Abstract
Cyanobacteria are photosynthetic organisms and are the only prokaryotes known to have a circadian lifestyle. Unicellular diazotrophic cyanobacteria such as Cyanothece sp. ATCC 51142 produce oxygen and can also fix atmospheric nitrogen, a process exquisitely sensitive to oxygen. To accommodate such antagonistic processes, the intracellular environment of Cyanothece oscillates between aerobic and anaerobic conditions during a day–night cycle. This is accomplished by temporal separation of the two processes: photosynthesis during the day and nitrogen fixation at night. Although previous studies have examined periodic changes in transcript levels for a limited number of genes in Cyanothece and other unicellular diazotrophic cyanobacteria, a comprehensive study of transcriptional activity in a nitrogen-fixing cyanobacterium is necessary to understand the impact of the temporal separation of photosynthesis and nitrogen fixation on global gene regulation and cellular metabolism. We have examined the expression patterns of nearly 5,000 genes in Cyanothece 51142 during two consecutive diurnal periods. Our analysis showed that ≈30% of these genes exhibited robust oscillating expression profiles. Interestingly, this set included genes for almost all central metabolic processes in Cyanothece 51142. A transcriptional network of all genes with significantly oscillating transcript levels revealed that the majority of genes encoding enzymes in numerous individual biochemical pathways, such as glycolysis, oxidative pentose phosphate pathway, and glycogen metabolism, were coregulated and maximally expressed at distinct phases during the diurnal cycle. These studies provide a comprehensive picture of how a physiologically relevant diurnal light–dark cycle influences the metabolism in a photosynthetic bacterium.
Keywords: circadian, cyanobacteria, microarray, nitrogen fixation, photosynthesis
Many organisms, including animals, plants, fungi, and algae, have evolved an internal timing system to anticipate daily variations in their environment. Circadian behavior, the endogenous oscillation of processes with a period length of ≈1 day, is a fundamental aspect of biology that allows organisms to respond to their environment. The circadian clock controls a wide variety of biological activities, including sleep–wake cycles in mammals, leaf movements of plants, conidiation of Neurospora, and bioluminescence in dinoflagellates (1–4). In contrast, prokaryotes were long thought incapable of daily biological rhythms, because their generation times are typically shorter than a circadian period (5). During the past 15 years, numerous studies have shown that cyanobacteria, photosynthetic microbes, are the only prokaryotes known to have a circadian clock (6, 7). Although components of the circadian clock have been identified and analyzed in detail in cyanobacteria (8–10), the interactions between the clock and cellular physiology and metabolism have not been well elucidated.
In cyanobacteria, oxygenic photosynthesis is a central metabolic process. In addition, many cyanobacteria are able to reduce atmospheric dinitrogen to ammonia. These oxygenic diazotrophs are challenged with maintaining obligate anaerobic conditions because the nitrogenase enzyme is highly sensitive to oxygen. In filamentous cyanobacteria such as Anabaena and Nostoc, nitrogen fixation is spatially separated and occurs in specialized heterocysts (11, 12), cells whose differentiation involves the specific expression of numerous genes (13). In contrast, unicellular diazotrophic cyanobacteria such as Cyanothece sp. ATCC 51142 must separate these processes temporally within the same cell, so that oxygenic photosynthesis occurs during the day and dinitrogen fixation during the night (14). This cycling of photosynthesis and nitrogen fixation provides an opportunity to examine in detail the diurnal rhythms of general cellular processes in a unicellular bacterium.
Details of the circadian clock in cyanobacteria have been most extensively studied in Synechococcus elongatus PCC 7942 and Synechocystis sp. PCC 6803, species that do not perform nitrogen fixation. These studies focused on the identification of circadian-controlled genes using promoter trap analyses and identified ≈100% and ≈77% of genes, respectively, with circadian promoter activity (15, 16). However, global DNA microarray analysis of circadian gene expression in Synechocystis 6803 identified only 2–7% of genes exhibiting circadian rhythm (17). Recently, in situ expression analyses of a limited number of individual genes associated with photosynthesis, respiration, nitrogen fixation, and fermentation in the unicellular thermophilic Synechococcus OS-A and OS-B′ demonstrated that the corresponding transcript levels cycled during the diurnal period, tracking both oxic and anoxic conditions (18).
In this study, we investigated global transcriptional changes in Cyanothece 51142 under nitrogen-fixing conditions in alternating light–dark cycles to identify genes with diurnal rhythms in their expression patterns. We found that nearly 30% of all Cyanothece genes exhibited cycling behavior. Importantly, oscillations in transcript levels were observed at specific times during the diurnal cycle for genes representing nearly all central metabolic pathways. A transcriptional network, generated by combining expression profiles from a 48-h time course, showed four distinct subnetworks of coregulated genes corresponding to central cellular functions, including nitrogen fixation and photosynthesis. Cyanothece cells were found to have increased transcriptional activity at night, suggesting that the demands of nitrogen fixation influence major metabolic activities.
Results
Identification of Genes with Oscillating Transcript Abundance.
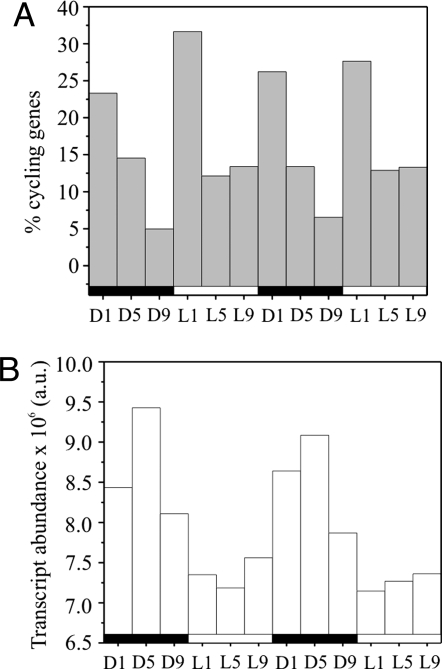
To investigate the effects of temporal separation of oxygenic photosynthesis and nitrogen fixation in detail, we analyzed changes in the differential transcript abundance in Cyanothece 51142 using a whole-genome DNA microarray. Samples from cells grown in alternating 12-h light–dark cycles were collected every 4 h over a 48-h time course, starting with the time point D1 (cells harvested after 1 h of darkness). The patterns of gene expression observed during both the light and dark periods were similar in two consecutive 24-h periods, demonstrating the robustness and reproducibility of the expression profiles (Fig. 1). Our analyses identified 1,445 genes with cycling transcripts, equivalent to ≈30% of the genes represented on the microarray [Table 1 and supporting information (SI) Table S1].
Fig. 1.
Cycling of gene expression in Cyanothece 51142. (A) The percentage of cycling genes that peak at dark–light and light–dark transitions is nearly twice as high as at other times. Percentages are of all cycling genes. (B) Relative cellular transcript abundance over time shows a maximum at D5, corresponding with peaks in nitrogenase-related gene expression (see Fig. 2A). The alternating dark–light cycles are indicated as black and white bars below the x axis.
Table 1.
Peak expression of cycling genes in Cyanothece 51142 by functional category
| Functional category | D1 |
D5 |
D9 |
L1 |
L5 |
L9 |
Total |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. | % | No. | % | No. | % | No. | % | No. | % | No. | % | No. | % | |
| Amino acid biosynthesis (96) | 23 | 24 | 7 | 7 | 2 | 2 | 5 | 5 | 3 | 3 | 3 | 3 | 43 | 45 |
| Biosynthesis of cofactors, prosthetic groups, carriers (130) | 9 | 7 | 13 | 10 | 5 | 4 | 16 | 12 | 5 | 4 | 4 | 3 | 52 | 39 |
| Cell envelope (81) | 3 | 4 | – | – | – | – | 16 | 19 | 10 | 12 | 9 | 11 | 38 | 46 |
| Cellular processes (109) | 5 | 5 | 11 | 10 | – | – | 18 | 17 | 5 | 5 | 3 | 3 | 42 | 39 |
| Central intermediary metabolism (59) | 9 | 15 | 10 | 17 | – | – | 14 | 24 | – | – | 1 | 2 | 34 | 58 |
| DNA replication, modification, recombination, repair (90) | 1 | 1 | 1 | 1 | – | – | – | – | 1 | 1 | 1 | 1 | 4 | 4 |
| Energy metabolism (114) | 23 | 20 | 9 | 8 | 2 | 2 | 10 | 9 | 4 | 4 | 3 | 3 | 51 | 45 |
| Fatty acid, phospholipid, sterol metabolism (52) | – | – | 4 | 8 | 2 | 4 | 11 | 21 | 1 | 2 | 3 | 6 | 21 | 40 |
| Other categories (438) | 37 | 8 | 13 | 3 | 6 | 1 | 27 | 6 | 13 | 3 | 14 | 3 | 110 | 25 |
| Photosynthesis and respiration (157) | 19 | 12 | 3 | 2 | 5 | 3 | 58 | 36 | 21 | 13 | 8 | 5 | 114 | 72 |
| Purines, pyrimidines, nucleosides, nucleotides (50) | 3 | 6 | 1 | 2 | 1 | 2 | 5 | 10 | – | – | 1 | 2 | 11 | 21 |
| Regulatory functions (198) | 11 | 6 | 5 | 3 | 4 | 2 | 16 | 8 | 7 | 4 | 2 | 1 | 45 | 23 |
| Transcription (39) | 8 | 21 | 5 | 13 | 1 | 3 | 4 | 10 | 1 | 3 | 1 | 3 | 20 | 51 |
| Translation (202) | 12 | 6 | 50 | 25 | 10 | 5 | 18 | 9 | 4 | 2 | 2 | 1 | 96 | 48 |
| Transport and binding proteins (254) | 16 | 6 | 1 | – | 2 | 1 | 20 | 8 | 8 | 3 | 6 | 2 | 53 | 21 |
| Unassigned (3,235) | 182 | 6 | 62 | 2 | 50 | 2 | 187 | 6 | 92 | 3 | 138 | 4 | 711 | 22 |
Genes were assigned to functional categories based on sequence homology to proteins in CyanoBase (http://bacteria.kazusa.or.jp). Total number of genes in the Cyanothece genome in each functional category is in parentheses. Number (No.) of cycling genes with peak expression at the given time point was calculated as described in Methods. Percentages (%) are of peaking genes as a percent of the entire Cyanothece genome. Data from two 24-h time courses were averaged.
We found that the majority of the cycling transcripts peaked at L1 (Fig. 1A); this presumably includes genes under circadian control as well as those oscillating in response to light. The transition from light to dark and vice versa [i.e., time points (Tp) D1 and L1] involved the activation of roughly twice as many genes as at other Tp and included a significant increase in the number of transcripts encoding regulatory proteins (Table 1), indicating the importance of these transitions to the cells. Most of the genes up-regulated during the dark period were maximally expressed at D1 and D5, with only ≈5% at D9. In contrast, L5 and L9 contained about the same percentage of highly expressed genes. In total, ≈10% more genes were maximally expressed during the light compared with the dark. Interestingly, the distribution of cellular transcript abundance over time showed the opposite pattern (Fig. 1B), being highest during the dark, with a maximum at D5, and lowest during the light. Apparently, fewer genes were transcribed at a much higher level during the dark, consistent with the known high expression of nitrogenase genes early in the dark period (19).
Chromosomal Locations of Cycling Genes with Related Functions.
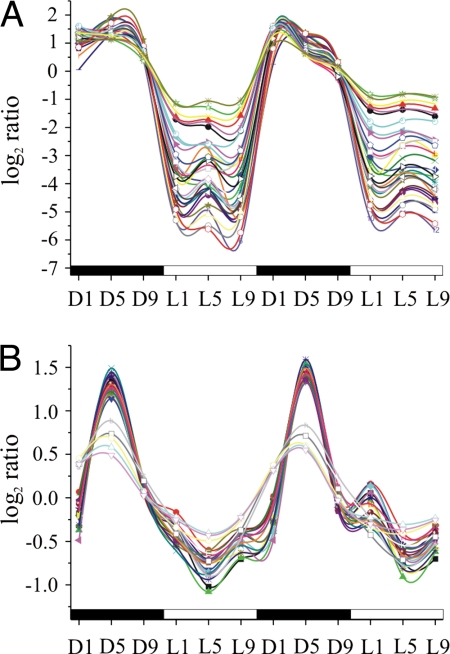
We found that genes proximally located on the genome had remarkably similar expression patterns (Fig. 2). For example, most of the genes involved in nitrogen fixation are located in a single 28-kb gene cluster, or transcriptional regulon, of 34 genes, all of which exhibited strong coregulation in their expression (Fig. 2A and Fig. S1A). For 16 of these coregulated genes, functions have not yet been determined, but their involvement in nitrogen fixation is suggested by their location. Our data showed that all 34 genes were maximally expressed during the dark period between D1 and D9, then down-regulated at L1, and remained so throughout the light period. Although regulated similarly, these genes comprise at least two operons, because nine genes at the beginning of the cluster are transcribed in a divergent direction compared with the rest of the genes (Fig. S1A).
Fig. 2.
Expression profiles of genes involved in (A) nitrogen fixation and (B) genes encoding ribosomal proteins. Dark–light cycles are indicated as black and white bars below the x-axis. Time points are designated as hours exposed to dark or light. The log2 ratios of transcript abundance to the pooled sample control are plotted on the y axis.
Similar expression profiles were also observed for a region of 32 genes encoding 30S and 50S ribosomal proteins and the preprotein translocase SecY subunit (secY) and RNA polymerase alpha subunit (rpoA) (Fig. S1B). This 16-kb gene cluster is highly conserved in all prokaryotes (20). All 28 of the genes from this group represented on the microarray were coregulated and maximally expressed at D5 (Fig. 2B). Interestingly, while still peaking at D5, the last five genes in this gene cluster exhibited a slightly different expression profile, suggesting their organization in a separate operon (Fig. S1B).
Network Analysis.
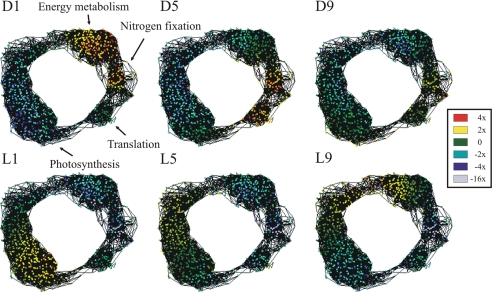
To visualize and group the temporal connections between various genes, we generated a coexpression network (Fig. 3) using the Cytoscape program (21). This network contained 734 genes, ≈15% of the genes present on the microarray (Table S2). In such a network, the genes with similar expression profiles over the entire time course clustered together in space, so that four major subnetworks (SN I–IV) of coregulated genes were identified. Each SN was significantly enriched in functionally related genes and was therefore classified according to the foremost functional category: energy metabolism and respiration, nitrogen fixation, protein translation and folding, and photosynthesis (Table S3), respectively. All four SNs were connected and formed a circle, with up- and down-regulation of the SNs progressing clockwise throughout the 24-h period (Fig. 3, Movie S1).
Fig. 3.
Coexpression network of strongly cycling genes. Genes whose transcript levels changed by at least 2-fold over the entire time course were visualized by using Cytoscape version 2.3.2 (21). Genes with Pearson correlation coefficients ≥0.9 are connected, and each node corresponds to one gene. Genes are colored according to their transcript abundance at each time point: red and yellow, genes up-regulated by 4- and 2-fold, respectively; green, genes not differentially expressed; cyan, blue, and light blue, genes down-regulated by 2-, 4-, and 16-fold, respectively. The first 24 h of the 48-h time course are shown. The four different subnetworks are labeled according to the most prominent functional category.
Most genes in SN I, with their expression highest at D1, are involved in respiration and energy metabolism, including glycolysis, the oxidative pentose phosphate pathway (OPP), the tricarboxylic acid (TCA) cycle and amino acid biosynthesis, and biogenesis of the bidirectional hydrogenase complex. Earlier studies have shown that the expression of a limited number of genes for the OPP and TCA cycle, such as 6-phosphogluconate dehydrogenase (gnd), transaldolase (tal), glyceraldehyde 3-phosphate dehydrogenase (gap), and 6-phosphofructokinase (pfkA), are under circadian control in Synechocystis 6803 (17), whereas here we find the concerted transcriptional regulation of these entire pathways in Cyanothece. Cytochrome c oxidase (coxA1, B1, C1 genes), the terminal acceptor and key enzyme of the cyanobacterial respiratory chain (22), is up-regulated at D1, consistent with observed increased respiratory activity early in the dark period (19). Interestingly, carB, the gene encoding the large subunit of carbamoyl-phosphate synthase, is one of the strongest temporally connected genes in this SN (Table S3). This enzyme is rate-limiting in the biosynthesis of arginine (23), one of the two amino acid components of cyanophycin, an inclusion granule for nitrogen storage synthesized during the dark period (24).
The majority of the genes in SN II are associated with nitrogen fixation, iron uptake, and iron–sulfur cluster biogenesis, and this SN is composed largely of the genes within the nif gene cluster (Fig. 2A). Genes encoding the uptake hydrogenase hupS and hupL are also located in SN II. Both the nitrogenase Fe protein NifH and the small subunit of the uptake hydrogenase HupS contain an iron–sulfur cluster that mediates electron transfer from the active sites to the respective redox partners. Interestingly, nifB and cysE2, the first genes in both transcriptional units, show the strongest temporal connectivity in this SN (Table S3). The genes in SN I and SN II are up-regulated during the entire dark period and down-regulated during the entire light period.
The genes in SN III are also up-regulated during the dark period, with maximal expression at D5. This SN contains, in addition to the ribosomal protein cluster (Fig. 2B), other genes involved in protein translation and protein folding, such as elongation factor P and the chaperonins GroES and GroEL. The gene with the strongest temporal connectivity to other genes is rps3, the first gene in the ribosomal protein cluster (Table S3).
SN IV contains 376 differentially expressed genes, primarily involved in photosynthesis and light harvesting, but also associated with cellular processes such as cell division, glycogen biosynthesis, and carbon fixation. This is the only SN that is up-regulated during the light and down-regulated during the dark period. Interestingly, all genes encoding subunits of photosystem I are maximally expressed at L5, except for psaL1 and psaK2, which are maximally expressed at D9 and L1, respectively. Genes for most of the photosystem II subunits, and the light-harvesting phycobiliproteins, peak at L1. Notably, the psb27 gene exhibits a strong temporal connectivity to other genes in SN IV (Table S3). The Psb27 protein is involved in the assembly of the water oxidation complex of photosystem II (25).
Regulation of Genes Encoding Enzymes of Central Metabolic Pathways.
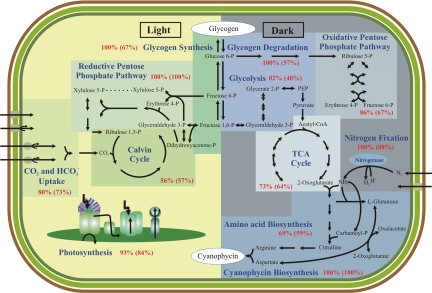
Because we found that genes associated with particular metabolic functions were likely to exhibit similar temporal expression patterns, we investigated the extent to which genes in well characterized pathways were coregulated during the diurnal cycle. Specifically, we examined whether multiple gene isoforms found in these pathways have similar expression patterns. By using the pathway information from the KEGG database (26), we analyzed the enzymes involved in glycogen degradation, glycolysis, pentose phosphate pathway, TCA cycle, amino acid biosynthesis, cyanophycin synthesis, photosynthesis, and CO2 fixation. We also examined both the total number of genes and the number of enzymes associated with different metabolic activities to determine the coregulation of the entire pathways (Fig. 4, Table S4).
Fig. 4.
Schematic organization of a Cyanothece cell showing central metabolic pathways. Pathways are divided according to the diurnal period in which they are most up-regulated. Double arrows show pathways that involve reversible reactions. The percentage of up-regulated enzymes in each pathway is in red, with the percentage of all up-regulated isoforms in parentheses.
The majority of the enzymes (82%) associated with glycolysis were up-regulated during the dark period. Although several enzymes were represented by more than one isoform, we found that multiple isoforms were typically not coregulated. For example, one of the genes encoding a glucose 6-phosphate isomerase (pgi2) was expressed at a constant level throughout the diurnal cycle, whereas the other gene (pgi1) had peak expression at D1. A similar pattern was observed for the isoforms of pyruvate kinase, pykF1-F4. The pykF1 gene exhibited maximal expression at D1, whereas the other isoforms did not cycle.
Cyanothece contains only one gene copy for each enzyme in the TCA cycle. However, succinate dehydrogenase has two subunits, a flavoprotein, sdhA, and a Fe-S protein, sdhB, but only sdhA is differentially expressed, peaking at D1. With the exception of succinyl-CoA synthetase (sucC/D), all remaining enzymes of the TCA cycle are coregulated and peak during the early dark period. Similarly, the OPP and nitrogen metabolism, including the biosynthesis of amino acids derived from glutamate, are coregulated. Six of the seven enzymes of OPP are maximally expressed during the dark: only the enzyme ribulose-phosphate (Ru-P) 3-epimerase (rpe) is maximally expressed at L1. In Prochlorococcus MED4 under nitrogen starvation, TCA cycle genes are up-regulated, whereas genes involved in bicarbonate transport (sbtA), CO2 fixation (Ru1,5-P carboxylase, rbcLS), and glycogen storage (glgA-C) are repressed (27), consistent with our findings. We found that, except for bicA, all transporters involved in CO2 and bicarbonate uptake are maximally expressed during the light period. BicA is a sodium bicarbonate symporter (28) and is maximally expressed during the dark period in Cyanothece. Approximately 84% of the photosynthetic subunits and phycobiliproteins are maximally expressed at L1 or L5, whereas the remaining 16% are largely noncoregulated isoforms or nondifferentially expressed genes.
Discussion
Diurnal oscillations have been studied in great detail in many eukaryotic systems. The discovery of a circadian clock in cyanobacteria established a new view of dynamics and regulation in a prokaryotic cell. The current study has significantly broadened our understanding of temporal regulation in unicellular oxygenic diazotrophic bacteria by investigating the global transcriptional changes under nitrogen fixing conditions in alternating light–dark cycles.
The relationship between the circadian cycle, diurnal processes, and metabolism in cyanobacteria is of great interest. Our analysis revealed that ≈30% of all genes in Cyanothece oscillate during a 24-h light–dark period. Significantly, these are primarily genes involved in core metabolic processes, such as photosynthesis, respiration, energy metabolism, and amino acid biosynthesis (Table 1). We found that more than half of all genes in these categories were differentially expressed. In contrast, most genes involved in transport, DNA replication, modification, and repair, and genes encoding regulatory proteins, were not differentially expressed, suggesting that those genes are not necessary to maintain diurnal processes. Interestingly, only 22% of the unassigned genes cycled, even though these 3,235 unassigned genes comprise more than half of the annotated genes in the Cyanothece genome. This implies that the diurnal lifestyle of Cyanothece, under our growth conditions, mainly impact the known and functionally assigned genes from major metabolic pathways. We propose that genes in the unassigned category may function preferentially in conditions other than our standard growth conditions, such as nutrient, redox, salt, or osmotic stresses.
We found that the abundance of transcripts was highest early in the dark period (Fig. 1B). This suggests an increase in cellular activity during the early dark period and likely a higher turnover of proteins throughout the dark compared with the light period. We also observed an induction of genes encoding ribosomal proteins at this time (Fig. 2B). Because translation essentially occurs cotranscriptionally in such a bacterium, this implies an increased rate of de novo protein synthesis. In fact, this has been observed: the nitrogenase proteins are newly synthesized in the early dark period and subsequently degraded at approximately D9 (29).
Both the nif and the ribosomal gene clusters in Cyanothece 51142 are organized as regulons of more than one transcriptional unit. Previous studies observed that neighboring genes show similarities in their expression pattern (17), but our analyses show a strong coregulation of functionally associated but distantly located genes (Fig. 2, Fig. S1). Furthermore, we found an interconnection between distinct metabolic processes and, more importantly, entire pathways. In many pathways, we found that different isoforms, or structural subunits, of the same proteins are not necessarily coregulated. For instance, only 48% of all genes, but 82% of all enzymes involved in glycolysis, are up-regulated during the dark period (Fig. 4), suggesting that expressions of only specific isoforms oscillate, and that expressions of different isoforms depend on the diurnal period or cellular condition. Apparently, Cyanothece uses these multiple gene copies to regulate specific pathways under different conditions.
The generation of a transcriptional network revealed four different SN, significantly enriched for genes with similar functions. We found that genes in each SN are coregulated, peak at distinct phases of the circadian cycle, and in many cases are regulated in anticipation of biological activities (Fig. 3, Movie S1). This suggests a tight transcriptional regulation of different cellular processes. Highly connected genes in each SN represent the temporal expression profile that is most representative for all genes in the SN (Table S3). It has been shown that such genes might play a central role in the regulation of cellular processes significant for the entire SN (30).
Previous studies in Cyanothece uncovered that inclusion body accumulation and degradation, particularly of glycogen and cyanophycin granules, occur concomitantly with photosynthesis and nitrogen fixation (14). Our analyses elucidate this at the transcriptional level, in that the degradation of glycogen through glycolysis, OPP, and the TCA cycle occurs during the dark to provide the cell with ATP, pyridine nucleotides, and cell intermediates. Respiratory electron transport scavenges oxygen to establish anaerobic intracellular conditions necessary for N2 fixation. Fixed nitrogen is incorporated into arginine and aspartate and eventually stored as cyanophycin granules inside the cell. Ribosomal proteins are up-regulated during this period to accommodate elevated levels of transcript abundance, likely due to nitrogen fixation-related proteins, as transcripts for the nif cluster comprise 16% of the cellular transcript abundance during the dark. Thus, the demands of nitrogen fixation have a significant impact on the diurnal cycle in Cyanothece.
Our analyses provide previously undescribed insights into the temporal separation of oxygenic photosynthesis, nitrogen fixation, and regulation of other central metabolic pathways during the diurnal cycle in Cyanothece 51142. From this analysis, the impact of a diurnal cycle on metabolism and physiology in a bacterial cell begins to emerge.
Materials and Methods
Cell Cultures.
Cyanothece cultures were grown in ASP2 medium without NaNO3 (14) at 30°C with air bubbling in alternating 12-h light–dark cycles with 50 μmol photons·m−2·s−1 of white light. Initially, cultures were inoculated with 1/10 volume of cells grown in ASP2 medium with NaNO3 and continuous light. After 7 d, 150-ml samples were taken every 4 h for 2 d, starting with 1 h into the dark period (Tp D1). In total, 12 samples were collected.
Microarrays.
The microarray was designed based on an early draft of the Cyanothece genome sequence. Single 60-mer oligonucleotide probes were designed for each of the 5,048 ORFs and were duplicated on the microarray to provide an estimate of intraarray variance. Of the original 5,480 probes, 4,930 mapped onto the final assembly; the remaining 55 were designated as negative controls. The probes covered 4,660 of the final annotated genes, with an additional 176 probes that mapped only to ORFs predicted in the draft assembly, for a total of 4,420 genes. Agilent Technologies fabricated the arrays. RNA was isolated by using RNAwiz reagent (Ambion) according to the manufacturer's instructions and quantified with the NanoDrop ND-1000 UV-VIS Spectrophotometer (NanoDrop Technologies). The quality of the RNA was verified by using the Bioanalyzer 2100 (Agilent Technologies). Total RNA (2.5 μg) was labeled directly with Cy3 and Cy5 dyes by using the Micromax ASAP RNA labeling kit (Perkin–Elmer Life and Analytical Sciences) according to the manufacturer's instruction. To remove the unbound dye from the reaction mixture, the RNA clean up kit-5 (Zymo Research) was used, and the specific activity for each dye was quantified with the NanoDrop UV-VIS Spectrophotometer. The microarrays were hybridized at MoGene by using 750 ng of RNA and a specific activity of 50 pmol for each dye. Six replicate microarrays were analyzed per Tp, with two biological replicates, two technical and one dye swap per biological replicate, for a total of 24 genome equivalent microarrays per Tp. An equimolar mixture of RNA from all Tp was used as the control.
Quantitative Real-Time PCR.
We used quantitative real-time PCR to validate our microarray analysis. The primer sequences and the results obtained from real-time PCR are presented in Table S5 and Table S6. Primer pairs for each gene were designed for 100- to 150-bp PCR products. Real-time PCR was performed by using QRT-PCR SYBR green dUTP Mix (Abgene) on an ABI 7500 system (Applied Biosystems). Twenty-five nanograms of DNase-treated total RNA was mixed with 1 μl of 0.1 mM gene-specific primers in a final volume of 11 μl, heat denatured for 5 min at 70°C, and chilled in ice water. Cycling conditions were 50°C for 45 min, 15 min at 95°C for activation of the hot start Taq polymerase, followed by 40 cycles of 15 s at 95°C and 30 s at 60°C, and by a dissociation curve. Each reaction was performed in four replicates, and the average CT was used to calculate the ratios for the amount of RNA obtained compared with the control.
Data Analyses.
The data from each microarray were independently normalized to correct for variations in labeling intensities between channels. The background signal for each measurement was subtracted from the raw signal. Points with background-subtracted signals ≤2 SD from the average background signal were flagged as below the noise level. There was no saturated point. For each channel, the signals were sorted by rank order, and probes that differed in rank order by ≥10% the number of probes were kept as the training set for normalization, discarding any probes below the noise level. A LOWESS curve was fit to the training set by using a window length of 25% the number of points fit. The signals were then normalized with the fit LOWESS curve by using linear interpolation to correct points that fell between two points in the training set. After normalization, log2 (experiment/control) ratios were calculated for each probe. With six replicate arrays for each Tp and two replicates per microarray, most log2 ratios were calculated from a minimum of 12 replicates, with additional replicates for genes with multiple unique probes per gene. Pearson correlations were calculated between all genes by using their log2 ratio expression profiles. Connections were made between genes with Pearson coefficients ≥0.90. Genes that changed by less than a given fold-change cutoff (2- or 1.3-fold) over the time course were discarded. Fold-change over the time course was calculated by adding the error (+/−2 SD) to each log2 ratio, then subtracting the lowest from the highest conservative error-adjusted values. The network was visualized by using Cytoscape version 2.3.2 (21), discarding any genes not connected to the central cyclic network. Cycling genes were identified by generating a network as above, by using a 1.3-fold cutoff. The numbers of cycling genes that peak at each Tp were counted by dividing the time course into 2 d, and for each gene, determining the Tp with maximal abundance on each day. Normalized control channels were scaled to a common median of 1,000, and replicate probes averaged to give the average control intensities for each gene. These were then scaled by the average log2 ratios to yield the transcript abundance at each Tp. The relative transcript abundances were calculated according to ref. 31 derived from global spot intensities on the Agilent microarrays.
Supplementary Material
Acknowledgments.
We thank all members of the Pakrasi laboratory for collegial discussions. This work is part of a Membrane Biology EMSL Scientific Grand Challenge project at the W. R. Wiley Environmental Molecular Sciences Laboratory, a national scientific user facility sponsored by the U.S. Department of Energy's Office of Biological and Environmental Research (BER) program located at Pacific Northwest National Laboratory (PNNL). PNNL is operated for the Department of Energy by Battelle. We thank MoGene for processing the microarrays and the initial data analysis.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The genome sequence reported in this paper has been deposited in the GenBank database (accession nos. CP000806–CP000811). The microarray data have been deposited in the European Bioinformatics Institute ArrayExpress database (accession nos. A-MEXP-864 and E-TABM-337).
This article contains supporting information online at www.pnas.org/cgi/content/full/0711068105/DCSupplemental.
References
- 1.Herzog ED. Neurons and networks in daily rhythms. Nat Rev Neurosci. 2007;8:790–802. doi: 10.1038/nrn2215. [DOI] [PubMed] [Google Scholar]
- 2.Hotta CT, et al. Modulation of environmental responses of plants by circadian clocks. Plant Cell Environ. 2007;30:333–349. doi: 10.1111/j.1365-3040.2006.01627.x. [DOI] [PubMed] [Google Scholar]
- 3.Dunlap JC, Loros JJ. How fungi keep time: circadian system in Neurospora and other fungi. Curr Opin Microbiol. 2006;9:579–587. doi: 10.1016/j.mib.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 4.Morse DS, Fritz L, Hastings JW. What is the clock? Translational regulation of circadian bioluminescence. Trends Biochem Sci. 1990;15:262–265. doi: 10.1016/0968-0004(90)90050-l. [DOI] [PubMed] [Google Scholar]
- 5.Johnson CH, Golden SS, Ishiura M, Kondo T. Circadian clocks in prokaryotes. Mol Microbiol. 1996;21:5–11. doi: 10.1046/j.1365-2958.1996.00613.x. [DOI] [PubMed] [Google Scholar]
- 6.Kondo T, et al. Circadian rhythms in prokaryotes: luciferase as a reporter of circadian gene expression in cyanobacteria. Proc Natl Acad Sci USA. 1993;90:5672–5676. doi: 10.1073/pnas.90.12.5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Woelfle MA, Ouyang Y, Phanvijhitsiri K, Johnson CH. The adaptive value of circadian clocks: an experimental assessment in cyanobacteria. Curr Biol. 2004;14:1481–1486. doi: 10.1016/j.cub.2004.08.023. [DOI] [PubMed] [Google Scholar]
- 8.Iwasaki H, Kondo T. Circadian timing mechanism in the prokaryotic clock system of cyanobacteria. J Biol Rhythms. 2004;19:436–444. doi: 10.1177/0748730404269060. [DOI] [PubMed] [Google Scholar]
- 9.Woelfle MA, Johnson CH. No promoter left behind: global circadian gene expression in cyanobacteria. J Biol Rhythms. 2006;21:419–431. doi: 10.1177/0748730406294418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mackey SR, Golden SS. Winding up the cyanobacterial circadian clock. Trends Microbiol. 2007;15:381–388. doi: 10.1016/j.tim.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 11.Haselkorn R. Heterocysts. Annu Rev Plant Physiol. 1978;29:319–344. [Google Scholar]
- 12.Wolk CP. Heterocyst formation. Annu Rev Genet. 1996;30:59–78. doi: 10.1146/annurev.genet.30.1.59. [DOI] [PubMed] [Google Scholar]
- 13.Campbell EL, Summers ML, Christman H, Martin ME, Meeks JC. Global gene expression patterns of Nostoc punctiforme in steady-state dinitrogen-grown heterocyst-containing cultures and at single time points during the differentiation of akinetes and hormogonia. J Bacteriol. 2007;189:5247–5256. doi: 10.1128/JB.00360-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reddy KJ, Haskell JB, Sherman DM, Sherman LA. Unicellular, aerobic nitrogen-fixing cyanobacteria of the genus Cyanothece. J Bacteriol. 1993;175:1284–1292. doi: 10.1128/jb.175.5.1284-1292.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Y, et al. Circadian orchestration of gene expression in cyanobacteria. Genes Dev. 1995;9:1469–1478. doi: 10.1101/gad.9.12.1469. [DOI] [PubMed] [Google Scholar]
- 16.Aoki S, Kondo T, Ishiura M. A promoter-trap vector for clock-controlled genes in the cyanobacterium Synechocystis sp. PCC 6803. J Microbiol Methods. 2002;49:265–274. doi: 10.1016/s0167-7012(01)00376-1. [DOI] [PubMed] [Google Scholar]
- 17.Kucho K, et al. Global analysis of circadian expression in the cyanobacterium Synechocystis sp. PCC 6803. J Bacteriol. 2005;187:2190–2199. doi: 10.1128/JB.187.6.2190-2199.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Steunou AS, et al. In situ analysis of nitrogen fixation and metabolic switching in unicellular thermophilic cyanobacteria inhabiting hot spring microbial mats. Proc Natl Acad Sci USA. 2006;103:2398–2403. doi: 10.1073/pnas.0507513103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meunier PC, Colon-Lopez MS, Sherman LA. Temporal changes in state transitions and photosystem organization in the unicellular, diazotrophic cyanobacterium Cyanothece sp. ATCC 51142. Plant Physiol. 1997;115:991–1000. doi: 10.1104/pp.115.3.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Makarova KS, Ponomarev VA, Koonin EV. Two C or not two C: recurrent disruption of Zn-ribbons, gene duplication, lineage-specific gene loss, and horizontal gene transfer in evolution of bacterial ribosomal proteins. Genome Biol. 2001;2:research0033.1–0033.13. doi: 10.1186/gb-2001-2-9-research0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hart SE, Schlarb-Ridley BG, Bendall DS, Howe CJ. Terminal oxidases of cyanobacteria. Biochem Soc Trans. 2005;33:832–835. doi: 10.1042/BST0330832. [DOI] [PubMed] [Google Scholar]
- 23.Devroede N, Huysveld N, Charlier D. Mutational analysis of intervening sequences connecting the binding sites for integration host factor, PepA, PurR, and RNA polymerase in the control region of the Escherichia coli carAB operon, encoding carbamoylphosphate synthase. J Bacteriol. 2006;188:3236–3245. doi: 10.1128/JB.188.9.3236-3245.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H, Sherman DM, Bao S, Sherman LA. Pattern of cyanophycin accumulation in nitrogen-fixing and nonnitrogen-fixing cyanobacteria. Arch Microbiol. 2001;176:9–18. doi: 10.1007/s002030100281. [DOI] [PubMed] [Google Scholar]
- 25.Roose JL, Pakrasi HB. Evidence that D1 processing is required for manganese binding and extrinsic protein assembly into photosystem II. J Biol Chem. 2004;279:45417–45422. doi: 10.1074/jbc.M408458200. [DOI] [PubMed] [Google Scholar]
- 26.Kanehisa M, Goto S, Kawashima S, Nakaya A. The KEGG databases at GenomeNet. Nucleic Acids Res. 2002;30:42–46. doi: 10.1093/nar/30.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tolonen AC, et al. Global gene expression of Prochlorococcus ecotypes in response to changes in nitrogen availability. Mol Sys Biol. 2006;53:1–15. doi: 10.1038/msb4100087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Price GD, Woodger FJ, Badger MR, Howitt SM, Tucker L. Identification of a SulP-type bicarbonate transporter in marine cyanobacteria. Proc Natl Acad Sci USA. 2004;101:18228–18233. doi: 10.1073/pnas.0405211101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sherman LA, Meunier P, Colon-Lopez MS. Diurnal rhythms in metabolism: A day in the life of a unicellular, diazotrophic cyanobacterium. Photosynth Res. 1998;58:25–42. [Google Scholar]
- 30.Aurora R, Hihara Y, Singh AK, Pakrasi HB. A network of genes regulated by light in cyanobacteria. OMICS. 2007;11:166–185. doi: 10.1089/omi.2007.4323. [DOI] [PubMed] [Google Scholar]
- 31.Held GA, Grinstein G, Tu Y. Relationship between gene expression and observed intensities in DNA microarrays - a modeling study. Nucleic Acids Res. 2006;34:e70. doi: 10.1093/nar/gkl122. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.