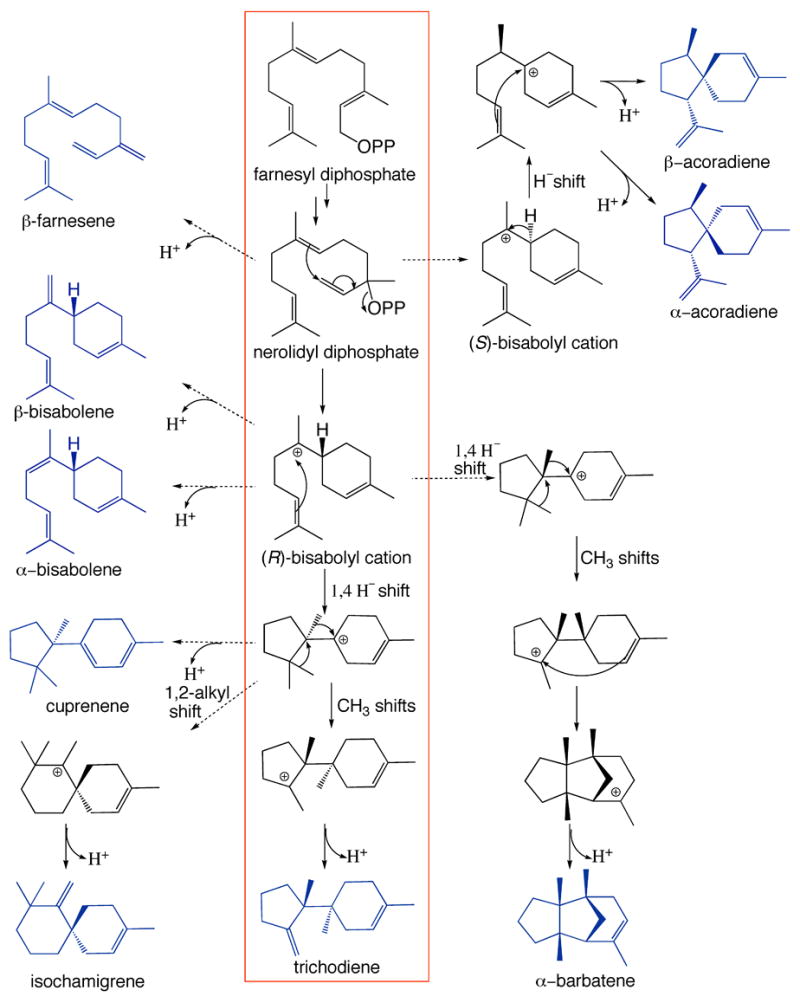
Red box: Postulated mechanism for the cyclization of farnesyl diphosphate (FPP) to trichodiene by trichodiene synthase; OPP = diphosphate, NPP = nerolidyl diphosphate [12]. For a proposed mechanistic variation, see the recently reported work of Hong and Tantillo [34]. Proposed mechanisms leading to alternative sesquiterpene products are also indicated.
To date, the dissection of the NSE/DTE-Mg2+B coordination motif in terpene cyclases by site-directed mutagenesis has not been accompanied by X-ray crystal structure analysis. Accordingly, we now report the first exploration of structure-function relationships for this motif in trichodiene synthase. In particular, we probe the apparent preference for NSE in microbial and fungal cyclases and DTE in plant cyclases. We also report the structure and function of the Y295F mutant to test a proposed role for this residue as a general base during catalysis [16]. Surprisingly, although the wild-type and mutant enzymes generate predominantly trichodiene, all active enzymes are found to generate up to 16 different sesquiterpene products, based on gas chromatographic and mass spectrometric (GC/MS) analysis of product mixtures.
Materials and Methods
Site-directed mutagenesis
Single site-specific mutations were introduced in the wild-type expression vector pZW03, using Stratagene’s QuikChange Site-Directed Mutagenesis kit. Optimal PCR conditions were previously described [17]. The plasmid for the N225D/S229T double mutant was generated by using the N225D plasmid as template.
Expression, purification, and crystal structure determination of trichodiene synthase mutants
The plasmid containing the gene encoding each trichodiene synthase variant was transformed into Escherichia coli BL21 (DE3), overexpressed, purified, and crystallized by the hanging drop vapor diffusion method as described for wild-type trichodiene synthase [14]. The complexes between mutant trichodiene synthases and pyrophosphate (PPi) were prepared using the same crystal soaking protocol used for the preparation of the complex with wild-type trichodiene synthase [14]. The mutant trichodiene synthases crystallized essentially isomorphously with the wild-type enzyme (space group P3121, a = b = 122.2 Å, c = 151.2 Å) [14]. We were unable to grow crystals of S229T trichodiene synthase. Crystals were prepared for data collection by cryoprotection in 25% ethylene glycol and flash cooling in liquid nitrogen.
Diffraction data were collected at the Advanced Light Source, Berkeley (beamline 5.0.2), National Synchrotron Light Source, Brookhaven National Laboratory (beamline X12C) and Cornell High Energy Synchroton Source (beamline F1). Data were indexed and merged using the program MOSFLM/Scala [18, 19] or HKL2000 [20]. The structures were solved by the difference Fourier technique. The programs CNS [21] and O [22] were used in refinement and rebuilding, respectively. Noncrystallographic symmetry constraints were used in the initial stages of refinement and subsequently relaxed into appropriately weighted restraints as judged by Rfree as refinement progressed. Molecular models shown in the figures were prepared with Bobscript version 2.4 or Raster3D version 2.7d [23–25]. Data collection and refinement statistics are reported in Table 1.
Table I.
Data Collection and Refinement Statistics
| Mutant/Complex | N225D | N225D-Mg2+3-PPi | N225D/S229T | Y295F | Y295F-Mg2+3-PPi |
|---|---|---|---|---|---|
| Resolution Range (Å) | 87.4-2.46 | 50-2.1 | 61.2-2.6 | 50-2.35 | 50-2.67 |
| Reflections (measured/unique) | 176908/46906 | 308536/76255 | 194477/38791 | 336863/54251 | 396431/37344 |
| Completeness (%) (overall/outer shell) | 95.9/98 | 99.7/100 | 96/90.8 | 98.2/97.8 | 100/100 |
| Rmergea (overall/outer shell) | 0.082/0.399 | 0.117/0.465 | 0.133/0.325 | 0.109/0.574 | 0.099/0.577 |
| <I/σ> (overall/outer shell) | 12.3/2.0 | 9.4/2.2 | 9.5/2.8 | 14.1/3.0 | 29.3/3.3 |
| Protein atoms (no.)b | 5735 | 5847 | 5750 | 5767 | 5858 |
| Solvent atoms (no.)b | 93 | 319 | 80 | 117 | 199 |
| Metal ions (no.)b | 2 | 3 | 0 | 2 | 3 |
| Ligand atoms (no.)b | 0 | 9 | 0 | 0 | 9 |
| Reflections used in refinement (work/free) | 44511/2338 | 74333/1910 | 36817/1940 | 51720/2169 | 35442/1867 |
| R/Rfreec | 0.244/0.282 | 0.224/0.248 | 0.223/0.256 | 0.234/0.258 | 0.203/0.245 |
| r.m.s. deviations | |||||
| bonds (Å) | 0.007 | 0.006 | 0.006 | 0.007 | 0.006 |
| angles (deg.) | 1.2 | 1.1 | 1.1 | 1.1 | 1.1 |
| dihedral angles (deg.) | 18.8 | 18.2 | 18.5 | 18.4 | 18.6 |
| improper dihedral angles (deg.) | 0.8 | 0.7 | 0.8 | 0.7 | 0.8 |
Rmerge = Σ|Ij −〈Ij〉|/ΣIj, where Ij is the observed intensity for reflection j and 〈Ij〉 is the average intensity calculated for reflection j from replicate data.
per asymmetric unit.
R = Σ||Fo| − |Fc||/Σ|Fo|, where R and Rfree are calculated by using the working and test reflection sets, respectively.
Determination of kinetic parameters of mutant trichodiene synthases
Trichodiene synthase mutants were assayed as previously described [26] in 10 mM Tris (pH 7.8), 5 mM MgCl2, 15 % glycerol, and 5 mM β-mercaptoethanol. Each series of assays was performed 2–4 times using concentrations of [1-3H]FPP (80–100 mCi/mmol) ranging from 0.025–60 μM. Optimal enzyme concentration was determined by incubating a fixed amount of radiolabeled substrate with varying concentrations of each mutant enzyme. A concentration at which the enzyme concentration dependence of product formation was linear, and where <10 % of substrate was turned over, was used for determination of kinetic parameters. The mixture was overlaid with 0.75 mL of hexane immediately after addition of enzyme and incubated for 7–10 min at 27–30 °C. The reaction was quenched by addition of 75 μL of 100 mM EDTA (pH 8.0) and vortexed for 25 s. The mixture was vortexed with an additional 0.75 mL of hexane before extraction of products. Hexane was removed using a Pasteur pipette. The hexane extract was passed through a silica gel column directly into a scintillation vial containing 5 mL of scintillation fluid. The aqueous phase was extracted with an additional 2 X 0.75 mL of hexane and passed through the same silica gel column. Finally, the column was washed with an additional 0.75 mL hexane. The scintillation counts were measured using a Beckman scintillation counter.
Alternatively, the reaction mixture was overlaid with 1 mL ethyl acetate in a screw-cap glass vial immediately after addition of enzyme and the cap was tightly screwed on before incubation at 30 °C. After 7–10 min, the reaction mixture was vortexed for 30 s. Ethyl acetate denatures the enzyme to quench the reaction. After the organic and aqueous layers separated, 0.4 mL of ethyl acetate was removed and added to scintillation fluid. For each substrate concentration, a blank was run to account for buffer-catalyzed FPP solvolysis. The counts for each blank were subtracted from the corresponding counts for the reaction mixture and corrected for the total volume.
Incubation of mutant trichodiene synthases with farnesyl diphosphate
Analysis of cyclization products generated from FPP was performed using gas chromatography–mass spectroscopy (GC/MS) as previously described [13, 15]. All glassware was washed and baked in an oven prior to use to ensure the removal of any possible trace contaminants. Farnesyl diphosphate (50–500 μM) was incubated with or without purified wild-type or mutant trichodiene synthase (1 mg) in 4 mL buffer (10 mM Tris (pH 7.8), 5 mM MgCl2, 15% glycerol, and 5 mM β-mercaptoethanol) and overlaid with HPLC-grade n-pentane (Fisher P399-1) in a glass test tube at 30 °C for 3–20 h. Reaction products were extracted with HPLC-grade n-pentane and the extracts were purified on a 3-cm 230–400 mesh silica gel column. The purified extract was concentrated on an ice-water mixture under reduced pressure until the volume decreased to <50 μL and the concentrate was analyzed by GC/MS. Reaction products were initially analyzed using a Hewlett–Packard 6890 gas chromatograph (GC) coupled to a 5973 mass selective detector (MSD) operating in a CI mode, and equipped with an HP-5MS capillary column (0.25 mm i.d. x 30 m with 0.25μm film) (Agilent Technologies). 2 μL of analyte was injected in splitless mode into the gas chromatograph-mass spectrometer. The oven temperature was maintained at 35 °C for 1 min, increased at the rate of 5 °C/min to 230 °C, followed by a 20 °C/min increase to 280 °C. The temperature was held at 280 °C for 20 min.
Identification of products generated by D100E trichodiene synthase
GC/MS (electron ionization or EI) was performed on a Hewlett-Packard Series 2 GC-MSD and equipped with an HP-5MS capillary column (30 m x 0.25 mm) at 70 eV EI, operating in a positive ion mode. The oven temperature was increased from 50 °C to 280 °C, at a rate of 5 °C/min. Each of the products was identified by direct comparison with the spectra of the corresponding reference compounds in the MassFinder 3.0 Library (http://massfinder.com) or with those of authentic standards.
Results
Kinetic parameters of trichodiene synthase mutants
Interestingly, relatively conservative amino acid substitutions in the NSE/DTE motif result in significant changes in catalytic efficiency. The N225D mutation results in only a 6-fold increase in KM, but with kcat decreased by 28-fold, the overall catalytic efficiency (kcat/KM) is diminished 177-fold relative to that of the wild-type enzyme (Table 2). The S229T mutation results in a 77-fold increase in KM, but with kcat decreased by only 9-fold the overall catalytic efficiency is diminished 708-fold (Table 2). Notably, S229T trichodiene synthase has a short half-life and 90 nM protein samples become inactive in less than a day. The N225D/S229T mutant is completely inactive under the conditions of the assay. Finally, the kinetic parameters for Y295F trichodiene synthase (Table 2) are not significantly different from those observed for the wild-type enzyme [26].
Table II.
Kinetic Parameters of Mutant Trichodiene Synthases
| Trichodiene synthase | kcat (s−1) | KM (μM) | kcat/KM (M−1s−1) (x 106) |
|---|---|---|---|
| Wild-typea | 0.138 ± 0.004 | 0.078 ± 0.0056 | 1.77 |
| N225Db | 0.005 ± 0.0003 | 0.485 ± 0.016 | 0.01 |
| S229Tb | 0.015 ± 0.004 | 6.0 ± 0.8 | 0.0025 |
| N225D/S229Tb | inactive | - | - |
| Y295Fb | 0.111 ± 0.005 | 0.069 ± 0.005 | 1.59 |
From reference 26.
Standard deviations for kcat and Km are the calculated statistical errors for the nonlinear least-squares regression fit of the data to the Michaelis-Menten expression, with R values typically >0.98.
Product array analysis of wild-type and mutant trichodiene synthases
Incubation of wild-type and mutant trichodiene synthases with FPP generates trichodiene as the major product along with up to 15 additional products [13, current studies]. Five of the additional products were identified in previous studies of D100E trichodiene synthase: β-farnesene, α-bisabolene, β-bisabolene, cuprenene, and isochamigrene [15]. Three more of the additional products were identified as α-barbatene, α-acoradiene, and β-acoradiene; proposed cyclization mechanisms are shown in Fig. 1. These sesquiterpene products were identified by comparison with authentic standards or by comparison of retention index ± 5) and EI-mass spectrum with terpenes in the MassFinder 3.0 Terpenoids Library of Dr. Detlev H. Hochmuth (http://www.massfinder.com). The remaining seven minor products could not be identified, however, due to low concentrations or absence of matching compounds in the MassFinder and other MS data libraries. Product arrays of known products of wild-type and mutant trichodiene synthases are summarized in Table 3 and both EI and CI mass spectra of identified products can be found in the Supplementary Material.
Table III.
Product Arrays of Wild-Type and Mutant Trichodiene Synthasesa
| TRb (min) | WTc (%) | D100E (%) | N225D (%) | S229T (%) | Y295F (%) | |
|---|---|---|---|---|---|---|
| α-Barbatene | 22.70 | 3.2 | 7.6 | 3.6 | 1.7 | n.o. |
| β-Farnesene | 23.81 | 0.5 | 2.5 | n.o.d | n.o. | n.d.e |
| α-Acoradiene | 24.06 | n.o. | 0.9 | n.o. | n.o. | n.d. |
| β-Acoradiene | 24.12 | 0.5 | 1.2 | n.o. | 0.5 | n.d. |
| Isochamigrene | 24.23 | 3.0 | 5.7 | 5.0 | 1.8 | n.d. |
| α-Bisabolene | 24.40 | 1.0 | 1.6 | 0.7 | 0.9 | n.d. |
| Cuprenene | 24.95 | 2.4 | 4.5 | 10.4 | 2.1 | 8.8 |
| β-Bisabolene | 25.12 | 0.9 | 1.4 | 3.1 | 0.9 | 17.5 |
| Trichodiene | 25.54 | 83.9 | 66.4 | 67.3 | 84.3 | 63.9 |
| Unknown | - | 4.6 | 8.2 | 9.9 | 7.8 | 9.8 |
| (# unknown products) | (6) | (7) | (3) | (6) | (3) |
Estimated errors in the percentages listed is generally less than 5% for major products but can range as high as 10–20% for minor products due to possible errors associated with baseline estimation in gas chromatograms.
TR: Retention time in n-pentane, using GC/MS (CI); TR for unknown products range from 22.80 to 26.00 min.
WT: Wild-type trichodiene synthase.
n. o.: Not observed under experimental conditions.
n. d.: Ratio not determined due to low concentration of product.
N225D trichodiene synthase structures
The crystal structure of unliganded N225D trichodiene synthase is identical to that of the wild-type enzyme [14] with only minor differences evident in side chain conformations (Fig. 2). The r.m.s. deviation for 349 Cα atoms between the two structures is 0.55 Å. Interestingly, as also observed in unliganded R304K trichodiene synthase [17], unliganded N225D trichodiene synthase binds one Mg2+ ion coordinated by D100 and E164 in the active site of monomer B (Fig. 2a). This metal ion most closely corresponds to Mg2+C in the structure of the wild-type enzyme complexed with Mg2+3-PPi [14].
Fig. 2.
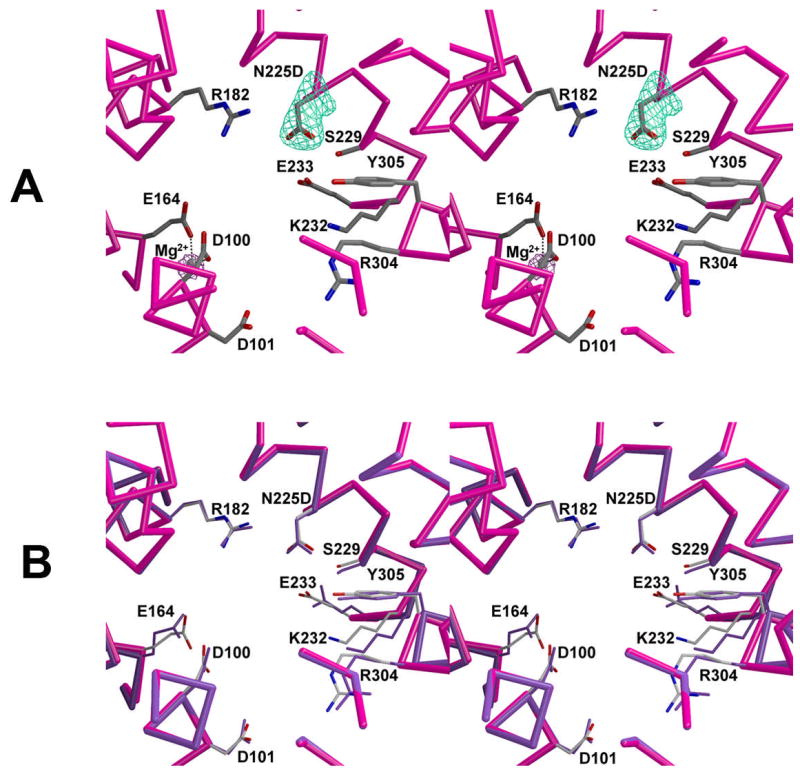
Active site of unliganded N225D trichodiene synthase. (a) Simulated annealing omit maps of D225 and magnesium ion are shown in cyan (4.7Å) and maroon (6.0Å), respectively. The Mg2+ ion is shown as a gray sphere. Metal ion coordination interactions are shown in black. (b) Superposition with the active site of unliganded trichodiene synthase (purple).
In the N225D trichodiene synthase-Mg2+3-PPi complex, the Mg2+3-PPi cluster binds to the active site of monomer B of the dimer (Fig. 3). This monomer undergoes diphosphate-induced conformational changes, with D101 and R304 forming a salt link to cap the active site cavity, as observed for the PPi-bound wild-type enzyme [14]. The r.m.s. deviation between the unliganded and liganded forms of this mutant is 1.4 Å for 349 Cα atoms, and the r.m.s. deviation between the Mg2+3-PPi complexes of the mutant and wild-type enzymes is 0.19 Å for 349 Cα atoms. These r.m.s. deviations indicate that N225D trichodiene synthase undergoes complete active site closure upon the binding of Mg2+3-PPi. Notably, the Mg2+B coordination polyhedron is essentially identical to that observed in the wild-type enzyme, with D225 occupying the coordination site formerly occupied by N225 (Fig. 3). As a consequence of the N225D substitution, however, the Mg2+B-O coordination distance for residue 225 shortens from 2.5 Å to 2.3 Å, and the Mg2+B-O coordination distance for E233 (which is opposite residue 225) lengthens from 2.0 Å to 2.6 Å due in part to a 36° conformational change in sidechain torsion angle χ3. These structural changes may be due to the additional negative charge introduced by the N225D substitution in the Mg2+B coordination polyhedron. Additionally, Mg2+C and Mg2+A move by 0.4 Å and 0.3 Å relative to their positions in the PPi-bound wild-type enzyme [14], even though these metal ions do not interact directly with D225.
Fig. 3.
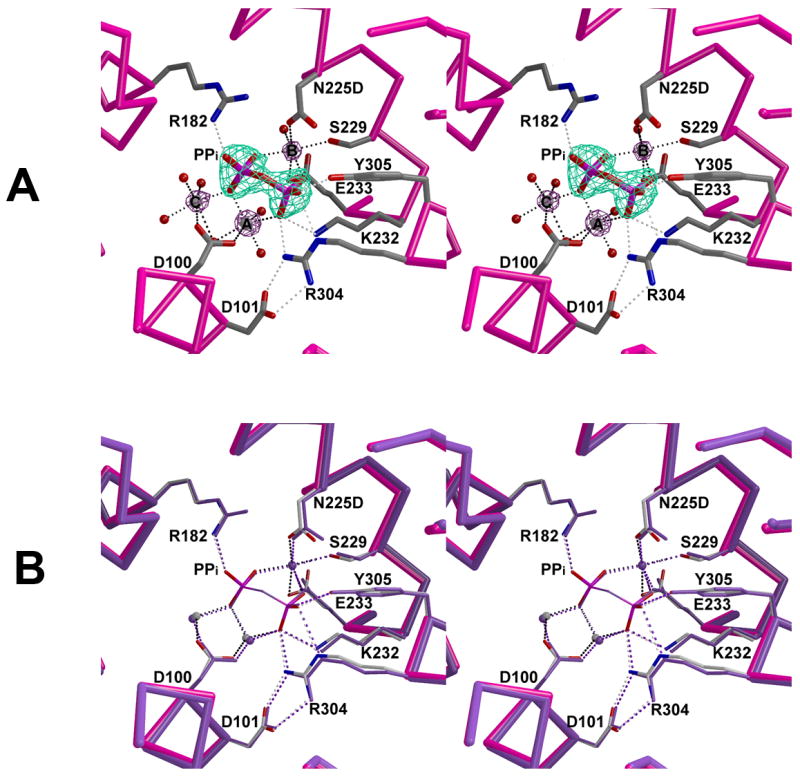
Active site of N225D trichodiene synthase complexed with Mg2+3-PPi. (a) Simulated annealing omit maps of PPi and metal ions are shown in cyan (7.5Å) and maroon (6.8σ), respectively. Metal ion coordination and hydrogen bond interactions are shown in black and gray, respectively. (b) Superposition of the active sites of PPi-bound wild-type (purple) and N225D (magenta) trichodiene synthases. Note that the metal ion coordination and hydrogen bond interactions are similar in the wild-type (purple) and N225D trichodiene synthases (black and gray).
N225D/S229T trichodiene synthase structure
The structure of this mutant is generally similar to that of the wild-type enzyme, but some differences are observed for the conformations of certain active site residues, e.g., E233 and R304 (Fig. 4) [14]. The r.m.s. deviation of 349 Cα atoms with the wild-type enzyme is 0.87 Å. The double mutation may affect binding of co-product PPi, since we were unable to prepare crystals of the complex with Mg2+3-PPi.
Fig. 4.
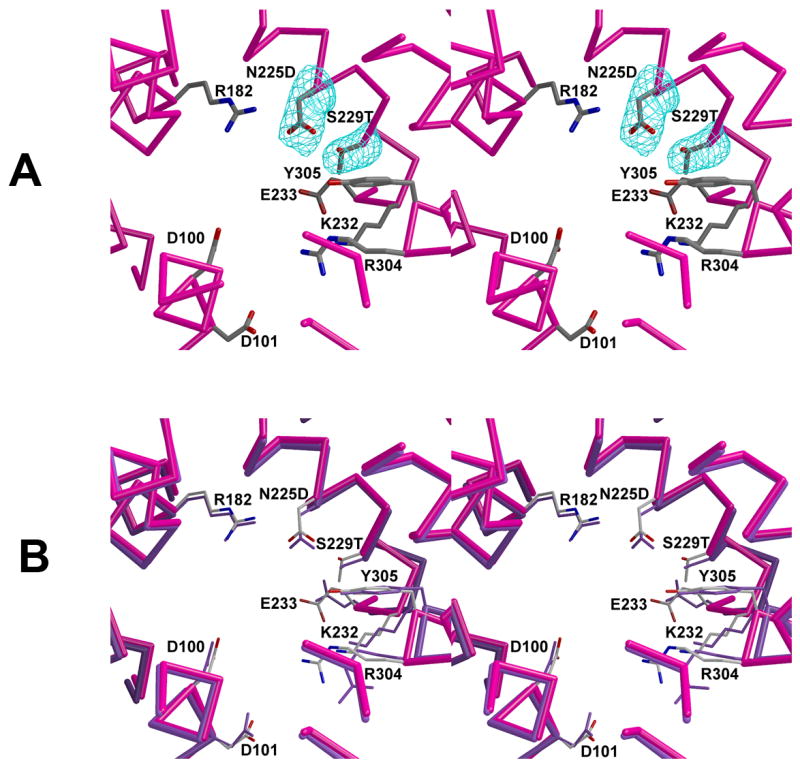
Active site of N225D/S229T trichodiene synthase. (a) Simulated annealing omit maps of the side chains of D225 and T229 are shown in cyan (5.0Å). (b) Superposition with the active site of unliganded wild-type trichodiene synthase.
Y295F trichodiene synthase structures
The structure of this mutant is generally similar to that of wild-type trichodiene synthase (Fig. 5) [14]. Although the r.m.s. deviation with the wild-type enzyme of 349 Cα atoms of 0.94 Å is relatively high in comparison with r.m.s. deviations of other mutant trichodiene synthase structures determined to-date [14, 16, 17], this is largely due to differences in some poorly ordered segments in the mutant, e.g., the N-terminal P5-T28 segment and the L307-Y329 loop. Regardless, it is clear that the Y295F substitution does not introduce significant structural perturbations in the active site.
Fig. 5.
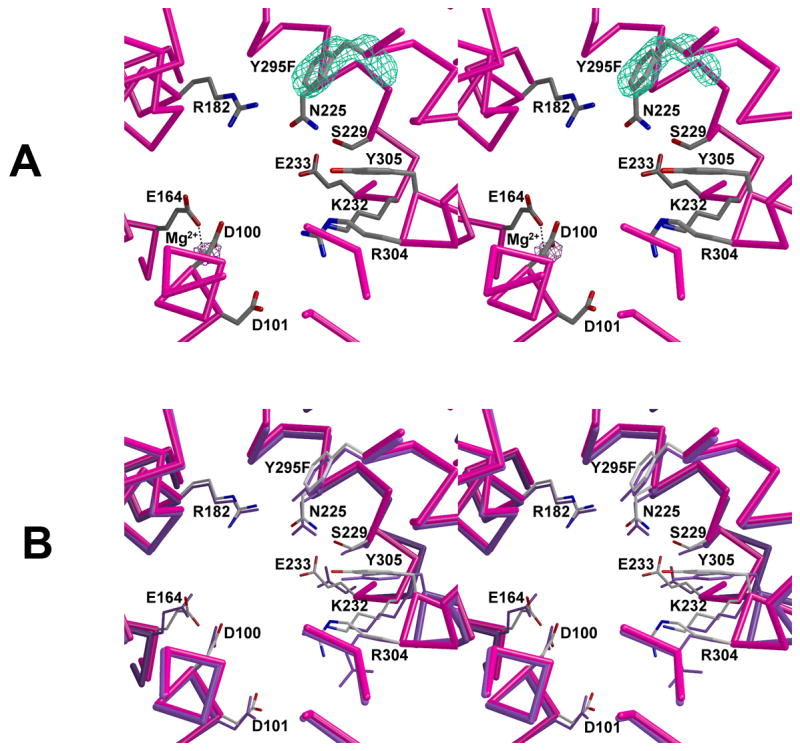
Active site of Y295F trichodiene synthase. (a) Simulated annealing omit maps of F295 and the Mg2+ ion are shown in cyan (6.0Å) and maroon (4.5Å), respectively. The Mg2+ ion is shown as a gray sphere. (b) Superposition with the active site of unliganded wild-type trichodiene synthase (purple).
Interestingly, as with R304K [17] and N225D trichodiene synthases, unliganded Y295F trichodiene synthase binds one Mg2+ ion in the active site of monomer B (Fig. 5a). This metal ion is coordinated by D100 and E164, as observed in R304K and N225D trichodiene synthases, and it most closely corresponds to Mg2+C in the structure of the wild-type enzyme complexed with Mg2+3-PPi [14].
Finally, the structure of Y295F trichodiene synthase complexed with Mg2+3-PPi is identical to that of the Mg2+3-PPi complex with the wild-type enzyme (Fig. 6) [14], with an r.m.s. deviation of 0.23 Å for 349 Cα atoms. The metal ion-binding site appears to be slightly perturbed, however, such that the r.m.s. deviation of the metal ion positions in this mutant complex with that of the wild-type enzyme complex is 0.6 Å. The binding of the Mg2+3-PPi cluster results in complete active site closure, with an r.m.s. deviation of 0.99 Å for 349 Cα atoms in comparison with the uncomplexed mutant enzyme.
Fig. 6.
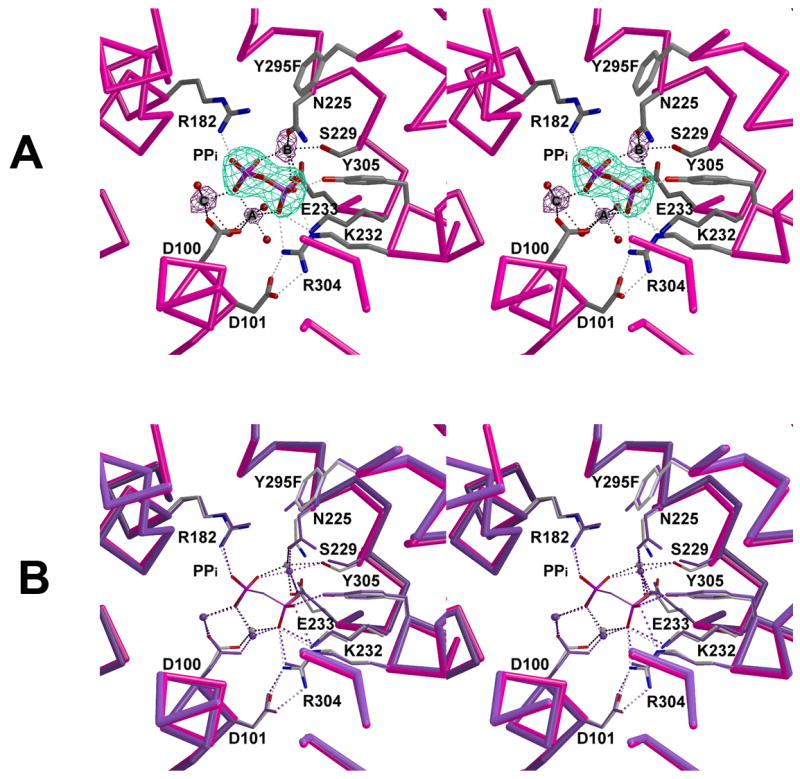
Active site of Y295F trichodiene synthase complexed with Mg2+3-PPi. (a) Simulated annealing omit maps of PPi and Mg2+ ions are shown in cyan (9.8Å) and maroon (7.5Å), respectively. (b) Superposition of the active sites of PPi-bound wild-type (purple) and Y295F (magenta) trichodiene synthases. Note that the metal ion coordination and hydrogen bond interactions are similar in the wild-type (purple) and Y295F trichodiene synthases (black and gray).
Discussion
Catalytic importance of the NSE/DTE motif
Substrate affinity is not substantially attenuated in N225D trichodiene synthase, as indicated by only a 4-fold increase in KM (Table 2). Moreover, since the structure of the Mg2+3-PPi complex is essentially identical for the N225D mutant and the wild-type enzyme, it is reasonable to expect that the binding of the Mg2+3-FPP complex is also identical in the mutant and wild-type enzymes. The apparently slight attenuation of substrate affinity may result from the introduction of an additional negative charge from the protein to the Mg2+B coordination polyhedron, which in turn would weaken the charge-charge interaction between Mg2+B and FPP. The 28-fold reduction in kcat may similarly result from Mg2+B coordination phenomena, for example if the additional negative charge in the metal ion coordination polyhedra compromises the metal ion-dependent activation of the diphosphate leaving group in the first step of catalysis, or if it compromises the release of Mg2+B that may accompany rate-determining product dissociation [27].
A more significant effect on catalysis is observed for S229 mutants of trichodiene synthase. The 77-fold increase in KM measured for S229T trichodiene synthase is the highest observed thus far for a trichodiene synthase mutant. This may be a consequence of the energetic cost for the conformational change of this side chain between the metal ion-free and metal ion-bound structures. In the wild-type enzyme, S229 undergoes a 142° rotation around side chain torsion angle χ1 in order to achieve Mg2+B coordination. Analysis of the wild-type enzyme structure [14] suggests that the nearby side chain of Y305 (which donates a hydrogen bond to PPi in the wild-type enzyme) may sterically hinder this conformational change in S229T trichodiene synthase. Additionally, the Cγ atom of the T229 side chain would clash with the main chain atoms of residues D225 or D226, or the side chain of E233. Thus, the S229T substitution may perturb this region of the protein structure. In bornyl diphosphate synthase, the corresponding residue, T500, does not undergo a conformational change upon the binding of ligands [28].
In comparison, residue Y305 of trichodiene synthase corresponds to Y572 of bornyl diphosphate synthase, a plant monoterpene cyclase. However, Y572 does not interact with the diphosphate moiety or with T500 in the DTE motif of bornyl diphosphate synthase [28], so the specific intramolecular and intermolecular interactions of residues in the NSE/DTE motif and their neighbors appear to have divergently evolved in fungal and plant cyclases based on these two examples. Accordingly, it is not straightforward to swap the NSE-Mg2+B motif of a microbial or fungal cyclase with the DTE-Mg2+B motif of a plant cyclase. The complete inactivity of N225D/S229T trichodiene synthase (Table 2) is probably due to a combination of the additional negative charge in the Mg2+B coordination polyhedron as well as the hindered conformational changes required for the bulkier T229 residue to achieve Mg2+B coordination.
Catalytic importance of Y295
Previous modeling studies of carbocation intermediates in the trichodiene synthase active site had suggested that Y295 could conceivably serve as the catalytic base responsible for the deprotonation of the final carbocation intermediate (Fig. 1) [16]. GC/MS analysis of the reaction products from both wild-type and Y295F trichodiene synthases reveals, however, that the major product of each enzyme is trichodiene (Table 3). Moreover, since Y295F trichodiene synthase exhibits nearly identical kinetic parameters to those of the wild-type enzyme (Table 2), we conclude that Y295 does not function as a general base in the trichodiene synthase mechanism.
If Y295 is not the general base that catalyzes the final proton abstraction in the trichodiene synthase mechanism, then what alternatives remain? Structural studies of farnesyl diphosphate synthase and aristolochene synthase indicate that the distal oxygen of the substrate diphosphate moiety is ideally positioned to abstract a proton from the final carbocation intermediate [29, 30]. Moreover, structural studies of wild-type and Y305F trichodiene synthases complexed with Mg2+3-PPi reveal a water molecule trapped in the active site that coordinates to Mg2+C [14, 31], and structural studies of bornyl diphosphate synthase reveal a water molecule trapped in the active site along with the cyclization product [28]. Therefore, we speculate that either the PPi anion or a trapped water molecule serves as the final general base in the trichodiene synthase mechanism.
Mechanistic inferences regarding product diversity
The hydrophobic and aromatic residues lining the active site cleft of trichodiene synthase form a template that enforces the proper conformation and orientation of the flexible FPP substrate and subsequently formed carbocation intermediates leading to formation of trichodiene. This template, however, is somewhat permissive in the wild-type enzyme as indicated by the observation of low percentages of alternative cyclization products [13] (Table 3). Significantly, this permissiveness is exaggerated in most active site mutants, and it is suggested that the structural basis of such product promiscuity is simply the increased volume of the active site cleft [16, 31]. It is also notable that a single-point mutation can cause a preferential increase in the generation of one particular alternative product. For example, the Y295F mutation causes a 19-fold increase in the generation of β-bisabolene relative to the fraction of this sesquiterpene side product generated by the wild-type enzyme (Table 3). The Y295F mutant exhibits an active site contour and kinetic parameters that are generally similar to those of the wild-type enzyme [14], so it is clear that even subtle changes to the active site template for FPP cyclization – the substitution of a hydrogen atom for a hydroxyl group – can result in significantly altered product arrays. This phenomenon is reminiscent of Keasling’s results with γ-humulene synthase mutants, in which 3–5 simultaneous mutations in the active site template alter and refocus product selectivity without compromising overall activity [32]. It is possible that multiple mutations in the active site template of trichodiene synthase may similarly lead to the formation of new sesquiterpene products. The alteration of residues that buttress the active site can also dramatically refocus the cyclization specificity of a sesquiterpene cyclase [33], demonstrating that subtle changes in the active site contour caused by mutation of neighboring residues rather than residues defining the cleft itself can result in altered product arrays.
Strikingly, the observation of the two acoradiene diastereomers in the product arrays of both wild type and mutant trichodiene synthases provides the first evidence that trichodiene synthase is capable of generating and stabilizing both (R)- and (S)-bisabolyl cations in its active site: the (R)-bisabolyl carbocation is an intermediate in trichodiene biosynthesis, and the (S)-bisabolyl carbocation is an intermediate in acoradiene biosynthesis (Fig. 1).
Similarly striking is the identification of α-barbatene in the product array of wild-type and mutant trichodiene synthases (Table 3), the biosynthesis of which must proceed through several new intermediates derived from the (R)-bisabolyl cation (Fig. 1). This is the first tricyclic sesquiterpene product identified for trichodiene synthase and its generation clearly requires a permissive active site template to accommodate the steric demands of the third cyclization reaction illustrated in Fig. 1. Future mutagenesis and structural studies of trichodiene synthase will allow us to probe and define the structural basis of specificity in the formation of alternative cyclization products.
Supplementary Material
Gas chromatogram of pentane extract of D100E trichodiene synthase reaction with FPP. The corresponding mass spectra were recorded by a mass selective detector operating in a positive ion EI mode.
Mass spectra of α-barbatene. (a) CI mass spectrum with inlaid structure. (b) EI mass spectrum (retention time = 22.87 min).
Mass spectra of (E)- β-farnesene. (a) CI mass spectrum with inlaid structure. (b) EI mass spectrum (retention time = 23.77 min).
Mass spectra of α-acoradiene. (a) CI mass spectrum with inlaid structure. (b) EI mass spectrum (retention time = 24.15 min).
Mass spectra of β-acoradiene. (a) CI mass spectrum with inlaid structure. (b) EI mass spectrum (retention time = 24.22 min).
Mass spectra of isochamigrene. (a) CI mass spectrum with inlaid structure. (b) EI mass spectrum (retention time = 24.34 min).
Mass spectra of α-bisabolene. (a) CI mass spectrum with inlaid structure. (b) EI mass spectrum (retention time = 24.95 min).
Mass spectra of cuprenene. (a) CI mass spectrum with inlaid structure.(b) EI mass spectrum (retention time = 25.04 min).
Mass spectra of β-bisabolene. (a) CI mass spectrum with inlaid structure. (b) EI mass spectrum (retention time = 25.12 min).
Mass spectra of trichodiene. (a) CI mass spectrum with inlaid structure. (b) EI mass spectrum (retention time = 25.6 min).
Acknowledgments
We thank the Advanced Light Source at Lawrence Berkeley National Laboratory, National Synchrotron Light Source at Brookhaven National Laboratory, and Cornell High Energy Synchrotron Source for access to X-ray crystallographic data collection facilities, and we thank Dr. Barry Cooperman for access to his laboratory scintillation counter.
Abbreviations
- FPP
farnesyl diphosphate
- PPi
inorganic pyrophosphate
- r.m.s.
root mean square
- CI
chemical ionization
- EI
electron ionization
- GC
gas chromatography
- MS
mass spectrometry
- MSD
mass selective detector
Footnotes
This work was supported by NIH grants GM56838 (D.W.C.) and GM30301 (D.E.C.).
Atomic coordinates and structure factors for N225D trichodiene synthase, the N225D trichodiene synthase-Mg2+3-PPi complex, N225D/S229T trichodiene synthase, Y295F trichodiene synthase, and the Y295F trichodiene synthase-Mg2+3-PPi complex have been deposited in the Research Collaboratory for Structural Bioinformatics (http://www.rcsb.org/pdb) with the following accession codes: 2PS4, 2PS5, 2PS6, 2PS7, and 2PS8, respectively.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cane DE. Acc Chem Res. 1985;18:220–226. [Google Scholar]
- 2.Croteau R, Cane DE. Methods in Enzymology. Steroids and Isoprenoids (Part A) Vol. 110. Academic Press; New York: 1985. pp. 383–405. [Google Scholar]
- 3.Cane DE. Chem Rev. 1990;90:1089–1103. [Google Scholar]
- 4.Wendt KU, Schulz GE. Structure. 1998;6:127–133. doi: 10.1016/s0969-2126(98)00015-x. [DOI] [PubMed] [Google Scholar]
- 5.Christianson DW. Chem Rev. 2006;106:3412–3442. doi: 10.1021/cr050286w. [DOI] [PubMed] [Google Scholar]
- 6.Teoh KH, Polichuk DR, Reed DW, Nowak G, Covello PS. FEBS Lett. 2006;580:1411–1416. doi: 10.1016/j.febslet.2006.01.065. [DOI] [PubMed] [Google Scholar]
- 7.Kaspera R, Croteau R. Phytochem Rev. 2006;5:433–444. doi: 10.1007/s11101-006-9006-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.You Z, Omura S, Ikeda H, Cane DE. J Am Chem Soc. 2006;128:6566–6567. doi: 10.1021/ja061469i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hohn TM, Plattner RD. Arch Biochem Biophys. 1989;272:137–143. doi: 10.1016/0003-9861(89)90204-x. [DOI] [PubMed] [Google Scholar]
- 10.Hohn TM, VanMiddlesworth F. Arch Biochem Biophys. 1986;251:756–761. doi: 10.1016/0003-9861(86)90386-3. [DOI] [PubMed] [Google Scholar]
- 11.Cane DE, Kang I. Arch Biochem Biophys. 2000;376:354–364. doi: 10.1006/abbi.2000.1734. [DOI] [PubMed] [Google Scholar]
- 12.Cane DE, Swanson S, Murthy PPN. J Am Chem Soc. 1981;103:2136–2138. [Google Scholar]
- 13.Vedula LS, Zhao Y, Coates RM, Koyama T, Cane DE, Christianson DW. Arch Biochem Biophys. 2007;466:260–266. doi: 10.1016/j.abb.2007.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rynkiewicz MJ, Cane DE, Christianson DW. Proc Natl Acad Sci USA. 2001;98:13543–13548. doi: 10.1073/pnas.231313098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cane DE, Xue Q, Fitzsimons BC. Biochemistry. 1996;35:12369–12376. doi: 10.1021/bi961344y. [DOI] [PubMed] [Google Scholar]
- 16.Rynkiewicz MJ, Cane DE, Christianson DW. Biochemistry. 2002;41:1732–1741. doi: 10.1021/bi011960g. [DOI] [PubMed] [Google Scholar]
- 17.Vedula LS, Cane DE, Christianson DW. Biochemistry. 2005;44:12719–12727. doi: 10.1021/bi0510476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leslie AGW. Joint CCP4 + ESF-EAMCB Newsletter on Protein Crystallography. 1992;26 [Google Scholar]
- 19.Collaborative Computational Project, Number 4. Acta Crystallogr. 1994;D50:760–763. [Google Scholar]
- 20.Otwinowski Z, Minor W. Methods in Enzymology. Macromolecular Crystallography (Part A) Vol. 276. Academic Press; San Diego: 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 21.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Acta Crystallogr. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 22.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Acta Crystallogr. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 23.Bacon DJ, Anderson WF. J Molec Graphics. 1988;6:219–220. [Google Scholar]
- 24.Merritt EA, Murphy MEP. Acta Crystallogr. 1994;D50:869–873. doi: 10.1107/S0907444994006396. [DOI] [PubMed] [Google Scholar]
- 25.Merritt EA, Bacon DJ. Methods in Enzymology. Macromolecular Crystallography (Part B) Vol. 277. Academic Press; San Diego: 1997. pp. 505–524. [Google Scholar]
- 26.Cane DE, Yang G, Xue Q, Shim JH. Biochemistry. 1995;34:2471–2479. doi: 10.1021/bi00008a010. [DOI] [PubMed] [Google Scholar]
- 27.Cane DE, Chiu HT, Liang PH, Anderson KS. Biochemistry. 1997;36:8332–8339. doi: 10.1021/bi963018o. [DOI] [PubMed] [Google Scholar]
- 28.Whittington DA, Wise ML, Urbansky M, Coates RM, Croteau RB, Christianson DW. Proc Natl Acad Sci USA. 2002;99:15375–15380. doi: 10.1073/pnas.232591099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hosfield DJ, Zhang Y, Dougan DR, Broun A, Tari LW, Swanson RV, Finn J. J Biol Chem. 2004;279:8526–8529. doi: 10.1074/jbc.C300511200. [DOI] [PubMed] [Google Scholar]
- 30.Shishova EY, Di Costanzo L, Cane DE, Christianson DW. Biochemistry. 2007;46:1941–1951. doi: 10.1021/bi0622524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vedula LS, Rynkiewicz MJ, Pyun HJ, Coates RM, Cane DE, Christianson DW. Biochemistry. 2005;44:6153–6163. doi: 10.1021/bi050059o. [DOI] [PubMed] [Google Scholar]
- 32.Yoshikuni Y, Ferrin TE, Keasling JD. Nature. 2006;440:1078–1082. doi: 10.1038/nature04607. [DOI] [PubMed] [Google Scholar]
- 33.Greenhagen BT, O’Maille PE, Noel JP, Chappell J. Proc Natl Acad Sci USA. 2006;103:9826–9831. doi: 10.1073/pnas.0601605103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hong YJ, Tantillo DJ. Org Lett. 2006;8:4601–4604. doi: 10.1021/ol061884f. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Gas chromatogram of pentane extract of D100E trichodiene synthase reaction with FPP. The corresponding mass spectra were recorded by a mass selective detector operating in a positive ion EI mode.
Mass spectra of α-barbatene. (a) CI mass spectrum with inlaid structure. (b) EI mass spectrum (retention time = 22.87 min).
Mass spectra of (E)- β-farnesene. (a) CI mass spectrum with inlaid structure. (b) EI mass spectrum (retention time = 23.77 min).
Mass spectra of α-acoradiene. (a) CI mass spectrum with inlaid structure. (b) EI mass spectrum (retention time = 24.15 min).
Mass spectra of β-acoradiene. (a) CI mass spectrum with inlaid structure. (b) EI mass spectrum (retention time = 24.22 min).
Mass spectra of isochamigrene. (a) CI mass spectrum with inlaid structure. (b) EI mass spectrum (retention time = 24.34 min).
Mass spectra of α-bisabolene. (a) CI mass spectrum with inlaid structure. (b) EI mass spectrum (retention time = 24.95 min).
Mass spectra of cuprenene. (a) CI mass spectrum with inlaid structure.(b) EI mass spectrum (retention time = 25.04 min).
Mass spectra of β-bisabolene. (a) CI mass spectrum with inlaid structure. (b) EI mass spectrum (retention time = 25.12 min).
Mass spectra of trichodiene. (a) CI mass spectrum with inlaid structure. (b) EI mass spectrum (retention time = 25.6 min).


