Abstract
Polyploidy is an important driver of eukaryotic evolution, evident in many animals, fungi, and plants. One consequence of polyploidy is subfunctionalization, in which the ancestral expression profile becomes partitioned among duplicated genes (termed homoeologs). Subfunctionalization appears to be a common phenomenon insofar as it has been studied, at the scale of organs. Here, we use a high-resolution methodology to investigate the expression of thousands of pairs of homoeologs during the development of a single plant cell, using as a model the seed trichomes (“cotton fiber”) of allopolyploid (containing “A” and “D” genomes) cotton (Gossypium). We demonstrate that ≈30% of the homoeologs are significantly A- or D-biased at each of three time points studied during fiber development. Genes differentially biased toward the A or D genome belong to different biological processes, illustrating the functional partitioning of genomic contributions during cellular development. Interestingly, expression of the biased genes was shifted strongly toward the agronomically inferior D genome. Analyses of homoeologous gene expression during development of this cell showed that one-fifth of the genes exhibit changes in A/D ratios, indicating that significant alteration in duplicated gene expression is fairly frequent even at the level of development and maturation of a single cell. Comparing changes in homoeolog expression in cultivated versus wild cotton showed that most homoeolog expression bias reflects polyploidy rather than domestication. Evidence suggests, however, that domestication may increase expression bias in fibers toward the D genome, potentially implicating D-genome recruitment under human selection during domestication.
Keywords: cotton, polyploidy, subfunctionalization
Polyploidy is an important component of eukaryotic evolution, evident in many animal and fungal genomes (1) and particularly in plants, where whole-genome sequences, EST datasets, and high-density maps have demonstrated cyclical and sometimes recurrent episodes of genome doubling in the history of all angiosperms (2). The merger of two differentiated genomes in a common nucleus (allopolyploidization) is accompanied by myriad genomic alterations (3, 4) and gene expression changes (5) and is thought to provide the raw material for the origin of morphological novelty, adaptation, and speciation (6). The attendant genome doubling provides a reservoir of duplicated genes as substrates for potential evolutionary innovation (7, 8).
Theory suggests that duplicated genes are subject to a dynamic tension between mutational decay and fixation by selective or neutral processes, the choice of which is determined by the interplay among population size, mutation rates, and the selective environment (9, 10). A presumably common means of duplicate gene retention, or escape from mutational obliteration, is expression partitioning, or subfunctionalization (11). In this process, the expression of duplicated genes (termed homoeologs) becomes partitioned such that one copy is expressed in a subset of the aggregate ancestral expression space (cell lines, tissues, organs, or stage), whereas the other copy is expressed in the remaining portion. An increasing body of empirical evidence substantiates subfunctionalization as an important consequence of polyploidy for plant evolution and development (11, 12). Subfunctionalization may occur very rapidly and hence be an immediate and epigenetic consequence of polyploidy, as shown in newly synthesized cotton polyploids (13), or it may arise on an evolutionary time scale following the dynamics predicted by population genetic models (14, 15). It has recently been shown that subfunctionalization may even occur in the same plant organ during development or under different environmental conditions, such as abiotic stress (16). Thus, expression partitioning of homoeologous genes appears to be a widespread phenomenon, although its scale and scope remain poorly known.
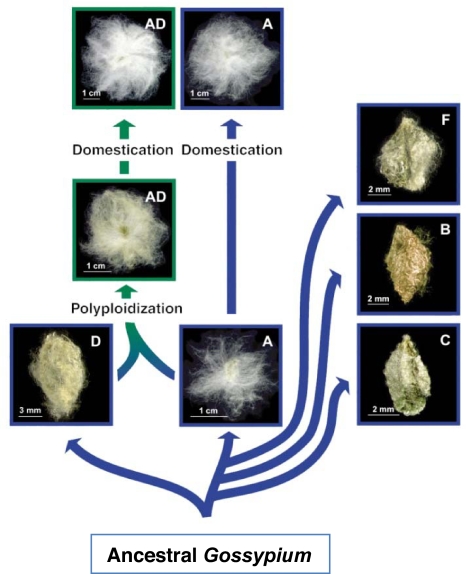
Here, we investigated the scale of expression partitioning of duplicated genes at a higher level of resolution than previously explored; that is, during development of a single polyploid cell, and using a high-throughput technology. We used the single-celled epidermal trichomes of cotton seeds (Gossypium), colloquially termed “cotton fiber,” which represent one of most distinct single cell types in the plant kingdom. A key step in the evolution of G. hirsutum (upland cotton) and G. barbadense (Pima cotton), which presently account for the majority of world cotton commerce, was an ancient [1–2 million years ago (mya)] hybridization between two diploid species, one from Africa–Asia (A genome) and the other from Central or South America (D genome), followed or accompanied by genome doubling leading to a new polyploid lineage (AD genome) (Fig. 1). Thus, modern polyploid cottons contain two ancestral genomes, A and D, which diverged from one another ≈7 mya (17) and which contributed a largely similar suite of genes to the nascent allopolyploid. Modern diploids considered most similar to the progenitors of allopolyploid Gossypium are G. herbaceum/G. arboreum (A2 genome) and G. raimondii (D5 genome) (17).
Fig. 1.
Evolutionary history of diploid and tetraploid Gossypium. Shown are the phylogeny of the genus (blue arrows) inferred from molecular data, the history of repeated domestication at both the diploid (n = 13) and polyploid (green arrows) levels, and representative examples of the morphology of the single-celled seed trichomes (“fiber”). A- and D-genome species groups are inferred to have diverged ≈7 mya, with these two genomes having become reunited with polyploid formation 1–2 mya (17). Species used in this study are: D, G. raimondii (D5); A, G. arboreum (A2; domesticated form); AD, G. hirsutum var. yucatanense (AD1; wild form)/G. hirsutum var. TM1 (AD1; domesticated form). Other species present in the figure (not used in the current study): A, wild G. herbaceum (A1; wild form); F, Gossypium longicalyx; B, Gossypium anomalum; C, Gossypium robinsonii.
Transcription profiling of cotton fibers has shown that the transcriptome of cotton fibers is extraordinarily complex, involving thousands of genes that vary in expression levels through the stages of cellular initiation, primary wall synthesis, secondary wall deposition, and maturation (18–22). Here, we simultaneously monitored transcript accumulation for 1,484 pairs of homoeologous genes by using custom short-oligonucleotide microarrays based on A- and D- genome-specific SNPs. These SNPs were identified in comparative analyses, enabling us to develop a platform capable of distinguishing homoeologous transcripts and diagnosing their origin from either the A or D genomes in a single allopolyploid sample (23). We show that duplicated genes in fibers are significantly biased toward one parent during development and that subfunctionalization is frequent and changeable during development of a single cell. By comparing cultivated cotton with a wild form of the same species, we also demonstrate how domestication may result in an increased level of homoeolog expression bias, potentially reflecting human selection.
Results and Discussion
Preferential Transcription of One of the Two Genomes.
The custom microarray platform included 11,350 genome-specific probe sets (corresponding a and d probes) representing 2,028 genes, of which 1,484 genes were investigated further by following a rigorous probe-selection process. Analyses were performed on pure fiber cells across a developmental time course from a few days after anthesis (DPA) through primary and secondary wall-synthesis stages. Four cotton accessions were analyzed, including domesticated and wild forms of allotetraploid (AD1 genome) G. hirsutum (TM1 and G. hirsutum var. yucatanense, respectively) and G. raimondii (D5) and G. arboreum (A2) as modern representatives of the ancestral genome donors. mRNAs, isolated from three developmental time points, 5, 10, and 20 DPA, were hybridized to the microarrays. In addition, an equimolar mixture (hereafter, mix) of RNAs from A2 and D5 was prepared and hybridized to the same microarray platform. For each gene, the log(a probe) − log(d probe) expression values [hereafter, log(a/d)] were calculated for the AD1, A2, and D5 genomes and for the reference mix. Comparison of the log(a/d) ratios between the allotetraploid (AD) and the reference mix showed that unequal accumulation of the two homoeologous transcripts in the tetraploid from the midparent value is common. For the nearly 1,500 genes for which we had diagnostic power, 25.3%, 37.0%, and 37.1% of the genes were significantly biased toward one of the two genomes in the allotetraploid (AD1) at 5, 10, and 20 DPA, respectively, whereas the majority of genes expressed at midparent values. These results parallel other microarray studies (24–26) showing that, in general, polyploidy appears to stabilize expression of genes toward the midparent levels. The expression of duplicated genes was found to be biased toward the D genome in all three time points studied [Table 1 and supporting information (SI) Fig. S1]. This preference for D-genome transcript accumulation increased during development, as shown by plotting the data for all 1,484 genes, in which the fraction of D-biased genes is 63%, 66%, and 76% for the three time points during cellular development (Fig. S1), or by estimating the fraction by using only significantly biased genes (Table 1), where the corresponding percentages of D-biased genes are 67, 72, and 84, respectively.
Table 1.
Number of A- and D-biased genes during three stages of fiber development in allopolypoid G. hirsutum and their putative biological roles
| DPA | Genome bias | No. of genes (%) | Significant biological processes |
|---|---|---|---|
| 5 | A | 93 (25) | Microtubule-based movement (GO:0007018) flavonoid biosynthetic process (GO:0009813) L-ascorbic acid binding (GO:0031418) dioxygenase activity (GO:0007018) glyoxysome (GO:0009514)cellular localization (GO:0051641) vesicle coat (GO:0030120) organ senescence (preventing) (GO:0010260) GTPase regulator activity (GO:0030695) |
| D | 282 (75) | Structural constituent of ribosome (GO:0003735) manganese ion binding (GO:0030145) fatty acid biosynthetic process (GO:0006633) amino acid biosynthetic process (GO:0008652) organic acid biosynthetic process (GO:0016053) disulfide oxidoreductase activity (GO:0015036) | |
| 10 | A | 154 (28) | Structural constituent of ribosome (GO:0003735) microtubule-based movement (GO:0007018) coenzyme A metabolic process (GO:0015936) GTPase regulator activity (GO:0030695) organ morphogenesis (GO:0009887) enzyme activator activity (GO:0008047) miRNA-mediated gene silencing (GO:0035195) |
| D | 394 (72) | Actin filament (GO:0005884) sucrose metabolic process (GO:0005985)fatty acid metabolic process (GO:0006631) structural constituent of ribosome (GO:0003735) manganese ion binding (GO:0030145) | |
| 20 | A | 87 (16) | Adventitious root development (GO:0048830) defense response to virus (GO:0051607)miRNA binding (GO:0035198) L-malate dehydrogenase activity (GO:0030060) glyoxysome (GO:0009514) |
| D | 463 (84) | Manganese ion binding (GO:0030145)structural constituent of ribosome (GO:0003735) ser/threonine phosphatase complex (GO:0008287) fatty acid metabolic process (GO:0006631) water transport (GO:0006833) |
Stages were 5, 10, and 20 DPA. For each stage, the number of differentially biased genes [log (a/d) values] were calculated (false discovery rate = 0.05). Significant biological processes are for P < 0.05
To address whether A-biased and D-biased genes differed in their levels of overall expression, we compared overall expression using a set of seven non-SNP probes that were also spotted on the microarray for each of the 1,484 genes. As shown in Fig. S2, there was no visual difference in normalized log expression values between the two groups, indicating that level of homoeolog bias is unrelated to global gene expression levels. Statistical analysis, however, showed that the D-biased genes were slightly over-expressed compared with A-biased genes, with means of 1.52 and 1.13, respectively [P(t) = 0.021].
To confirm the microarray-based interpretations, we used an application of a MALDI-TOF-based validation technique (see SI Text). The robustness of this technique was recently demonstrated (27), showing an R2 value of 0.64 compared with real-time PCR-based analysis. Cotton fiber cDNAs were PCR-amplified with multiplex primer sets, which targeted 35 randomly selected genes from the homoeolog-specific microarray. Amplified multiplex products were subjected to homoeolog-specific MALDI-TOF mass-spectrometry quantification. The mean value for each of nine replicates (three technical reps of three independent biological samples) was determined and compared with the estimates derived from the homoeolog-specific microarray. As shown in Fig. S3, the correlations between the SNP-specific microarray and the MALDI-TOF-based techniques were very high, with R2 values of 0.82, 0.62, and 0.63 for 5, 10, and 20 DPA, respectively (P value for all correlations is <0.001). Matching previous microarray-based studies (25, 27–29), these results demonstrate the robustness of our SNP-specific microarray platform.
Our results demonstrate extraordinary variation among duplicated genes in their contributions to the transcript pool of single-celled fibers from allopolyploid cotton, ranging from near-complete silencing of the A copy to the same for the D copy. The distribution of ratios deviated from normal, but approximated normality (Fig. S1). The mean of the distributions are not equal A and D expression but, instead, are shifted toward preferential transcription of D-genome homoeologs. From the standpoint of morphology, this might be considered an unexpected result, given that A-genome species have relatively long, spinnable fibers, whereas D-genome species have short, tightly adherent seed trichomes that would not be recognized as “cotton” by a casual observer (29) (Fig. 1). Hence, one might expect, a priori, that any biased expression would favor the A genome, as suggested by Yang et al. (30) in a recent bioinformatic analysis of ESTs from cotton ovaries. The incongruence between the aforementioned study and ours may reflect differences in tissues sampled (ovules vs. fibers), developmental stages studied, or methodology. With respect to the latter, Yang et al. based their interpretations on bioinformatic analysis of a nonnormalized cDNA library; it is possible that the use of a nonnormalized library may have biased their results toward highly expressed genes, which may be maternally biased in the ovular tissue they studied. Particularly intriguing in light of the D-genome dominance demonstrated here are results of numerous quantitative trait loci analyses, which have shown that a majority of loci for important fiber traits are located on chromosomes derived from the D-genome parent (30). Our data may reflect transcript-level evidence of this possibility of “recruitment” of D-genome homoeologs after polyploid formation, manifested as novel or enhanced expression levels and thereby potentially contributing to evolutionary innovation or, in this case, superior commercial cotton.
Functional Partitioning of Duplicate Genes During Fiber Development.
The significant biological processes for A- and D-biased genes during fiber development are presented in Table 1. In general, the two genomes are biased toward emphasizing different biological processes. The A-biased group is enriched for genes involved in microtubule-based movement, antioxidant and senescence-preventing process (e.g., l-ascorbic acid binding and glyoxysome building), vesicle coat transport, and GTPase-regulator activity, all shown as processes involved in fiber elongation and development (21, 31). In contrast, genes from the D-biased group are associated with “housekeeping” processes, including fatty acid biosynthesis, manganese ion binding, amino acid biosynthesis and organic acid biosynthesis, in addition to biological processes tightly connected with fiber development like actin filament biosynthesis (at 10 DPA) and water transport (at 20 DPA). One process, the structural constituent of ribosomes (GO:0003735), identified as connected with fiber development, particularly in fiber initials (20), is shared between A- and D-biased groups of genes. These results illustrate the partitioning of processes between duplicated genes originating from polyploidy in this developing cell. Also, this result may indicate D-genome processes that were up-regulated after tetraploid formation.
Changes in Homoeolog Bias in Wild and Domesticated Cotton Fibers.
To provide a temporal component to expression partitioning of duplicated genes after genome doubling, we included additional expression profiling data on a wild form of tetraploid cotton, namely, G. hirsutum var. yucatanense, the latter selected based on prior analyses of diversity within the species (32). Analyses were performed on fiber cells across the same developmental time course as described above (5, 10, and 20 DPA). Log(a/d) ratios for var. yucatanense mirrored those for domesticated cotton. Across the three time points, 25.1%, 32%, and 36.9% of the genes were significantly biased toward one of the two genomes (compared with 25.3%, 37.0%, and 37.1%, respectively, in the domesticated form), with a mostly similar set of biased genes and biological processes (data not shown). These results indicate that most differences in gene-expression bias resulted from polyploid formation and not from domestication. However, direct comparison of the changes in biased genes between the wild and the domesticated forms (Fig. 2) showed that additional expression alteration accompanied the transition to domestication. Moreover, this human-mediated shift was accompanied by increasing bias toward the D genome. We note that because the wild form used may not be wholly representative of the progenitor lineage of domesticated G. hirsutum, differences in gene expression between the two forms may reflect factors in addition to domestication.
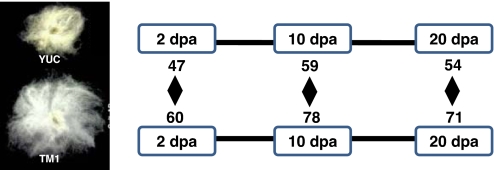
Fig. 2.
Cotton fiber domestication involved homoeolog expression changes biased toward the D genome. Ratio values of the log(a/d) were contrasted between the wild (YUC) and domesticated (TM1) species at 5, 10, and 20 DPA. The presented numbers are for the significantly D-biased genes for each comparison. Note that D-biased genes for one direction are actually A-biased for the other direction. For example, in the temporal transition from wild to domesticated forms, at 5 DPA, 47 became more A-biased and 60 became more D-biased.
Changes in Homoeolog Bias During Fiber Development.
To better appreciate patterns of change in homoeolog-specific expression during fiber development, we studied the temporal component of homoeolog-specific transcript accumulation from 2 to 25 DPA, using microarrays from additional two time points during fiber development (2 and 25 DPA). This analysis, which was performed only for the domesticated AD1, demonstrated that duplicate gene-expression patterns are dynamic even during development of a single cell (Table 2), with most changes reflecting gradual adjustments; that is, adjacent time points typically exhibited less dramatic alterations in homoeolog ratios than did more distant developmental stages. Overall, 22% (317 genes; false discovery rate <0.05) of the gene pairs studied exhibited changed ratios of contribution to the transcript pool during fiber development. Four genes displayed reciprocal silencing of alternative homoeologs during development, each changing from A to D expression. Thus, the pattern described among floral organs (11) and for organ development (16) is extended here to the level of a single cell.
Table 2.
Number of differentially biased genes between any two developmental time points in polyploid cotton fiber cell
| 2 DPA | 5 DPA | 10 DPA | 20 DPA | 25 DPA | |
|---|---|---|---|---|---|
| 2 DPA | |||||
| 5 DPA | 9 (44, 56) | ||||
| 10 DPA | 74 (52, 48) | 0 (0, 0) | |||
| 20 DPA | 118 (49, 51) | 62 (55, 45) | 57 (57, 43) | ||
| 25 DPA | 162 (45, 55) | 147 (47, 53) | 164 (52, 48) | 4 (50, 50) |
Percentages of D-biased genes; percentages of A-biased genes are given in parentheses.
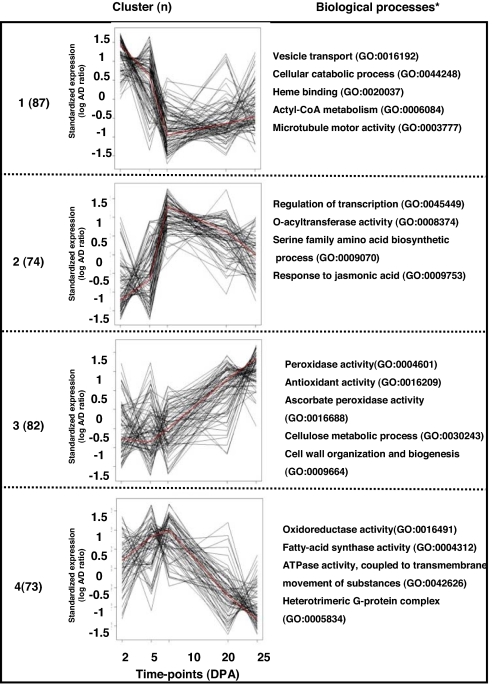
Cluster analysis of the 317 genes showing developmentally regulated change in homoeolog bias led to recognition of four statistically significant clusters (Fig. 3). Cluster 1 comprises 87 genes that exhibit bias toward D-genome expression at a time when rates of fiber elongation are high (33). Some of the processes in this cluster have been connected to fiber elongation, such as vesicle-mediated transport and microtubule motor activity, hinting once again at the hypothesized contribution of the “inferior” D genome to fiber elongation. Clusters 2 and 3 show genes that were D-biased at the beginning of development but were changed toward the A genome. Overrepresented genes at these clusters belong to processes like regulation of transcription, stress prevention, and hormone response. Cluster 4 shows genes that were A-biased early in development and that belong to, among other processes, oxidoreductase activity, fatty-acid synthase activity, ATPase activity, and transmembrane movement of substances.
Fig. 3.
Cluster analysis of differentially biased genes during fiber development. The log(A/D) ratios between five time points during fiber development were standardized and clustered, as described (21). Time points studied are: 2 DPA, fiber initiation; 5 DPA, early elongation; 10 DPA, rapid elongation; 20 DPA, transition for secondary cell biosynthesis; 25 DPA, halt in elongation, only secondary cell biosynthesis. Shown are the number of genes and the significant biological processes (P < 0.05) for each cluster. Red lines indicate the mean values for each time point. The data presented here are for the AD1 domesticated form.
Our results indicate that changes in duplicate gene expression in polyploids is a common phenomenon, occurring even at the single-cell level and fluctuating at a rate comparable with that which has been observed for entire tissues and organs in cotton and in other systems. Even though our platform permitted discrimination among homoeologs for perhaps only≈5% of the duplicate gene pairs in the genome, our analyses suggests that temporal partitioning of duplicate gene expression may, in aggregate, contribute significantly to processes important in fiber development. By extension, the data point to a hitherto unformulated dimension to the adaptive significance or functional relevance of polyploidy, namely, the coordinated and newly combined transcriptional networks that may lead to physiological and/or morphological innovation at the level of a single cell.
Materials and Methods
Plant Materials, Experimental Design, and RNA Isolation and Preparation.
Three replicate blocks of four Gossypium accessions, G. arboreum (A2), G. raimondii (D5), G. hirsutum var. TM1 (AD1), and G. hirsutum var. TX2094 (AD1 wild) were grown in the Pohl Conservatory at Iowa State University. These four accessions include, respectively, representatives of progenitor diploid genomes (A and D genomes), a domesticated allopolyploid and a wild-occurring allopolyploid (Fig. 1). For the diploid-cultivated G. arboreum (A2), no wild form has ever been discovered, and hence, by necessity, we used the domesticated form. Fiber tissues from all accessions were harvested and purified as described (21). We found that collecting pure fibers from wild species with very short fiber, like that found in the D genome, is technically challenging before 5 DPA. In addition, to optimize expenses associated with microarrays, we sampled all four taxa (A, D, and AD1 wild and domesticated) at 5, 10, and 20 DPA (representing fiber early elongation, rapid elongation, and transition for secondary cell biosynthesis, respectively). To gain additional information, we added two more time points in the domesticated tetraploid (2 and 25 DPA, representing fiber initiation and end of elongation, respectively). The three biological replicates were generated by pooling tissues from a minimum of five flowers obtained from three individuals. RNA extractions and amplifications were performed as described (21). From each pair of A and D replicates, an equimolar RNA mixture was made. RNA samples were sent to NimbleGen Systems for cDNA synthesis, labeling, and hybridization to 42 microarrays by following their proprietary protocols.
Microarray Platform and Data Analysis.
We have designed and implemented a microarray platform capable of measuring homoeolog-specific expression in Gossypium species (23). This microarray features two classes of probes, ≈35-mer probe pairs differing by an A- or D-genome homoeolog-specific SNP at their middle base, and ≈60-mer generic probes (not specific to either homoeolog). Thus, this microarray platform has the ability to measure expression from both homoeologs, detected by the corresponding ≈35-mer homoeolog-specific probes, and total gene expression, detected by the ≈60-mer generic probes designed in areas of common sequence between both homoeologs. The utility of this design has been demonstrated (23).
Raw data values for each microarray were natural-log transformed, median centered, and scale normalized across all arrays before performing a mixed linear model:
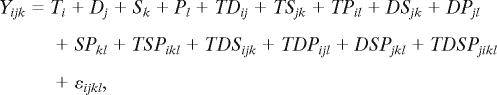 |
where T is the treatment effect for the ith biological treatment (species A2 or D5), D is the time-point effect for the jth time point (5, 10, 25), S is the strand effect for kth strand (+ and − strand probes were designed for homoeolog-specific probes), P is the homoeolog-specific probe type effect for the lth probe type (A or D genome-specific probe type), and the other 12 terms are interactions and the error term (ε).
The linear model was used to find diagnostic, homoeolog-specific probe sets by identifying those probe sets for which the expression level for a given A-genome probe was significantly greater (false discovery rate ≥ 0.05) than the corresponding D-genome probe when hybridized with A-genome RNA and vice versa when hybridized with D-genome RNA. Only probes that met these conditions for all three time points were considered as diagnostic and were used further for diagnosing expression levels from the mix and from allopolyploid Gossypium. Of the 22,798 probes representing 2,028 contigs, 5,078 probes representing 1,484 contigs were analyzed further. For each contig, a Tukey biweight correction was calculated. The difference between corrected natural logs of the A and D values from allopolyploid and the mix samples were calculated for each of 1,484 contigs by using this linear model:
where T is the treatment effect for the ith biological treatment (AD1 or mix), D is the time point effect for the jth time point (5, 10, 25), and TD and ε are the interaction and error, respectively.
Values of the least-square means and errors for all 1,484 genes and all treatments (species: AD1, AD1 wild, A2, D5, and Mix; time points: 5, 10, and 20 DPA) can be found in Table S1.
To analyze gene bias changes during development, a linear model that included only one effect (time point, with five levels: 2, 5, 10, 20, and 25) and an error was used.
The 1,484 P values from each comparison were converted to q values by using the method of Storey and Tibshirani (34). These q values were used to identify the number of differentially biased genes for a given comparison when controlling the false discovery rate at various levels.
Blast2GO (www.blast2go.de/) was used to identify biochemical pathways involved in a given comparison and to calculate the statistical significance of each pathway. Blast2GO includes the Gossip package (35) for statistical assessment of annotation differences between two sets of sequences, by using Fisher's exact test for each GO term. P values (P < 0.05) were used for the assessment of differentially significant metabolic pathways.
Supplementary Material
Acknowledgments.
We thank Anna Krush and Einat Hovav for technical assistance and Alan Gingle for database management. This work was supported by the U.S. National Science Foundation Plant Genome Program, the U.S. Department of Agriculture National Research Initiative, and the Department of Biotechnology, India.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/short/0711569105/DCSupplemental.
References
- 1.Otto SP, Whitton J. Polyploid incidence and evolution. Annu Rev Genet. 2000;34:401–437. doi: 10.1146/annurev.genet.34.1.401. [DOI] [PubMed] [Google Scholar]
- 2.Leitch IJ, Bennett MD. Polyploidy in angiosperms. Trends Plant Sci. 1997;2:470–476. [Google Scholar]
- 3.Rieseberg LH. Polyploid evolution: Keeping the peace at genomic reunions. Curr Biol. 2001;11:R925–R928. doi: 10.1016/s0960-9822(01)00556-5. [DOI] [PubMed] [Google Scholar]
- 4.Liu B, Wendel JF. Epigenetic phenomena and the evolution of plant allopolyploids. Mol Phylogenet Evol. 2003;29:365–379. doi: 10.1016/s1055-7903(03)00213-6. [DOI] [PubMed] [Google Scholar]
- 5.Adams KL, Wendel JF. Novel patterns of gene expression in polyploid plants. Trends Genet. 2005;21:539–543. doi: 10.1016/j.tig.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 6.Lynch M. The Origins of Genome Architecture. Sunderland, MA: Sinauer; 2007. [Google Scholar]
- 7.Ohno S. Evolution by Gene Duplication. New York: Springer; 1970. [Google Scholar]
- 8.Lynch M. Genomics—Gene duplication and evolution. Science. 2002;297:945–947. doi: 10.1126/science.1075472. [DOI] [PubMed] [Google Scholar]
- 9.Force A, et al. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- 11.Adams KL, Cronn R, Percifield R, Wendel JF. Genes duplicated by polyploidy show unequal contributions to the transcriptome and organ-specific reciprocal silencing. Proc Natl Acad Sci USA. 2003;100:4649–4654. doi: 10.1073/pnas.0630618100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mochida K, Yamazaki Y, Ogihara Y. Discrimination of homoeologous gene expression in hexaploid wheat by SNP analysis of contigs grouped from a large number of expressed sequence tags. Mol Genet Genomics. 2004;270:371–377. doi: 10.1007/s00438-003-0939-7. [DOI] [PubMed] [Google Scholar]
- 13.Adams KL, Percifield R, Wendel JF. Organ-specific silencing of duplicated genes in a newly synthesized cotton allotetraploid. Genetics. 2004;168:2217–2226. doi: 10.1534/genetics.104.033522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lynch M, Force A. The probability of duplicate gene preservation by subfunctionalization. Genetics. 2000;154:459–473. doi: 10.1093/genetics/154.1.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rachel WA, Durrett R. Subfunctionalization: How often does it occur? How long does it take? Theor Popul Biol. 2004;66:93–100. doi: 10.1016/j.tpb.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 16.Liu Z, Adams KL. Expression partitioning between genes duplicated by polyploidy under abiotic stress and during organ development. Curr Biol. 2007;17:1699–1674. doi: 10.1016/j.cub.2007.08.030. [DOI] [PubMed] [Google Scholar]
- 17.Wendel JF, Cronn RC. Polyploidy and the evolutionary history of cotton. Adv Agron. 2003;78:139–186. [Google Scholar]
- 18.Arpat AB, et al. Functional genomics of cell elongation in developing cotton fibers. Plant Mol Biol. 2004;54:911–929. doi: 10.1007/s11103-004-0392-y. [DOI] [PubMed] [Google Scholar]
- 19.Shi YH, et al. Transcriptome profiling, molecular biological, and physiological studies reveal a major role for ethylene in cotton fiber cell elongation. Plant Cell. 2006;18:651–664. doi: 10.1105/tpc.105.040303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taliercio EW, Boykin D. Analysis of gene expression in cotton fiber initials. BMC Plant Biol. 2007 doi: 10.1186/1471-2229-7-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hovav R, et al. A majority of genes are expressed in the single-celled seed trichome of cotton. Planta. 2008;227:319–329. doi: 10.1007/s00425-007-0619-7. [DOI] [PubMed] [Google Scholar]
- 22.Wu Y, Liewellyn DJ, Ruggiero K, Al-Ghazi Y, Dennis ES. Laser capture microdissection and cDNA microarrays used to generate gene expression profiles of rapidly expanding fibre initials cells on surface of cotton ovules. Planta. 2007;226:1475–1490. doi: 10.1007/s00425-007-0580-5. [DOI] [PubMed] [Google Scholar]
- 23.Udall JA, Swanson JM, Nettleton D, Percifield RJ, Wendel JF. A novel approach for characterizing expression levels of genes duplicated by polyploidy. Genetics. 2006;173:1823–1827. doi: 10.1534/genetics.106.058271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang JL, et al. Genomewide nonadditive gene regulation in Arabidopsis allotetraploids. Genetics. 2006;172:507–517. doi: 10.1534/genetics.105.047894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hegarty MJ, et al. Transcriptome shock after interspecific hybridization in Senecio is ameliorated by genome duplication. Curr Biol. 2006;16:1652–1659. doi: 10.1016/j.cub.2006.06.071. [DOI] [PubMed] [Google Scholar]
- 26.Stupar RM, et al. Phenotypic and transcriptomic changes associated with potato autopolyploidization. Genetics. 2007;176:2055–2067. doi: 10.1534/genetics.107.074286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Springer NM, Stupar RM. Allele-specific expression patterns reveal biases and embryo-specific parent-of-origin effects in hybrid maize. Plant Cell. 2007;19:2391–2402. doi: 10.1105/tpc.107.052258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baudo MM, et al. Transgenesis has less impact on the transcriptome of wheat grain than conventional breeding. Plant Biotechnol J. 2006;4:369–380. doi: 10.1111/j.1467-7652.2006.00193.x. [DOI] [PubMed] [Google Scholar]
- 29.Poole R, Barker G, Wilson ID, Coghill JA, Edwards KJ. Measuring global gene expression in polyploidy; a cautionary note from allohexaploid wheat. Funct Integr Genomics. 2007;7:207–219. doi: 10.1007/s10142-007-0046-7. [DOI] [PubMed] [Google Scholar]
- 30.Yang SS, et al. Accumulation of genome-specific transcripts, transcription factors and phytohormonal regulators during early stages of fiber cell development in allotetraploid cotton. Plant J. 2006;47:761–775. doi: 10.1111/j.1365-313X.2006.02829.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rong J, et al. Meta-analysis of polyploid cotton QTL shows unequal contributions of subgenomes to a complex network of genes and gene clusters implicated in lint fiber development. Genetics. 2007;176:2577–2588. doi: 10.1534/genetics.107.074518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brubaker CL, Wendel JF. Reevaluating the origin of domesticated cotton (Gossypium hirsutum Malvaceae) using nuclear Restriction-Fragment-Length-Polymorphisms (RFLPs) Am J Bot. 1994;81:1309–1326. [Google Scholar]
- 33.Applequist WL, Cronn R, Wendel JF. Comparative development of fiber in wild and cultivated cotton. Evol Dev. 2001;3:3–17. doi: 10.1046/j.1525-142x.2001.00079.x. [DOI] [PubMed] [Google Scholar]
- 34.Storey JD, Tibshirani R. SAM thresholding and false discovery rates for detecting differential gene expression in DNA microarrays. In: Parmigiani G, Garrett ES, Irizarry RA, Zeger SL, editors. The Analysis of Gene Expression Data: Methods and Software. New York: Springer; 2003. pp. 272–290. [Google Scholar]
- 35.Bluthgen N, et al. Biological profiling of gene groups utilizing Gene Ontology. Genome Inform Ser Workshop Genome Inform. 2005;16:106–115. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.