Abstract
Of the five echinoderm classes, only the modern sea urchins (euechinoids) generate a precociously specified embryonic micromere lineage that ingresses before gastrulation and then secretes the biomineral embryonic skeleton. The gene regulatory network (GRN) underlying the specification and differentiation of this lineage is now known. Many of the same differentiation genes as are used in the biomineralization of the embryo skeleton are also used to make the similar biomineral of the spines and test plates of the adult body. Here, we determine the components of the regulatory state upstream of these differentiation genes that are shared between embryonic and adult skeletogenesis. An abrupt “break point” in the micromere GRN is thus revealed, on one side of which most of the regulatory genes are used in both, and on the other side of which the regulatory apparatus is entirely micromere-specific. This reveals the specific linkages of the micromere GRN forged in the evolutionary process by which the skeletogenic gene batteries were caused to be activated in the embryonic micromere lineage. We also show, by comparison with adult skeletogenesis in the sea star, a distant echinoderm outgroup, that the regulatory apparatus responsible for driving the skeletogenic differentiation gene batteries is an ancient pleisiomorphic aspect of the echinoderm-specific regulatory heritage.
Keywords: embryonic and adult skeletogenesis, gene network evolution, sea urchin, sea star
The geometrical, elegantly formed bilateral skeleton of the sea urchin embryo is one of its most prominent features. Its vertex underpins the aboral vertex of the embryo body wall, producing its triangular shape, and its rods extend four arms that project anteriorly of the mouth, mounting rows of cilia that assist in transfer of food microorganisms for ingestion. Its exact form is species-specific, but an embryonic skeleton is a canonical character of all indirectly developing modern sea urchins (i.e., the “thin spined” or euechinoid sea urchins). The skeleton is constructed during and immediately after the gastrula stage of development, by dedicated cells all belonging to a polyclonal lineage consisting of all descendants of four fifth-cleavage large micromeres (micromere lineage). These micromeres arise at the vegetal pole of the egg in a stereotypic series of unequal cleavages. The micromere lineage undergoes a set number of further cleavages during blastula stage while they reside in the center of the vegetal plate, and then, well before gastrular invagination of the archenteron, these cells acquire motility, and singly ingress into the blastocoel. In Strongylocentrotus purpuratus, the subject of most of the work in this article, there are 16 skeletogenic micromeres at time of ingression, and this number just doubles as skeletogenesis begins. The skeletal rods or spicules are deposited on the inner wall of the blastocoel within syncytial cables formed by the now mesenchymal descendants of the micromere lineage and are positioned according to signals produced by the ectoderm (ref. 1; for review of embryonic skeletogenesis, see ref. 2).
Even the nearest outgroup to the euechinoids, the “pencil urchins” or cidaroids, lack a precociously invaginating skeletogenic micromere lineage (3–5). Their skeletogenic cells ingress with other kinds of mesodermal cell types at gastrulation, and they form the skeleton much later, at the very end of embryogenesis just before feeding begins. This distinction provides a time line for the evolutionary origin of the euechinoid type of skeletogenic micromere lineage, because the only sea urchin-like survivors of the Permian-Triassic extinction 250 mya were two cidaroid lineages (6); the first euechinoids appear only later, in the Triassic. No other class of echinoderms produces a skeletogenic lineage either [for phylogeny of the echinoderm classes, see supporting information (SI) Fig. S1]. Indirectly developing crinoid, sea star, and sea cucumber embryos have no skeleton at all, and, although ophiouroids do, they have no micromere lineage and build the skeleton differently (7, 8). The euechinoid skeletogenic micromere lineage and its embryonic skeletal structures thus may be considered a derived euechinoid character that appeared in the euechinoid common ancestor probably not long after 250 mya.
How could this character have arisen, in terms of the heritable genomic regulatory program that controls development? More precisely, what regulatory innovations could have been responsible for inserting skeletogenesis into a micromere embryological address? A starting point is at the differentiation gene batteries that generate the biomineral matrix. Earlier evidence indicated that many specific protein components of the biomineral matrix are present in both demineralized embryonic skeletal spicules and demineralized adult endoskeleton, i.e., spines and test plates (9). Analysis of biomineralization genes in the S. purpuratus genome sequence (10) proves this is the case, although, for some families of these genes, certain paralogues are used in adult skeletons, some in embryonic and some in both. Thus, to begin, we may consider that our problem resolves at base into the regulatory changes that might have been required to cause mobilization of what were adult skeletogenic differentiation gene batteries in the blastula stage micromere lineage cells. This is when in development downstream skeletogenic differentiation genes begin to be expressed in the micromere lineage (for review, see ref. 11).
Recently the GRN was solved that embodies the program for development of the micromere lineage, all of the way from its initial specification, to expression of skeletogenic differentiation genes (12). Above the differentiation genes there are 23 genes in this network, all encoding transcription factors, except for some that encode signaling functions, another essential aspect of the developmental role of the micromere lineage. Interactions among these regulatory genes generate a progressive sequence of regulatory states, beginning with those that define the distinct functional identity of the micromeres, then enhancing and stabilizing the expression of the definitive set of regulators that in concert operate the differentiation gene batteries. We can now resolve our evolutionary question further: At exactly what level of this known regulatory cascade was the control system of the adult skeletogenic gene batteries joined to the developmental control system that defines the micromere lineage? A step toward answering this question can be taken by determining what subset of the 23 regulatory and signaling genes of the micromere GRN is also used in cells building the skeleton of the adult body plan.
Results
Development of Adult Skeletal Structures in Advanced Larvae.
The skeletal structures of the adult body plan begin to be formulated during larval life in indirectly developing echinoderms. These structures are the test plates of the adult endoskeleton and the primary spines (for review, see ref. 8). Microscopic visualizations of the larvae of S. purpuratus and Asterina miniata with newly formed skeletal elements of the juvenile adult body plan are displayed in Fig. 1. A recently demonstrated fact important for this study is that in sea urchins the larval cells producing the skeletal structures of the adult body plan are not descendants of the skeletogenic micromere lineage of the embryo but rather an independently arising postembryonic mesodermal cell population (13). This issue does not arise in sea stars, because they have no embryonic skeletogenic cells.
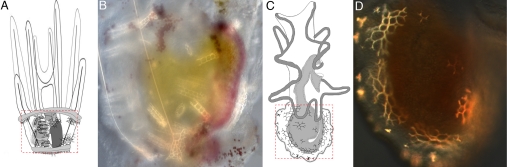
Fig. 1.
Adult skeletogenesis in sea urchin and sea star larvae. (A) Diagram of advanced rudiment stage larva of S. purpuratus (adapted from ref. 13, with permission from Elsevier). (B) Polarized light image of juvenile skeletogenic centers (red dashed area in A), showing spines, plates, and disks of the primary tube feet, elements of the adult skeleton. (C) Diagram of advanced rudiment stage A. miniata larva (modified from ref. 8). (D) Polarized light image of juvenile skeletogenic center (red dashed area in C), showing spines and plates.
The accessibility of adult skeletogenesis as shown in Fig. 1 made it possible to explore the utilization, in the juvenile skeletogenic centers, of the genetic regulatory apparatus contained in the micromere lineage GRN. Although it is not easy to establish functional linkages in larval skeletogenesis, we could determine first by QPCR measurements of staged larval RNA, and then by whole-mount in situ hybridization (WMISH), whether or not the genes that are known to be functionally linked in the micromere GRN are expressed in the specific juvenile skeletogenic centers of the larva.
Comparisons of Gene Expression in Adult and Embryonic Skeletogenesis of S. purpuratus.
The sea urchin micromere GRN (ref. 12; http://sugp.caltech.edu/endomes) includes 31 genes, of which 23 are regulatory and signaling genes (one yet unidentified) and the remainder are a sample of differentiation genes, i.e., genes encoding biomineral proteins and cell biology functions required for skeletal deposition (10). Every regulatory gene expressed specifically in the micromere lineage up through late blastula stage is included in the GRN. Here, we report the presence or absence of transcripts of each of the network genes encoding transcription factors in the juvenile skeletonization centers of advanced larvae. We also studied the genes encoding the Delta and VEGFR-II ligands, which are expressed in the micromere lineage, plus four of the differentiation genes (two biomineral matrix genes and two cell biology genes), a total of 24 genes. They are listed by name in Table S1 and can be seen in the comparative GRN map below. A sampling of WMISH images that reveal unequivocal expression in the forming spines and test plates is shown in Fig. 2, and the remainder are presented in Fig. S2.
Fig. 2.
Representative WMISH of probes for genes expressed in the micromere lineage to juvenile skeletonization centers of sea urchin larvae. (Upper) Larval WMISH viewed from the left sides of the larvae, displaying labeling in ventral and dorsal spines. (Lower) Sections made after WMISH, cut along the left–right axis, except B3, which is cut along the oral-aboral axis. Identity of probes is indicated in the figures. (A2, B2, and C2) Labeling in forming spines and plates, and, in C2, the tube foot discs as well. (D) The tbr gene is not expressed in the juvenile skeletogenic centers.
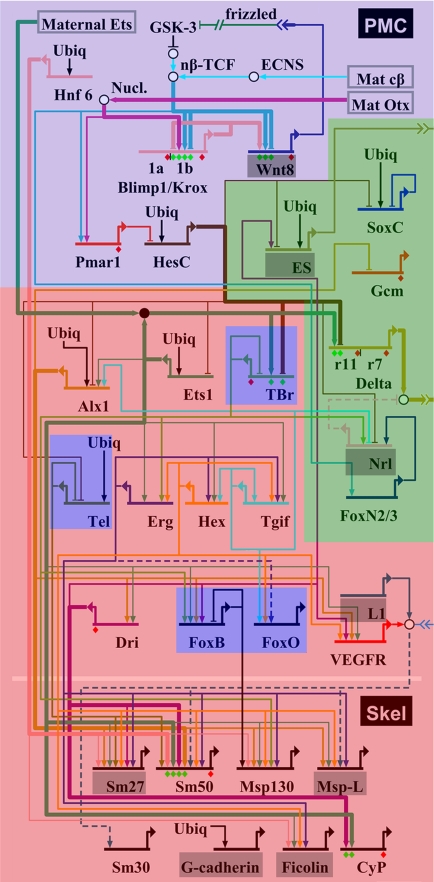
These data are superimposed on the micromere GRN in Fig. 3. The resulting pattern of overlap in use provides a clear general answer to our question. The part of the GRN the function of which is to install the initial state of micromere specification is particular to the micromeres and is not used in the juvenile skeletogenic centers (violet region in Fig. 3), nor is the part of the GRN the function of which is presentation of inductive signals to the adjacent cells (green in Fig. 3). But the expressed differentiation genes tested, and, with few exceptions, the complete repertoire of expressed regulatory genes upstream of these gene batteries and downstream of the initial specification system is shared with the juvenile skeletogenic centers (red in Fig. 3). The exact point at which the micromere-specific regulatory apparatus and the definitive skeletogenic regulatory systems are joined, or were joined in evolution, is just downstream of the pmar1-hesC linkage in the GRN (Fig. 3).
Fig. 3.
Map of gene expression data on micromere GRN. Genes expressed in juvenile skeletogenic centers and micromere lineage are shown over the red background. Genes used only in micromere lineage (<24–30 h) are shown over violet and green backgrounds. Green denotes genes used in micromere-specific signaling functions. The violet territory at the top denotes the portion of the GRN that executes the initial micromere specification function (see text); the four additional blue genes are individual cooptions, as discussed. Several genes not studied here are denoted over gray backgrounds.
Pleisiomorphic Regulatory System Controlling Skeletogenesis.
A definitive and unique feature of echinoderms is the calcite biomineral of their adult endoskeleton (stereom) and it is likely that the structural components of this are an ancient pleisiomorphic character (14). Stereom is evident even in fossil stem group echinoderms that had yet to acquire pentaradial lateral symmetry (15). Is the regulatory apparatus that controls skeletogenic gene batteries possibly also an ancient pleisiomorphy? Or, in the context of the present study, is that portion of the expressed regulatory gene repertoire shared between these two very different developmental addresses, the embryonic micromere lineage and the skeletogenesis centers of the forming adult body plan, also pleisiomorphic?
The sea stars (Asteroids) are a distant class within the echinoderm phylum (Fig. S1), and their last common ancestors with sea urchins existed at the end of the Cambrian, half a billion years ago. We tested the juvenile skeletogenesis centers of advanced sea star larvae for expression of eight key skeletogenic regulatory genes and one downstream gene (Table S1) and found results identical to those seen in sea urchin larvae. Representative examples are shown in Fig. 4, and the remainder are shown in Fig. S3. The skeletogenic regulatory apparatus may thus have existed as a modular GRN unit since early in echinoderm history, and it was this same unit that was placed into the embryological control system defining the micromere lineage regulatory state. There is presumptive evidence for another highly conserved GRN kernel, such as that responsible for endoderm specification discovered in a comparison of endomesoderm GRNs of the sea star and the sea urchin (16). But this proposition could only be substantiated only by determining whether the actual cis-regulatory linkages are as pleisiomorphic as is the skeletogenic gene expression complex.
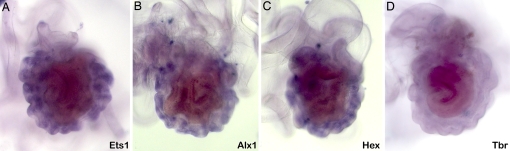
Fig. 4.
Representative WMISH of probes for genes expressed in the sea urchin micromere lineage to juvenile skeletonization centers of sea star larvae. Views are from the left side of the whole larvae. As in the sea urchin, ets1 (A), alx1 (B), and hex (C) are expressed in juvenile spines, whereas tbr (D) is not expressed.
Discussion
We show here the added value for evolutionary analysis of a solved developmental GRN. The GRN provides a map for orientation of expression data. In Fig. 3, we show a clear boundary on the GRN map between the micromere regulatory apparatus that is shared with the juvenile skeletogenic centers and the apparatus that is particular to this embryonic lineage, and, from this, some conclusions can be drawn. Although we cannot be certain that the genes in the shared region of the map are indeed linked similarly in the two developmental contexts, we can be fairly certain of the parts that are micromere-specific.
The Micromere “Address”: Past and Present.
In terms of embryological function the micromeres have first to interpret the anisotropic inputs sequestered uniquely within them and transform them into a zygotic regulatory state and then to perform two general classes of function. These are to express essential intercellular signals and to develop skeletogenic potential. As we discuss in ref. 12, the micromere lineage GRN provides a complete causal explanation of these functions. The first two of these tasks are executed respectively by the apparatus in the violet and green portions of the comparison map in Fig. 3, and the third by that in the red is the shared portion.
One interpretation is that an earlier evolutionary state in the echinoid line was the presence of micromeres that operated the violet and green portions of the current GRN; i.e., that established their initial unique regulatory state to serve as an embryonic signaling center. There are two essential micromere signals. The first is the “Early Signal” (ES in the GRN), still not defined biochemically, which in echinoids executes a derepression function needed to permit the endomesodermal development of the adjacent cells. The second is the Delta signal, used in echinoids for specification of mesoderm, particularly pigment cells. Significantly, both are under control of the pmar1-hesC regulatory system, as can be seen in Fig. 3. Both of these genes encode repressors and this system functions as a double-negative gate (17–19). This gate performs the function of transducing the initial anisotropies sequestered in the micromeres into a micromere-specific regulatory state, because the pmar1 gene is activated in response to these localized inputs. Thus, once presented in the micromeres, the Pmar1 repressor in turn represses the hesC gene, the source of an otherwise global zygotic transcriptional repressor that keeps the ES and Delta genes off outside the micromeres. Its target genes, here the es and delta genes, are thereby activated specifically in the micromeres. The micromere double-negative gate may have evolved initially as a controller of the micromere signaling apparatus.
The double-negative gate is also the precise junction between the rest of the micromere apparatus (blue in Fig. 3) and the part shared with the juvenile skeletogenic centers (red in Fig. 3). To effect the incorporation of the latter into the micromere GRN, two genes of the adult skeletogenic pathway need have been placed additionally under control of the double-negative gate, namely, alx1 and ets1. This requires providing them with cis-regulatory systems that respond to ubiquitously active inputs and hesC repression (ref. 19; genes in this subnetwork are all ubiquitously expressed whether pmar1 is overexpressed or hesC is prevented from being expressed).
Superimposed Cooptions.
The skeletogenic regulatory apparatus can be defined as GRN circuitry that provides the immediate or penultimate inputs into the differentiation genes. The skeletogenic regulatory apparatus of the micromeres differs from that of the juvenile skeletogenesis centers by at least four genes that are used in the micromeres but not in the juvenile centers (of course, there could be others used in the juvenile and not in the micromeres). These are foxB, foxO, tel, and tbrain (tbr) (blue regions in Fig. 3). The two fox genes are activated very late and may not be intrinsic components of the skeletogenesis system per se, i.e., they may control cell biology functions, as suggested for instance by the tel target gene msp130 (12), which encodes a cell surface glycoprotein. But tel and tbr are in the upstream region of the GRN and are under control of the double-negative gate. We know nothing of evolutionary significance for this story about tel, but tbr provides an explicit case of cooption, because this gene is expressed in endomesoderm in other echinoderms, including sea stars (20, 21), where it is obligatory for endomesoderm specification (21); in echinoids, tbr is expressed only in the skeletogenic micromere lineage. We may conclude that, perhaps typically, following the linkage of the pleisiomorphic skeletogenic regulatory apparatus into the micromere GRN, additional regulatory gene cooptions took place in the echinoid lineage. It is interesting that the route of these cooptions was the same: linkage into the double-negative gate (Fig. 3).
Skeletogenic Paleogenomics.
Finally, we may inquire about the evolutionary origin of the pleisiomorphic skeletogenic regulatory apparatus of echinoderms. Although it lies beyond the scope of this article, an essential point is that many of the genes of this GRN circuitry are also expressed in nonskeletogenic mesoderm. Cooption of some additional regulatory genes, erection of stabilizing intergenic circuitry, and formulation of biomineralization gene batteries by linking them into the modified mesodermal regulatory circuitry must have taken place in the remote preskeletogenic stem group echinoderm lineage. A similar sequence of events engaging other kinds of mesodermal structural genes, superimposed on a broadly distributed mesodermal regulatory state, must underlie the evolutionary diversification of mesoderm in Bilateria.
Heterochrony.
The precocious expression of skeletogenic functions in the micromere lineage is a classic case of evolutionary heterochrony in which a complex function pleisiomorphically larval and adult and, in some primitive echinoids, late embryonic has been moved forward in the life cycle to the early embryonic period (4, 13). Here, we can glimpse the mechanism of this heterochrony: Essentially, an entire adult tissue GRN subnetwork has been transferred to an embryonic developmental regulatory address. We show how this could have occurred by means of a few changes in the cis-regulatory linkages of a few genes at the top of the subnetwork hierarchy, plus a few cooptions. Of course, the subnetwork was inserted not into a vacuum but rather into an also-pleisiomorphic regulatory apparatus operating to define the vegetal endomesoderm of the egg. Unfortunately, we have little comparative evidence on the structure of the underlying vegetal specification GRN. But this could be a phylogenetically widespread system judging from clues such as the near universal eumetazoan nuclearization of β-catenin in embryonic cells at the vegetal poles of eggs in deuterostomes, lophotrochozoans, and cnidarians (22–25).
Concluding Remark.
Whether or not the evolutionary pathway sketched out here is an accurate reconstruction of events, it is a plausible explanation, couched in terms of what would be at root genomic cis-regulatory changes that affect GRN architecture. It is these kinds of change that must be responsible for features of morphology and function that arise physically during the life cycle through the operation of evolutionarily derived developmental pathways (26). Some specific and testable correlative predictions follow from the proposed pathway. For example, it might be predicted that, in cidaroids, the micromeres have the signaling capacities of euechinoid micromeres and that these could be under the control of the same double-negative regulatory gate as in the euechinoid, whereas the skeletogenic regulatory genes are not. A set of additional predictions that could be tested by reintroduction into sea urchin eggs, together with computational genomics, concerns the cis-regulatory modules of the sea star skeletogenic regulatory genes. If there is indeed a pan-echinoderm skeletogenic kernel, then the sea star regulatory modules thereof should display most of the same functional linkages and require most of the same inputs when introduced in reporter constructs into sea urchin eggs as are portrayed in the sea urchin GRN in Fig. 3.
Developmental GRNs provide a potent approach to definition of the mechanisms of evolutionary change in the body plans of embryos and adults. Only in the terms of GRN architecture can the actual nature of such changes be proposed and tested.
Materials and Methods
WMISH.
Sea urchin larvae were grown as described in refs. 27 and 28, and similar methods were used to culture sea star (A. miniata) larvae. The larvae were fed on Rhodomonas lens and/or R. salina. Adult skeleton was first visible from 2 and 4 weeks after fertilization in S. purpuratus and A. miniata, respectively. S. purpuratus and A. miniata larvae required, respectively, 6 and 9 weeks after fertilization to attain the stage competent for the metamorphosis.
Fixation and RNA extraction were done every week from the embryo through the late larval stages. Larvae were fixed in 4% paraformaldehyde as described in ref. 29. Twenty-four skeletogenic micromere genes from the sea urchin GRN and nine sea star orthologs isolated by library screening and degenerate PCR were studied by WMISH. Digoxigenin was used to label riboprobes of ≈1 kb. For genes with larger transcripts, several probes were made from different regions, and a mix of probes was used. Hybridization was for 1 week (29).
Viewing, Embedding, Sectioning, and Photography.
After the staining reaction, the specimens were observed in an inverted microscope to determine the profile of their expression at all stages. For expression patterns that were not clear in whole mount display, larvae were embedded and sectioned. Staining patterns were examined in serial 8-μm cryosections or in 4-μm Durcupan sections.
Supplementary Material
Acknowledgments.
We thank Paola Oliveri and Qiang Tu for their extraordinary efforts in constructing the micromere GRN in sea urchins, which provided the foundations for this evolutionary study; Andy Cameron and Pat Leahy for the instruction in culturing sea urchin larvae; Veronica Hinman for the instruction in working with sea star embryos and for the screening of sea star ets1 cDNA clone; and Andy Cameron, Haixia Huang, and Viveca Sapin for help with microtomes. This work was supported by National Science Foundation Grant IOS-0641398 and the Camilla Chandler Frost Fellowship.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0801201105/DCSupplemental.
References
- 1.Armstrong N, McClay DR. Skeletal pattern is specified autonomously by the primary mesenchyme cells in sea urchin embryos. Dev Biol. 1994;162:329–338. doi: 10.1006/dbio.1994.1090. [DOI] [PubMed] [Google Scholar]
- 2.Guss KA, Ettensohn CA. Skeletal morphogenesis in the sea urchin embryo: Regulation of primary mesenchyme gene expression and skeletal rod growth by ectoderm-derived cues. Development. 1997;124:1899–1908. doi: 10.1242/dev.124.10.1899. [DOI] [PubMed] [Google Scholar]
- 3.Tennent DH. The early influence of the spermatozoan upon the characters of echinoid larvae. Carnegie Inst Wash Publ. 1914;182:129–138. [Google Scholar]
- 4.Wray GA, McClay DR. The origin of spicule-forming cells in a “primitive” sea urchin (Eucidaris tribuloides) which appears to lack primary mesenchyme cells. Development. 1988;103:305–315. doi: 10.1242/dev.103.2.305. [DOI] [PubMed] [Google Scholar]
- 5.Schroeder TE. Development of a “primitive” sea urchin (Eucidaris tribuloides): Irregularities in the hyaline layer, micromeres, and primary mesenchyme. Biol Bull. 1981;161:141–151. [Google Scholar]
- 6.Smith AB, Hollingworth NTJ. Tooth structure and phylogeny of the Upper Permian echinoid, Miocidaris keyserlingi. Proc Yorkshire Geol Soc. 1990;48:47–60. [Google Scholar]
- 7.Nakano H, Hibino T, Oji T, Hara Y, Amemiya S. Larval stages of a living sea lily (stalked crinoid echinoderm) Nature. 2003;421:158–160. doi: 10.1038/nature01236. [DOI] [PubMed] [Google Scholar]
- 8.Hyman LH. The Invertebrates: Echinodermata. New York: McGraw–Hill; 1955. pp. 1–763. [Google Scholar]
- 9.Benson SC, Benson NC, Wilt F. The organic matrix of the skeletal spicule of sea urchin embryos. J Cell Biol. 1986;102:1878–1886. doi: 10.1083/jcb.102.5.1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Livingston BT, et al. A genome-wide analysis of biomineralization-related proteins in the sea urchin Strongylocentrotus purpuratus. Dev Biol. 2006;300:335–348. doi: 10.1016/j.ydbio.2006.07.047. [DOI] [PubMed] [Google Scholar]
- 11.Davidson EH. The Regulatory Genome: Gene Regulatory Networks in Development and Evolution. San Diego: Academic; 2006. pp. 1–303. [Google Scholar]
- 12.Oliveri P, Qiang T, Davidson EH. Global regulatory logic for specification of an embryonic cell lineage. Proc Natl Acad Sci USA. 2008;105:5955–5962. doi: 10.1073/pnas.0711220105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yajima M. A switch in the cellular basis of skeletogenesis in late-stage sea urchin larvae. Dev Biol. 2007;307:272–281. doi: 10.1016/j.ydbio.2007.04.050. [DOI] [PubMed] [Google Scholar]
- 14.Bottjer DJ, Davidson EH, Peterson KJ, Cameron RA. Paleogenomics of echinoderms. Science. 2006;314:956–960. doi: 10.1126/science.1132310. [DOI] [PubMed] [Google Scholar]
- 15.Smith AB. biomineralization in echinoderms. In: Carter JG, editor. Skeletal Biomineralization: Patterns, Processes and Evolutionary Trends. London: Van Nostrand Reinhold; 1990. pp. 413–443. [Google Scholar]
- 16.Hinman VF, Nguyen AT, Cameron RA, Davidson EH. Developmental gene regulatory network architecture across 500 million years of echinoderm evolution. Proc Natl Acad Sci USA. 2003;100:13356–13361. doi: 10.1073/pnas.2235868100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oliveri P, Carrick DM, Davidson EH. A regulatory gene network that directs micromere specification in the sea urchin embryo. Dev Biol. 2002;246:209–228. doi: 10.1006/dbio.2002.0627. [DOI] [PubMed] [Google Scholar]
- 18.Oliveri P, Davidson EH, McClay DR. Activation of pmar1 controls specification of micromeres in the sea urchin embryo. Dev Biol. 2003;258:32–43. doi: 10.1016/s0012-1606(03)00108-8. [DOI] [PubMed] [Google Scholar]
- 19.Revilla-i-Domingo R, Oliveri P, Davidson EH. A missing link in the sea urchin embryo gene regulatory network: hesC and the double-negative specification of micromeres. Proc Natl Acad Sci USA. 2007;104:12383–12388. doi: 10.1073/pnas.0705324104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maruyama YK. A sea cucumber homolog of the mouse T-Brain-1 is expressed in the invaginated cells of the early gastrula in Holothuria leucospilota. Zool Sci. 2000;17:383–389. doi: 10.2108/jsz.17.383. [DOI] [PubMed] [Google Scholar]
- 21.Hinman VF, Nguyen AT, Davidson EH. Evolutionary plasticity of developmental gene regulatory network architecture. Proc Natl Acad Sci USA. 2007;104:19404–19409. doi: 10.1073/pnas.0709994104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Logan CY, Miller JR, Ferkowicz MJ, McClay DR. Nuclear beta-catenin is required to specify vegetal cell fates in the sea urchin embryo. Development. 1999;126:345–357. doi: 10.1242/dev.126.2.345. [DOI] [PubMed] [Google Scholar]
- 23.Imai K, Takada N, Satoh N, Satou Y. β-catenin mediates the specification of endoderm cells in ascidian embryos. Development. 2000;127:3009–3020. doi: 10.1242/dev.127.14.3009. [DOI] [PubMed] [Google Scholar]
- 24.Schneider SQ, Bowerman B. Beta-catenin asymmetries after all animal/vegetal-oriented cell divisions in Platynereis dumerilii embryos mediate binary cell-fate specification. Dev Cell. 2007;13:73–86. doi: 10.1016/j.devcel.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 25.Wikramanayake AH, et al. An ancient role for nuclear beta-catenin in the evolution of axial polarity and germ layer segregation. Nature. 2003;426:446–450. doi: 10.1038/nature02113. [DOI] [PubMed] [Google Scholar]
- 26.Davidson EH, Erwin DH. Gene regulatory networks and the evolution of animal body plans. Science. 2006;311:796–800. doi: 10.1126/science.1113832. [DOI] [PubMed] [Google Scholar]
- 27.Leahy P. In: Methods in Cell Biology. Schroeder TE, editor. Orlando, FL: Academic; 1986. pp. 1–13. [Google Scholar]
- 28.Cameron RA, Britten RJ, Davidson EH. Expression of two actin genes during larval development in the sea urchin Strongylocentrotus purpuratus. Mol Reprod Dev. 1989;1:149–155. doi: 10.1002/mrd.1080010302. [DOI] [PubMed] [Google Scholar]
- 29.Arenas-Mena C, Cameron RA, Davidson EH. Spatial expression of Hox cluster genes in the ontogeny of a sea urchin. Development. 2000;127:4631–4643. doi: 10.1242/dev.127.21.4631. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.