Abstract
During meiosis, homologous chromosomes (homologues) recognize each other and then intimately associate. Studies exploiting species with large chromosomes reveal that chromatin is remodeled at the onset of meiosis before this intimate association. However, little is known about the effect the remodeling has on pairing. We show here in wheat that chromatin remodeling of homologues can only occur if they are identical or nearly identical. Moreover, a failure to undergo remodeling results in reduced pairing between the homologues. Thus, chromatin remodeling at the onset of meiosis enables the chromosomes to become competent to pair and recombine efficiently.
Keywords: Ph1, wheat, heterochromatin, recombination, telomere
Organisms exhibiting sexual reproduction carry two copies (homologues) of each chromosome. Before meiosis, each homologue is replicated, forming two sister chromatids that remain linked together. At the start of meiosis, each chromosome must recognize its homologue from among all of the chromosomes present in the nucleus. The homologues must then become intimately aligned or paired along their entire lengths and a proteinaceous structure known as the synaptonemal complex (SC) must be assembled between them, a process called synapsis, as reviewed by Zickler and Kleckner (1). In this way, meiotic recombination (the exchange of DNA strands) is completed, forming chiasmata, physical links that hold the homologues together after disassembly of the SC. The homologues are then segregated so that each gamete carries only a single copy of each chromosome.
Meiotic studies of species with large chromosomes reveal that chromosomes undergo extensive chromatin remodeling at the onset of meiosis (2, 3). On entry into meiosis just before chromosome pairing, the subtelomeric heterochromatin knobs visualized on Lilium, rye, and maize chromosomes “disappear” as a result of these conformational changes (3–6). In common with these structural changes in the subtelomeric regions, visualization of the overall chromosome structure by using dispersed repetitive sequences reveals that these repetitive regions also undergo extensive remodeling before pairing (7). Wheat centromeres also change conformation early in meiosis (8). At the same time as chromatin remodeling occurs, the telomeres cluster in a group termed “a bouquet” in many species (6, 9). It is believed that this telomere clustering facilitates homologue recognition (6, 9). Homologues then intimately align from the subtelomeric regions. Consistent with this observation, the subtelomeric regions have been shown to be important for pairing and recombination in wheat (10). However, neither these studies nor any other investigations of meiosis reveal whether chromatin remodeling is essential for chromosome pairing and recombination.
Recently, cell biological investigations have revealed that one of the effects of a major chromosome pairing locus (Ph1) on chromosome 5B in wheat is to control chromatin remodeling at the onset of meiosis. As with most polyploids, hexaploid wheat was generated by wide hybridization between three distinct species. Therefore, hexaploid wheat possesses three related genomes, totaling 16,000 Mb in size, composed of seven sets of six related chromosomes with similar gene orders and vast tracts of related and highly repetitive sequences. The Ph1 locus ensures that only true homologues pair at meiosis from among the six related chromosomes (11). At the onset of meiosis, homologues undergo synchronized chromatin remodeling in the presence of Ph1, when the telomeres cluster as a bouquet and engage in intimate pairing (7). In the absence of Ph1, homologues remodel their chromatin asynchronously; the homologues and related chromosomes are thus all in different conformational states, increasing the chance of interactions between related chromosomes rather than between true homologues.
Wheat cultivars carrying a 1BL.1RS wheat-rye translocation were developed in the 1930s. All of the chromosomes of these cultivars undergo regular pairing during meiosis, forming 21 bivalents with chiasmata, and segregate correctly (12, 13). Thus, the translocated chromosome behaves like the rest of the wheat chromosomes. In fact, initially this regular pairing led some researchers to suggest that the chromosome arms were of wheat not of rye origin. The 1RS chromosome arm in these cultivars is derived from the rye variety Petkus and carries important resistance genes that have been exploited in breeding programs worldwide. The lack of variation in the 1RS Petkus arm in these cultivars prompted breeders to create additional translocated 1BL.1RS chromosomes derived from other rye varieties that are distinct in their subtelomeric heterochromatin (13). The translocated chromosomes in these wheat cultivars again still form 21 bivalents at meiosis and segregate correctly (13). It is known that crossing more distantly related wheat varieties often leads to pairing failure between the divergent wheat chromosomes in the resulting progeny (14). The 1RS chromosome arms derived from the different rye varieties also exhibit a similar phenomenon in that recombination only occurs between closely related translocated chromosomes (15–18).
Thus, a key question is whether the observed pairing and recombination phenomena are a consequence of chromatin remodeling. By visualizing the behavior of homologues that are distinct in their subtelomeric heterochromatin, we show here that varying the degree of homology has a clear effect on the ability to remodel chromatin, resulting in effects on pairing and recombination.
Results and Discussion
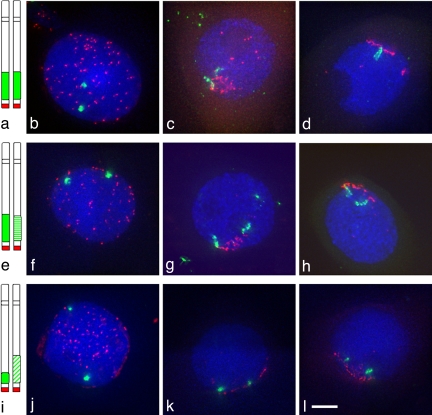
We used four wheat lines in which the short arms of a pair of wheat chromosomes have been substituted for the equivalent rye arms. In different lines these rye arms are either identical, similar, or distinct in their subtelomeric heterochromatin. Two lines carry homologues with identical heterochromatin regions, both arms being derived from the same rye variety, either King II or Petkus (Fig. 1a). The third line carries homologues with similarly sized but slightly different heterochromatin regions, one arm from the variety King II and the other from Petkus (Fig. 1e). The fourth wheat line carries a pair of homologues that differ in the size of their subtelomeric heterochromatin regions, one arm being derived from the rye variety Petkus and the other from Imperial (Fig. 1i).
Fig. 1.
Heterochromatin remodeling at meiosis. Subtelomeric heterochromatin, labeled in green, uses the pSc250 rye sequence as a probe and telomeres, in red, use a PCR product derived from primers (5′-TTTAGGG-3′)5 and (5′-CCCTAAA-3′)5 as the probe. In premeiotic nuclei (b, f, and j), the rye segments are condensed in all lines, and the behavior of the rye segments has been analyzed during telomere bouquet (c, d, g, h, k, and l). In the line King II/King II (a) with identical heterochromatin, the rye heterochromatin elongates before the full formation of the telomere cluster (c), and the homologues align in most of the cells (d). In the line Petkus/King II (e) with similar heterochromatin, the rye heterochromatin elongates before the full formation of the telomere cluster (g) and homologue alignment is slightly delayed (h). In the line Petkus/Imperial (i) with different heterochromatin, the rye heterochromatin does not elongate even at the telomere cluster (k and l). (Scale bar: ≈10 μm.)
The behavior of these lines was analyzed by 3D fluorescence in situ hybridization in meiocytes at various stages by using a probe to the rye heterochromatin. Before meiosis, the telomeres were dispersed around the nuclear periphery. In these premeiotic cells, no change in conformation of the subtelomeric heterochromatin was seen, with these regions remaining compact in all of the meiocytes examined from the different wheat lines (Fig. 1 b, f, and j). However, the subtelomeric heterochromatin behavior varied between the lines when the telomeres began to cluster in the meiocytes. When the subtelomeric heterochromatin regions were identical on the two homologues, they were localized together before the telomere bouquet formation in 50% (King II/King II or Petkus/Petkus) of the meiocytes examined (Fig. 2 b and c). During the telomere bouquet stage, these regions then underwent extensive remodeling in all of the meiocytes examined from this line (Fig. 1 c and d). The remodeled subtelomeric heterochromatin regions on the homologues extended up to 5 μm in length but differed by no more than 30% in length from each other (Table 1). The extended subtelomeric heterochromatin then formed a V-shaped paired structure with the telomere sites at the apex before “zipping up” (Figs. 1d and 2f). The subtelomeric heterochromatin regions were paired in 98% of the meiocytes examined from diplotene to metaphase I (Table 2). Similar results were obtained for the Petkus/Petkus line.
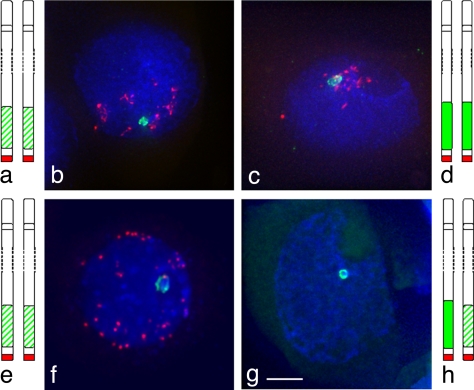
Fig. 2.
Heterochromatin colocalization and association at meiosis. Subtelomeric heterochromatin, labeled in green, uses the pSc250 rye sequence as a probe and telomeres, in red, use a PCR product derived from primers (5′-TTTAGGG-3′)5 and (5′-CCCTAAA-3′)5 as the probe. In the lines Petkus/Petkus (a and e) and King II/King II (d), with identical heterochromatin, the chromosomes can colocalize before the telomere bouquet (b and c) and can associate as a fork after telomere bouquet (f). In the line Petkus/King II (g and h), with similarly sized heterochromatin, the segments associate as a ring structure. (Scale bar: ≈10 μm.)
Table 1.
Heterochromatin remodeling during the telomere bouquet formation
| Lines |
||||||
|---|---|---|---|---|---|---|
| Identical heterochromatin |
Similar heterochromatin from different rye varieties |
|||||
| Rye segment 1 | Rye segment 2 | Ratio | Rye segment 1 | Rye segment 2 | Ratio | |
| Segment elongation | 5.269 | 4.509 | 1.2 | 4.425 | 4.093 | 1.1 |
| 4.217 | 3.948 | 1.1 | 3.893 | 2.652 | 1.5 | |
| 4.676 | 3.470 | 1.3 | 4.952 | 3.298 | 1.5 | |
| 5.023 | 4.899 | 1.0 | 5.040 | 3.938 | 1.3 | |
| 4.053 | 3.797 | 1.1 | 4.106 | 3.302 | 1.2 | |
| 4.021 | 3.195 | 1.3 | 3.823 | 3.567 | 1.1 | |
| 3.678 | 2.896 | 1.3 | 3.773 | 3.752 | 1.0 | |
| 4.333 | 3.809 | 1.1 | 3.034 | 2.539 | 1.2 | |
| 3.942 | 3.064 | 1.3 | 3.865 | 3.098 | 1.2 | |
| 3.451 | 3.451 | 1.0 | 5.313 | 3.524 | 1.5 | |
| 3.936 | 3.936 | 1.0 | 3.463 | 3.431 | 1.0 | |
| 3.677 | 3.677 | 1.0 | 3.175 | 2.965 | 1.1 | |
| 2.515 | 2.515 | 1.0 | 2.512 | 2.155 | 1.2 | |
| 2.143 | 2.143 | 1.0 | 4.359 | 3.558 | 1.2 | |
| 1.692 | 1.692 | 1.0 | 3.662 | 0.891 | 4.1 | |
| 4.082 | 2.263 | 1.8 | ||||
| 4.796 | 2.450 | 2.0 | ||||
| 3.599 | 3.238 | 1.1 | ||||
| 3.143 | 2.236 | 1.4 | ||||
| 2.956 | 1.766 | 1.7 | ||||
| 3.466 | 3.425 | 1.0 | ||||
| 4.915 | 3.509 | 1.4 | ||||
| 3.953 | 2.474 | 1.6 | ||||
| 4.424 | 3.941 | 1.1 | ||||
| 3.140 | 2.736 | 1.1 | ||||
| 4.115 | 3.022 | 1.4 | ||||
| 5.041 | 3.913 | 1.3 | ||||
| 4.100 | 1.843 | 2.2 | ||||
| 1.997 | 1.586 | 1.3 | ||||
| 2.390 | 1.803 | 1.3 | ||||
| 2.508 | 1.655 | 1.5 | ||||
| 3.697 | 3.799 | 1.0 | ||||
| 3.653 | 2.724 | 1.3 | ||||
| Mean | 3.775 | 3.400 | 3.799 | 2.883 | ||
| Difference of means | 0.375 | 0.916 | ||||
| Standard error of difference | 0.339 | 0.203 | ||||
| F test | 1.38 on | 1.00 on | ||||
| 14 df | 32 df | |||||
| Probability t test | 0.279 | <0.001 | ||||
The length of the remodeled heterochromatin was measured in 3D stacks of meiocytes exhibiting telomere clustering. Segment 1 is shown as the longest of the pair. The t test on the samples was performed by using Genstat 9th for the following Null hypothesis that the mean length of segment 1 is equal to the mean length of segment 2 for each genotype. The hypothesis was tested under a 95% confidence interval for difference in means (alpha level = 0.05). For the line with two identical segments, the probability is P = 0.279 (P value > alpha level). Therefore, the Null hypothesis is not rejected and the mean length of segment 1 is equal to mean length of segment 2. The two segments elongate at the same time. For the line with two similar segments, the probability is P < 0.001 (P value < alpha level). Therefore, the Null hypothesis is rejected and the mean length of segment 1 is not equal to the mean length of segment 2. The two segments do not elongate at the same time.
Table 2.
Percentage of meiocytes with paired heterochromatin sites during prophase I
| Homologous pairing during meiosis |  |
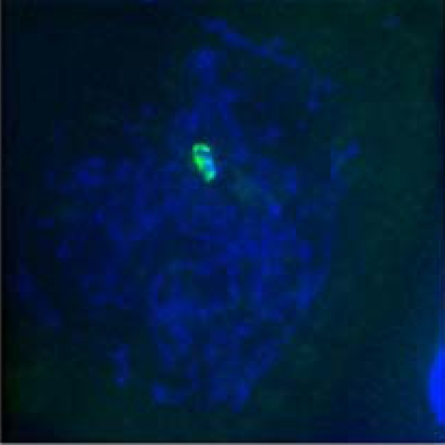 |
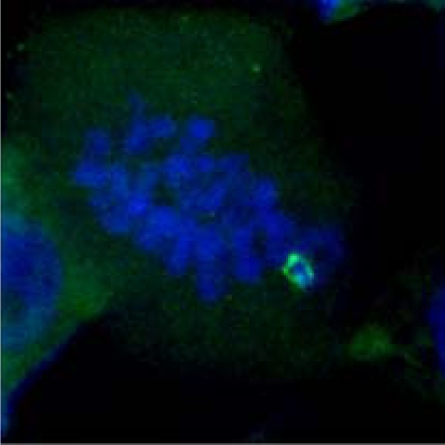 |
|---|---|---|---|
| Late Zygotene | Diplotene | Metaphase I | |
(a)*
|
98% | 98% | 98% |
(b)†
|
82% | 79% | 56% |
(c)‡
|
44% | 30% | 16% |
*King II/King II or Petkus/Petkus lines.
†Petkus/King II line.
‡Petkus/Imperial line.
In the line carrying similarly sized but slightly different subtelomeric heterochromatin (Petkus/King II), the subtelomeric heterochromatin regions were not colocalized before the telomere bouquet in the meiocytes. However, at the telomere bouquet stage in meiocytes from this line, both subtelomeric heterochromatin regions did undergo chromatin remodeling, but the remodeled regions differed from each other by up to 2-fold in length (Fig. 1 g and h, Table 1). Moreover, the extended heterochromatin regions then did not “zip up” as in the parental lines but paired at either end of the heterochromatin regions forming a loop structure (Fig. 2g). The loop structure then coalesced so that the remodeled heterochromatin regions were paired in 79% of the meiocytes at diplotene and 56% at metaphase I (Table 2). In this case, recombination has been observed between markers on the Petkus/King II chromosome arms in an F2 mapping population (15, 16).
In contrast to these observations, the subtelomeric heterochromatin remained compact during the telomere clustering and bouquet formation in the wheat line carrying homologues with differently sized subtelomeric heterochromatin regions (Petkus/Imperial), and these regions remained unassociated (Fig. 1 i–l). Subsequently, the subtelomeric heterochromatin regions were only paired with each other in 30% of the meiocytes at diplotene, which then reduced further to 16% of the meiocytes by metaphase I (Table 2).
Recombination has been assessed in these lines by using restriction fragment length polymorphism (RFLP) markers rga5.2 and iag95 and the rye seed storage protein locus Sec-1, which are predicted to span ≈50% of the physical rye arm. However, these markers cosegregated in a wheat-mapping population of 120 F2 lines derived from a 1BL-1RS (Petkus) and 1BL-1RS (Imperial) heterozygote, showing a lack of recombination between the Petkus and Imperial chromosome arms (13, 17, 18). Our results suggest that this lack of recombination is because of the failure of chromosome remodeling and pairing.
The present study shows that there can be a significant level of homologue association by telomeric regions before full formation of the telomere bouquet. However, this association only happens in those cases where the homologues are identical in their subtelomeric regions. If the homologues are identical then, as has been observed previously, they can intimately align in a “zipping up process” from the telomeres (7). If these regions are slightly different, as in Petkus/King II, then homologue association occurs within the telomere bouquet, but only after the chromatin remodeling has occurred, and the chromatin is remodeled to different lengths in the two homologues. Intimate alignment between the subtelomeric regions then occurs through “a pegging together and coalescing” process. Although there is a reduction in the overall level of pairing and subsequent recombination in the line where the regions are slightly different, in the Petkus/Imperial line where the rye arms are further diverged, there is a complete failure to remodel the subtelomeric heterochromatin. This has the most marked effect on the subsequent overall level of pairing and recombination. Thus, we conclude that the ability to remodel chromatin depends on overall relatedness of chromosomes and that this affects subsequent pairing and recombination.
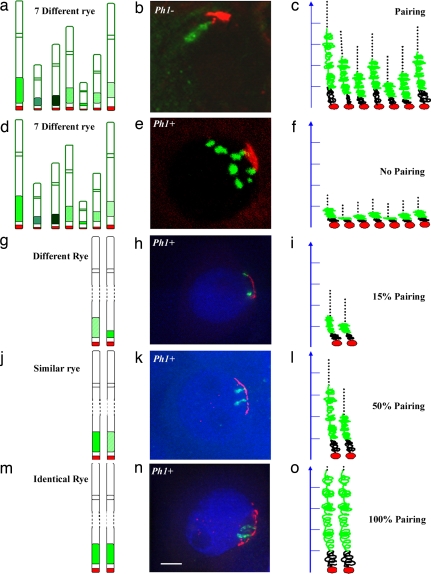
The data in the present study are complementary to studies exploiting wheat-rye hybrids (Fig. 3) (5, 7). Wheat-rye hybrids contain a haploid set of 21 wheat chromosomes and a haploid set of 7 rye chromosomes, producing 28 homoeologues and no homologues. Each of the rye chromosomes possesses a large subtelomeric heterochromatin region. In the absence of the Ph1 locus, the subtelomeric heterochromatin regions on all of the rye chromosomes remodel on entry into meiosis irrespective of their relatedness or the presence of a homologue (Fig. 3 a–c) (5, 7). The remodeled subtelomeric heterochromatin can colocalize together as a single diffuse structure (Fig. 3b). Recombination can occur in the absence of Ph1 between the wheat and rye chromosomes (19, 20). In contrast, in the presence of the Ph1 locus with no true homologues present, the subtelomeric heterochromatin cannot remodel (Fig. 3 d–f). Recombination does not occur between the wheat and rye chromosomes (19, 20). Even possessing two homologous chromosomes is in itself not sufficient to induce chromatin remodeling of both homologues in the presence of Ph1 (Fig. 3 g–i). Both homologues need to be identical or near identical for remodeling to occur (Fig. 3 j–o). Thus, Ph1 in wheat affects the ability to coordinate and control chromatin remodeling at meiosis (7). The chromatin remodeling enables chromosomes to become competent to pair and recombine. Moreover, the Ph1 locus in wheat is also able to block recombination from occurring between similar but distinct chromosome segments located within otherwise identical chromosomes (21, 22). Thus, the present study suggests that the lack of recombination between these regions in the presence of Ph1 reflects a failure to remodel the regions at the onset of meiosis.
Fig. 3.
Summary of the ability of chromatin to remodel at meiosis. Subtelomeric heterochromatin, labeled in green, uses the pSc250 rye sequence as a probe and telomeres, in red, use a PCR product derived from primers (5′-TTTAGGG-3′)5 and (5′-CCCTAAA-3′)5 as the probe. In wheat-rye hybrids carrying seven rye chromosomes in the absence of Ph1 (a), the rye heterochromatin can elongate (b and c) and the chromosomes can pair. In a wheat-rye hybrid carrying seven rye chromosomes (d), the rye heterochromatin does not elongate (e and f) and there is no pairing. In the presence of Ph1, when homologues carry different rye subtelomeric heterochromatin (g), the heterochromatin does not elongate at the telomere cluster (h and i). When homologues carry similar rye subtelomeric heterochromatin (j), the heterochromatin elongates at the telomere cluster (k) and the remodeled regions can differ from each other by up to 2-fold in length (l). When homologues carry identical rye subtelomeric heterochromatin (m), heterochromatin elongates synchronously at the telomere cluster (n and o) up to 5 μm in length. (Scale bar: b and e, ≈10 μm; h, k, and n, 5 μm).
The Ph1 locus has recently been defined to a cluster of cdk-like genes (23), of which Cdk2 from humans is the closest known homologue (24). Ph1 not only affects chromatin remodeling at meiosis, preventing nonhomologous synapsis of chromosomes, but also has effects at replication (7, 25). Overexpression of Cdk2 in humans results in nonhomologous synapsis, and Cdk2 is also involved in replication in humans (26–28). The Ph1 locus must be suppressing the expression of Cdk2 loci on other chromosomes because the loss of the Ph1 locus increases their expression level (24), which then results in homoeologous pairing. It remains unclear how homologues that are not paired or in intimate contact sense the level of homology they share among themselves to trigger the remodeling process. However, because both Cdk2 and Ph1 affect replication we hypothesize that the sensing mechanism occurs during premeiotic replication.
Methods
Plant Materials.
The following wheat-rye translocation lines were exploited in the present study: CS/Holdfast 1BL-1RS (King II 1RS), Federation/Kavkas 1BL-1RS (Petkus 1RS), Gabo 1BL-1RS (Imperial 1RS) × Veery 3 1BL-1RS (Petkus1RS) F1 line, CS/Holdfast 1BL-1RS (King II 1RS) × Federation/Kavkas 1BL-1RS (Petkus 1RS) F1 line in a Ph1 background and Chinese Spring/Secale cereale cv. Petkus F1 hybrids with and without the Ph1 locus (ph1b deficiency). Plants were grown in a controlled environmental room under optimized conditions for wheat. The wheat-rye translocation lines Gabo 1BL-1RS (Imperial 1RS) and CS/Holdfast 1BL-1RS (King II 1RS) were developed by K. W. Shepherd, Waite Agricultural Research Institute, University of Adelaide.
Tissue Preparation.
Meiosis was monitored by anther squashes in acetocarmine staining under light microscope. The anthers were fixed by vacuum infiltration of fresh 4% paraformaldehyde in 2× PEM [50 mM Pipes/KHO (pH 6.9), 5 mM EGTA, 5 mN MgSO4] for 1 h, and washed 15 min in 1× TBS (29). The anthers were sectioned in water with a vibratome, and the 50- to 100-μm sections were placed onto a γ-aminopropyl triethoxy silane (APTES)-coated slide that had been glutaraldehyde (2.5%)-activated (29). Slides were dried overnight at room temperature.
Probe Generation.
The telomere probes were labeled with biotin-16-dUTP (Boeringer Mannheim) by Nick Translation XE “Nick Translation” of the PCR product obtained by amplification of the oligomer primers (5′-TTTAGGG-3′)5 and (5′-CCCTAAA-3′)5 in the absence of template DNA (29). The rye probes were generated by labeling pSc250 sequence with digoxygenin-11-dUTP (Boeringer Mannheim) by Nick Translation (29) XE “Nick Translation.”
Fluorescence in Situ Hybridization.
Sections were dehydrated in a methanol series, permeabilized with a mixture of 2% cellulase, 1% pectolyase in TBS for 1 h at 37°C, and dehydrated again for hybridization. The denatured probe mixture (50% deionized formamide, 20% dextran sulfate, 1× Pipes/EDTA buffer (100:10), 0.3M NaCl, 500 ng of salmon sperm blocking DNA, and 50 ng of each probe) was applied to the tissue with a coverslip, and slides were moved into a modified Omnislide thermal cycler (Hybaid). Chromosome DNA denaturation occurs at 75°C and temperature is gradually brought to 37°C for an overnight hybridization. This was followed by a series of 10-min washes (42°C washes with 20% formamide in 0.1× SSC followed by 2× SSC and room temperature washes with 2× SSC and 4× SSC, 0.2% Tween 20). A blocking solution (5% BSA in 4× SSC, 0.2% Tween 20) was applied for 5min in a humidity chamber at room temperature before the detection reaction. Digoxygenin-labeled probes were detected by an anti-digoxygenin FITC-conjugated antibody (1 h incubation at 37°C) and biotin-labeled probes were detected with extravidin-cy3 (1 h incubation at 37°C). Slides were counterstained in 1 μg/ml DAPI (4′,6-diamino-2-phenyl-indole) and mounted in Vectashield (H-1000) medium.
Microscopy and Imaging.
Meiocytes were visualized by using a Nikon Eclipse E600 epifluorescence microscope equipped with a Hamamatsu Orca-ER cooled CCD camera and a Prior Proscan x–z stage. Stack images of individual cells were collected by using MetaMorph (Universal Imaging) software. Deconvolutions of images were processed with AutoDeblur (AutoQuant Imaging). Projections of 3D pictures were performed with the public domain program ImageJ written by Wayne Rasband and obtainable from http://rsb.info.nih.gov/ij/.
Statistics.
The two-tailed t test probability was performed with Genstat 9th software to evaluate the differences in the means between the line with identical heterochromatin and the line with similar heterochromatin. We postulated the Null hypothesis that the mean length of segment 1 is equal to the mean length of segment 2 for each genotype and that the two sets of data are independent. The Null hypothesis was tested for 95% confidence interval for the difference in means where the alpha level is equal to 0.05 (cutoff point). The P value represents the probability of error involved in accepting our hypothesis of the existence of a difference. If P value is more than the alpha level, the Null hypothesis is not rejected, and the mean length of segment 1 is equal to the mean length of segment 2. However, if P value is less than the alpha level, the Null hypothesis is rejected and the mean length of segment 1 is not equal to the mean length of segment 2.
Acknowledgments.
We thank Steve Reader for providing seeds and advice on meiosis and Grant Calder for help in microscopy and image processing. This work was supported by the Biotechnology and Biological Sciences Research of the U.K. and a Marie Curie fellowship from the Early Stage Training program (MEST-CT-2004-504273). M.W. was supported by a National Institutes of Health grant from the National Center for Research Resources IDeA Network of Biomedical Research Excellence program.
Footnotes
The authors declare no conflict of interest.
References
- 1.Zickler D, Kleckner N. Meiotic chromosomes: Integrating structure and function. Annu Rev Genet. 1998;32:619–697. doi: 10.1146/annurev.genet.33.1.603. [DOI] [PubMed] [Google Scholar]
- 2.Dawe RK, Sedat JW, Agard D, Cande WZ. Meiotic chromosome pairing in maize is associated with novel chromatin organisation. Cell. 1994;76:901–912. doi: 10.1016/0092-8674(94)90364-6. [DOI] [PubMed] [Google Scholar]
- 3.Dawe RK. Meiotic chromosome organisation and segregation in plants. Annu Rev Plant Phys Plant Mol Biol. 1998;49:371–395. doi: 10.1146/annurev.arplant.49.1.371. [DOI] [PubMed] [Google Scholar]
- 4.Stern H, Westergaard M, Von Wettstein D. Presynaptic events in meiocytes of Lilium longiflorum and their relation to crossing-over: A preselection hypothesis. Proc Natl Acad Sci USA. 1975;72:961–965. doi: 10.1073/pnas.72.3.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prieto P, Moore G, Reader S. Control of conformational changes asscoiated with homologue recognition at meiosis. Theor Appl Genet. 2005;111:505–510. doi: 10.1007/s00122-005-2040-6. [DOI] [PubMed] [Google Scholar]
- 6.Bass HW, Marshall WF, Sedat JW, Agard DA, Cande WZ. Telomeres cluster de novo before the initiation of synapsis: A three-dimensional spatial analysis of telomere positions before and during meiotic prophase. J Cell Biol. 1997;137:5–18. doi: 10.1083/jcb.137.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prieto P, Shaw P, Moore G. Homologue recognition during meiosis is associated with a change in chromatin conformation. Nat Cell Biol. 2004;6:906–908. doi: 10.1038/ncb1168. [DOI] [PubMed] [Google Scholar]
- 8.Martinez-Perez E, Shaw P, Aragon-Alcaide L, Moore G. Chromosomes form seven groups in hexaploid and tetraploid wheat as a prelude to meiosis. Plant J. 2003;36:21–39. doi: 10.1046/j.1365-313x.2003.01853.x. [DOI] [PubMed] [Google Scholar]
- 9.Chikashige Y, et al. Meiotic nuclear reorganisation switiching the position of centromeres and telomeres in the fission yeast Schizosaccaromyces pombe. EMBO J. 1997;16:193–202. doi: 10.1093/emboj/16.1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lukaszewski AJ. The development and meiotic behavior of asymmetrical isochromosomes in wheat. Genetics. 1997;145:1155–1160. doi: 10.1093/genetics/145.4.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Riley R, Chapman V. Genetic control of cytologically diploid behaviour of hexaploid wheat. Nature. 1958;182:713–715. [Google Scholar]
- 12.Berzonsky WA, Francki MG. Biochemical and cytogenetic technologies for characterising 1RS in wheat: A review. Eurphytica. 1999;108:1–19. [Google Scholar]
- 13.Ko JM, et al. Production of a new wheat line possessing the 1BL. 1RS wheat-rye translocation derived from Korean rye cultivar Paldanahomil. Theor Appl Genet. 2002;104:171–176. doi: 10.1007/s00122-001-0783-2. [DOI] [PubMed] [Google Scholar]
- 14.Person C. Some aspects of monosomic wheat breeding. Can J Bot. 1956;34:60–70. [Google Scholar]
- 15.Singh NK, Shepherd KW, McIntosh RA. Linkage mapping of genes for resistance to leaf, stem and stripe rust and w-secalins on short arm of the rye chromosome 1R. Theor Appl Genet. 1990;80:609–616. doi: 10.1007/BF00224219. [DOI] [PubMed] [Google Scholar]
- 16.Mago R, et al. High resolution mapping and mutation analysis separate the rust resistance genes Sr31, Lr26 and Lr9 on the short arm of rye chromosome 1. Theor Appl Genet. 2005;112:41–50. doi: 10.1007/s00122-005-0098-9. [DOI] [PubMed] [Google Scholar]
- 17.Spielmeyer W, et al. NBS-LRR sequence family is associated with leaf and strip rust resistance on the end of homoeologous chromosome group 1S of wheat. Theor Appl Genet. 2000;101:1139–1144. [Google Scholar]
- 18.Mago R, et al. Identification and mapping of molecular markers linked to rust restance located on chromosome 1RS of rye using wheat-rye translocation lines. Theor Appl Genet. 2002;104:1317–1324. doi: 10.1007/s00122-002-0879-3. [DOI] [PubMed] [Google Scholar]
- 19.Naranjo T, Fernandez-Rueda P. Pairing and recombination between individual chromosomes of wheat and rye hybrids carrying the ph1b mutation. Theor Appl Genet. 1996;93:242–248. doi: 10.1007/BF00225752. [DOI] [PubMed] [Google Scholar]
- 20.Benavente E, Fernandez-Calvin B, Orellana J. Relationship between the levels of wheat-rye metaphase I chromosomal pairing and recombination revealed by GISH. Chromosoma. 1996;101:365–373. doi: 10.1007/BF02509518. [DOI] [PubMed] [Google Scholar]
- 21.Dubcovsky J, Luo M, Dvorak J. Differentiation between homoelogous chromosome 1Am of Triticum monococcum and its recognition by the wheat Ph1 locus. Proc Natl Acad Sci USA. 1995;92:6645–6649. doi: 10.1073/pnas.92.14.6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luo MC, Dubcovsky J, Dvorak J. Recognition of homeology by the wheat Ph1 locus. Genetics. 1996;144:1195–2003. doi: 10.1093/genetics/144.3.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Griffiths S, et al. Molecular characterisation of Ph1 as a major chromosome pairing locus in hexaploid wheat. Nature. 2006;439:749–752. doi: 10.1038/nature04434. [DOI] [PubMed] [Google Scholar]
- 24.Al-Kaff N, et al. Detailed dissection of the chromosomal region containing the Ph1 locus in wheat Triticum aestivum:with deletion mutants and expression profiling. Ann Bot. 2007 doi: 10.1093/aob/mcm252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martinez-Perez E, Shaw P, Moore G. The Ph1 locus is needed to ensure specific somatic and meiotic centromere association. Nature. 2001;411:204–207. doi: 10.1038/35075597. [DOI] [PubMed] [Google Scholar]
- 26.Contreras A, et al. The dynamic mobility of histone H1 is regulated by cyclin/CDK phosphorylation. Mol Cell Biol. 2003;23:8626–8636. doi: 10.1128/MCB.23.23.8626-8636.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marston AL, Amon A. Meiosis: Cell-cycle controls shuffle and deal. Nat Rev Mol Cell Biol. 2004;5:983–997. doi: 10.1038/nrm1526. [DOI] [PubMed] [Google Scholar]
- 28.Cohen PE, Pollack SE, Pollard JW. Genetic analysis of chromosome pairing, recombination and cell cycle control during first meiotic prophase in mammals. Endocr Rev. 2006;27:398–426. doi: 10.1210/er.2005-0017. [DOI] [PubMed] [Google Scholar]
- 29.Prieto P, Moore G, Shaw P. Fluorescence in situ hybridisation on vibratome sections of plant tissues. Nat Protocols. 2007;2:1831–1838. doi: 10.1038/nprot.2007.265. [DOI] [PubMed] [Google Scholar]