Abstract
Adenovirus E1A drives oncogenesis by targeting key regulatory pathways that are critical for cellular growth control. The interaction of E1A with p400 is essential for many E1A activities, but the downstream target of this interaction is unknown. Here, we present evidence that the oncoprotein transcription factor Myc is the target of this interaction. We show that E1A stabilizes Myc protein via p400 and promotes the coassociation of Myc and p400 at Myc target genes, leading to their transcriptional induction. We also show that E1A requires Myc for its ability to activate Myc-dependent gene expression and induce apoptosis, and that forced expression of Myc is sufficient to rescue the activity of an E1A-mutant defective in p400 binding. Together, these findings establish that Myc, via p400, is an essential downstream target of E1A.
Keywords: proteolysis, transcription, transformation, apoptosis
One of the most important tools for exposing the mechanisms of oncogenic transformation are DNA tumor viruses. Because these viruses, such as adenovirus, depend on the cellular DNA replication machinery to propagate, they must drive host cells into the cell cycle. It is this release from cellular growth control that promotes oncogenic transformation. The utility of DNA tumor viruses for cancer research is based on the premise that they have evolved to target the minimum number of cellular pathways necessary for virus propagation and cellular transformation. Understanding how DNA tumor viruses promote oncogenesis, therefore, can reveal the most vulnerable cellular pathways and nodes that are linked to tumorigenesis.
Adenovirus E1A is perhaps the most widely studied oncogene from a DNA tumor virus. E1A encodes proteins that have a range of activities, including the ability to induce cell proliferation and transformation, inhibit differentiation, and promote apoptosis. E1A proteins exert these effects by binding to, and modifying the function of, key cell cycle regulators (1). The most prominent of these regulators is the tumor suppressor protein Rb, but interactions of E1A with chromatin remodeling factors such as p300/CBP (2) and p400 (3) also contribute to its biological activities. The interaction of E1A with p400 is particularly important because E1A fails to induce apoptosis in cells that do not express p400 (4), and an E1A mutant that is specifically defective for p400 binding (Δ26–35), but can still interact with p300/CBP, is impaired for both transformation and apoptosis (3, 4). Although p400 is clearly required for E1A's activities, the mechanism through which it functions in this capacity is unknown.
Recently, it was reported that E1A can inhibit the ubiquitin (Ub)-mediated destruction of Myc during the course of adenovirus infection (5). It has also been reported that E1A can interact with multiple subunits of the 19S proteasome to inhibit proteasomal proteolysis (6). Although global proteasome inhibition could account for the stabilization of Myc by E1A, the exact mechanism through which E1A stabilizes the Myc protein is unknown. We have investigated how E1A attenuates Myc proteolysis and find that, contrary to expectations, stabilization of Myc does not occur via widespread proteasome inhibition. Instead, E1A stabilizes Myc by promoting its association with p400, which in turn reduces Myc ubiquitylation and promotes formation of a Myc–p400 cocomplex on promoter DNAs. Consistent with these findings, we also show that E1A can activate Myc target genes and that Myc is an essential downstream effector of E1A. Together, these data reveal that stabilization of Myc by E1A is a specific targeted effect of the adenoviral protein and establish that the E1A–p400–Myc connection is important for oncogenesis.
Results and Discussion
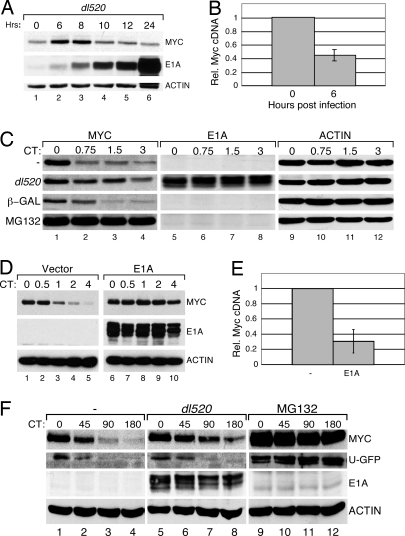
To examine the effects of E1A on Myc protein stability, human U2OS cells were infected with the Ad5 adenovirus dl520, which expresses WT 12S E1A, and endogenous Myc protein levels were assessed by Western blot (WB; Fig. 1A). Six to 8 h after infection, at the point at which E1A expression was first detected, we observed an increase in steady-state Myc levels, which gradually subsided over a 12- to 24-h period. Consistent with previous reports (5), the increase in Myc protein levels at 6 h was accompanied by a decrease in the levels of Myc mRNA (Fig. 1B). Although Baluchamy et al. (7) have reported that adenovirus activates Myc gene expression, these experiments were done in quiescent cells; our assays, and those of Lohr et al. (5), were performed in cycling cells, suggesting that regulation of Myc transcription by adenovirus is influenced by the growth status of the cells.
Fig. 1.
Adenovirus E1A stabilizes Myc. (A) U2OS cells were infected with adenovirus dl520 and harvested at the indicated time points. Levels of Myc, E1A, and actin were determined by WB. (B) U2OS cells were infected with adenovirus, as in A, and RNA was harvested. Levels of c-Myc cDNAs were analyzed by quantitative PCR and normalized to those of an actin control. (C) U2OS cells were either infected with dl520 or a control virus expressing β-gal, mock-infected, or treated with the proteasome inhibitor, MG132, and incubated for 6 h. CHX was added to inhibit protein synthesis, and protein samples were taken at the indicated times (CT; hours). WB analysis shows the levels of Myc, E1A, and actin. (D) U2OS cells were transduced with a retroviral expression construct encoding WT 12S E1A (pLPC E1A). CHX chase, followed by WB, was used to analyze Myc turnover in E1A expressing cells and control (vector) cells. (E) RNA was isolated from cells in D, and levels of c-Myc cDNAs relative to actin, determined by quantitative PCR. (F) U2OS cells were transiently transfected with a plasmid encoding unstable GFP (U-GFP) and incubated for ≈20 h. These cells were then either infected with dl520, mock-infected, or treated with MG132 and incubated for 6 h. CHX chase was used to analyze the levels of GFP, Myc, E1A, and actin.
The transient increase in Myc levels we observed, together with the decline in Myc mRNA, suggested that Myc is stabilized during the course of adenovirus infection. This notion was confirmed by treating infected cells with cyclohexamide (CHX), and monitoring Myc levels by WB (Fig. 1C). Under these conditions, adenovirus stabilized Myc considerably, and in a manner that depended on E1A; a virus that expresses LacZ (β-gal) instead of E1A did not induce Myc stability (Fig. 1C). Importantly, expression of E1A alone was sufficient to stabilize Myc; retroviral expression of 12S E1A in U2OS cells resulted in a potent stabilization of Myc (Fig. 1D) and a commensurate decrease in Myc mRNA levels (Fig. 1E). Thus, confirming earlier work (5), E1A promotes Myc stability.
E1A has been shown to interact with multiple subunits of the 19S proteasome, and this interaction has been proposed to inhibit proteasomal proteolysis (6). Because Myc proteolysis requires proteasome function (8), global inhibition of proteasome activity by E1A could account for its ability to stabilize Myc. It is important to note, however, that E1A has not been shown to elicit widespread proteasome inhibition in vivo. We therefore asked whether adenoviral infection stabilizes the synthetic protein U-GFP (9), a substrate that is widely used as an in vivo reporter of proteasome activity (Fig. 1F). Under conditions where adenovirus stabilized Myc, there was little, if any, change in the rate of destruction of U-GFP, demonstrating that proteasome function is not generally attenuated in adenovirus-infected cells. This notion was supported by comparing the effects of adenovirus infection and proteasome inhibition on Myc localization [supporting information (SI) Fig. S1]. Whereas proteasome inhibition results in the redistribution of Myc from the nucleoplasm to the nucleolus (10), infection of cells with adenovirus expressing E1A did not significantly alter the nuclear distribution of the Myc protein. Thus the consequences of stabilization of Myc by E1A are different from those of proteasome inhibition. Taken together, these data support a model in which the ability of E1A to stabilize Myc does not involve general inhibition of proteasome function.
E1A Stabilizes Myc via p400.
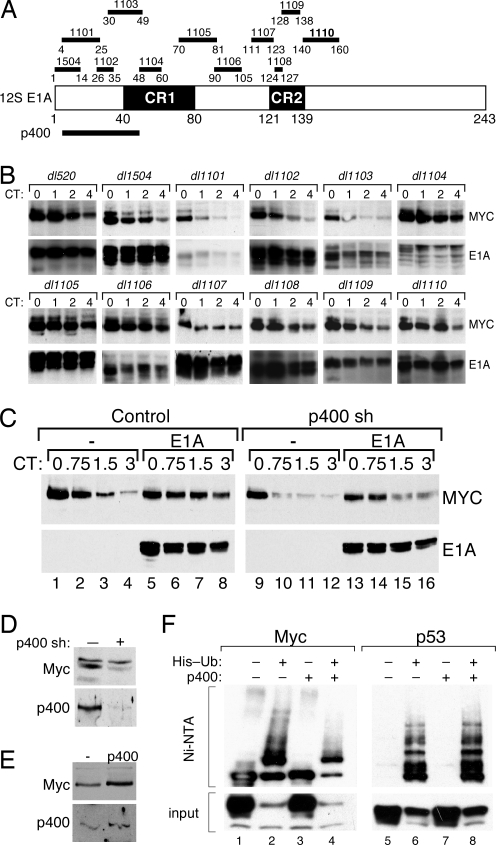
We next probed for the regions of E1A that are required to stabilize Myc. We analyzed a panel of adenovirus E1A deletion mutants, depicted in Fig. 2A, for their ability to stabilize Myc at 6 h after adenovirus infection (Fig. 2B). This experiment revealed that, of the 11 mutant viruses tested, all but three were able to stabilize Myc efficiently. The three viruses that failed to stabilize Myc (dl1101, dl1102, and dl1103) encode E1A proteins with deletions that span residues 4–49. This region of E1A is important for its interaction with p400, CBP/p300, TBP, and Rb, but the smallest deletion that disrupts Myc stabilization, Δ26–35 (dl1102), interacts with all of these proteins, with the exception of p400 (4). Although the Δ26–35 mutation also disrupts interaction of E1A with the coactivator TRRAP (11), interaction with TRRAP additionally requires CR1; disruption of CR1 (as in the dl1104 and dl1105 viruses) did not block the ability of E1A to stabilize Myc (Fig. 2B). Importantly, we also observed that WT E1A, but not the Δ26–35 mutant, stabilized Myc in Rat1 and IMR90 cells (Fig. S2), demonstrating that the ability of E1A to interact with p400 correlates with Myc stabilization in a variety of cell types. Together, these data indicate that adenoviral-mediated stabilization of Myc requires interaction of E1A with p400.
Fig. 2.
p400 is required for E1A to stabilize Myc. (A) 12S E1A is depicted, showing conserved regions 1 (CR1) and 2 (CR2). The positions of adenovirus E1A deletion mutants dl1504 and dl1101–dl1110 are shown above, and residues required for interaction with p400 are shown below. (B) U2OS cells were infected with adenovirus deletion mutants, or dl520, and incubated for 6 h, and CHX chase was performed. WB was used to monitor levels of E1A and endogenous Myc. (C) U2OS cells were transduced with retroviral expression constructs encoding (i) a short hairpin directed against p400 (sh p400), or vector control, and (ii) WT 12S E1A, or vector control (−). CHX chase followed by WB analysis was used to determine levels and stability of Myc and E1A. (D) WB analysis was used to determine levels of Myc and p400 in control (−) and p400 hairpin (+)-expressing cells. (E) U2OS cells were transiently transfected with HA-tagged Myc, in the absence (−) or presence of an expression construct encoding p400. Levels of Myc and p400 were determined by immunoblotting. (F) U2OS cells were transfected with expression vectors encoding either Myc or p53, together with FLAG-tagged p400 (or vector control), and a plasmid expressing His-tagged Ub (20). Ub conjugates were recovered by Ni-NTA chromatography, and ubiquitylated Myc and p53 proteins were detected by WB.
To determine whether p400 plays a role in E1A-mediated stabilization of Myc, we knocked down expression of p400 in U2OS cells by using short-hairpin-mediated gene silencing and examined the effects of E1A on Myc turnover. This analysis (Fig. 2C) showed that knockdown of p400 attenuates the ability of E1A to stabilize Myc. Moreover, we found that knockdown of p400, in the absence of E1A expression, increased the rate of Myc proteolysis and decreased steady-state Myc protein levels (Fig. 2D), indicating that p400 normally acts to promote Myc stability. Consistent with this idea, overexpression of p400 promoted accumulation of Myc protein (Fig. 2E) and specifically reduced the formation of high-molecular-weight Myc–Ub conjugates (Fig. 2F); p53–Ub conjugates, which we assayed as a control, were unaffected by p400 expression. Based on these results, we conclude that p400 acts to stabilize Myc by reducing the extent of Myc ubiquitylation and that E1A targets this process.
E1A Promotes Formation of a p400–Myc Complex.
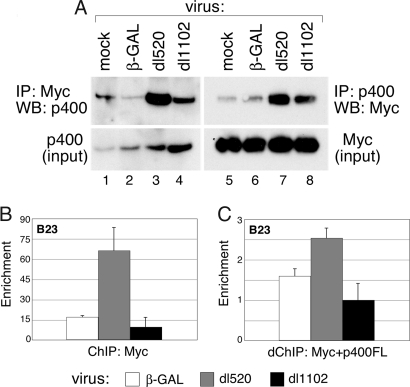
Given the ability of p400 to stabilize Myc, a simple model to explain our observations is that E1A promotes the association of Myc and p400. To test this model, U2OS cells expressing HA-tagged Myc and FLAG-tagged p400 were infected with various adenoviruses, and Myc–p400 complexes were detected by coimmunoprecipitation analysis (Fig. 3A). By recovering either Myc or p400 immune complexes, we found that WT E1A, but not the Δ26–35 mutant, promoted coassociation of both proteins. Importantly, WT E1A could also stimulate the association of Myc and p400 on promoter DNA, as assayed by ChIP. Because commercially available anti-p400 antibodies did not function for ChIP (data not shown), we expressed FLAG-tagged p400 in cells infected with various adenoviral vectors for these analysis. We first performed ChIP by using an antibody against Myc and found that expression of WT E1A (encoded by dl520), but not the Δ26–35 mutant (dl1101), promoted the association of Myc with the nucleophosmin (B23) promoter (Fig. 3B). We then recovered Myc–DNA complexes from the ChIP reaction and performed a second round of immunoprecipitation with anti-FLAG antibody to recover chromatin that was bound by both Myc and p400. Consistent with the ability of E1A to promote association of Myc and p400 in solution, expression of E1A also promoted an enrichment of Myc–p400 cocomplexes at the B23 promoter (Fig. 3C); as expected, this enrichment was not observed with the Δ26–35 E1A mutant. The ability of E1A to promote both the association of Myc with a target gene, and, beyond this, to promote corecruitment of Myc and a coactivator, supports a model in which E1A functionally targets Myc via its interactions with p400.
Fig. 3.
E1A expression promotes the formation of Myc–p400 complexes. (A) U2OS cells were transiently transfected with HA-tagged Myc and FLAG-tagged p400 for 48 h. Transfected cells were infected for 6 h with β-GAL, dl520, or dl1102 adenovirus. Coimmunoprecitation was performed as described (4) by using anti-Myc (N262) or anti-FLAG (M2) antibodies, and Myc and p400 were detected by WB. (B) U2OS cells were transfected with FLAG-p400 or control DNA for 48 h and then infected with β-GAL, dl520, or dl1102 adenovirus for 6 h. ChIP analysis was performed by using the anti-Myc antibody N262, and coprecipitating DNAs corresponding to the B23 E-box were detected by quantitative PCR. Enrichment is calculated relative to a non-Myc binding sequence. (C) Protein–DNA complexes recovered in B were eluted and subject to a second round of ChIP with anti-FLAG antibodies to recover p400-containing chromatin. Levels of coprecipitating DNA from the B23 E-box were determined by quantitative PCR. To control for nonspecific background, signal in the re-ChIP was normalized to that from an identical, parallel, experiment from cells not expressing FLAG-tagged p400.
Functional Interaction Between Myc and E1A.
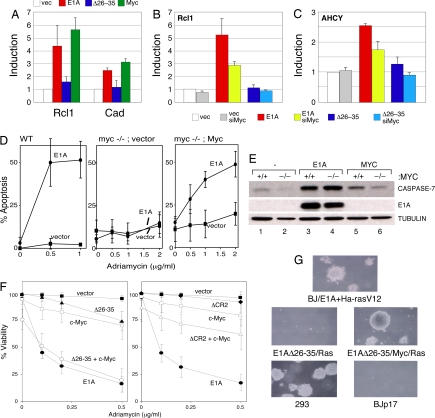
E1A and Myc share a number of functional similarities, including the ability to promote ectopic S-phase entry, sensitize cells to apoptosis, and collaborate with oncogenes like Ras to transform rat fibroblasts (12). Given our demonstration that E1A can induce both Myc levels and the Myc–p400 interaction, we speculated that the overlapping activities of these proteins may result, in part, from the ability of E1A to function through Myc. To probe this idea, we first asked whether E1A can stimulate Myc's transcriptional activity. We examined expression of two Myc target genes (Rcl1 and Cad) in diploid IMR90 cells transduced to express either WT E1A or the Δ26–35 E1A mutant (Fig. 4A). Compared with vector control, we found that both genes were induced by expression of WT E1A, to a level comparable to that observed upon overexpression of Myc. The Δ26–35 E1A mutant, in contrast, failed to activate either gene. Importantly, activation of Rcl1 (Fig. 4B) and AHCY (Fig. 4C) by E1A depended on Myc, as siRNA-mediated knockdown of Myc in these cells attenuated the ability of E1A to activate these genes. Similar behavior was observed at the B23 and PCNA genes (Fig. S3). Thus E1A can activate a Myc target gene in a Myc-dependent manner, supporting the notion that it functionally stimulates the Myc protein.
Fig. 4.
Myc is a downstream effector of E1A. (A) IMR90 cells were transduced by retroviral vectors expressing 12S E1A, the E1A Δ26–35 mutant, or Myc. RNA was harvested, and levels of Rcl1 and Cad cDNAs were determined by quantitative PCR. (B) IMR90 cells were transduced with the indicated retroviruses, and then transfected with either nontargeting siControl, or siMyc RNA, duplexes for 48 h. RNA was then harvested and levels of Rcl cDNA were analyzed by quantitative PCR. Fold induction is normalized to an actin control for each sample. (C) As in B but assaying for levels of AHCY cDNA. (D) WT (TGR-1 cells) or congenic myc−/− Rat-1 cells (HO.15.19 cells) were transduced with retroviruses expressing, E1A, E1A Δ26–35, or empty vector, and the resulting cell populations were treated with adriamycin and assessed for viability after 24 h by trypan blue exclusion. The myc−/−; Myc panel represents myc−/− cells where full-length Myc was reintroduced by retroviral gene transfer. (E) WB analysis of samples derived from C showing expression of caspase-7, E1A, or tubulin. (F) IMR90 cells were transduced with retroviral expression constructs encoding E1A (•), E1A Δ26–35 (▴), E1A ΔCR2 (♦), Myc (□), E1A Δ26–35 + Myc (○), E1A ΔCR2 + Myc (▵), or an empty vector (■). Cell populations were treated with the indicated doses of adriamycin for 24 h, and viability was determined by trypan blue exclusion. (G) BJ fibroblasts were retrovirally transduced with the indicated constructs. The resulting cells were assayed for colony formation in soft agar.
The E1A–p400 interaction has recently been found to be important for the ability of E1A to promote apoptosis (4). If, as our model predicts, the E1A–p400 interaction functions through Myc, we would expect that Myc will also be required for induction of apoptosis by E1A. We therefore asked whether E1A can induce apoptosis in Rat1 fibroblasts in which both copies of the c-myc gene were disrupted by homologous recombination (13). In Rat1 cells, E1A was a potent inducer of apoptosis triggered by adriamycin (Fig. 4D; see Fig. S4A for expression data on the E1A and Myc proteins). In congenic Myc-null cells, however, E1A was unable to induce apoptosis. This deficit was caused by a loss of Myc and not a secondary mutation, because reintroduction of Myc into Myc-null cells restored the ability of E1A to induce apoptosis. Importantly, this deficit was not caused by a general defect in E1A activity in Myc-null cells. Our previous studies have shown that binding of E1A to Rb is important for inducing the expression of several caspases, and that this induction potentiates cell death in E1A-expressing cells (14). When we examined caspase-7 levels in our system (Fig. 4E), we found that E1A was capable of inducing caspase-7, and that this induction was not diminished in Myc-null cells. This result demonstrates clearly that the ability of E1A to function via the Rb pathway does not depend on Myc expression and reveals that only a subset of E1A activities require Myc.
A key prediction of our model is that overexpression of Myc should rescue defects in E1A that are associated with loss of the p400 interaction. To challenge this prediction, we asked whether overexpression of Myc can restore the ability of the E1A Δ26–35 mutant to sensitize cells to apoptosis (4). As reported (15), expression of E1A in IMR90 fibroblasts sensitizes them to apoptosis in the presence of adriamycin (Fig. 4F Left) and results in the induction of both ARF and p53 (Fig. S4B). Under these conditions, E1A is a more potent inducer of apoptosis than Myc, and its proapoptotic activity (Fig. 4F), and ability to induce ARF and p53, is disrupted by the Δ26–35 mutation. As predicted from the model, overexpression of Myc in the presence of the Δ26–35 E1A mutant rescued both E1A's ability to trigger apoptosis (Fig. 4F) and induce ARF and p53 (Fig. S4B). The rescue of mutant E1A function was specific, because overexpression of Myc did not rescue the apoptotic defects of an E1A mutant (ΔCR2) that still interacts with p400 but fails to interact with Rb (Fig. 4F). Thus, increased expression of Myc can specifically rescue apoptotic defects associated with loss of the E1A–p400 interaction.
Finally, we asked whether the same phenomenon applied to the ability of E1A to drive human cell transformation. Our previous studies have shown that expression of E1A and activated Ha-RasV12 in early passage normal human foreskin fibroblasts (designated BJ) allows the formation of colonies in soft agar (Fig. 4G and ref. 16). Blocking the ability of E1A to interact with p400 also attenuates its ability to collaborate with Ras to drive human cell transformation in this assay (Fig. 4G). Importantly, this activity can be restored by overexpression of Myc. Thus, overexpression of Myc can rescue both the transformation and apoptotic defects that result when the ability of E1A to interact with p400 is blocked. These data are consistent with the idea that the critical function of the E1A–p400 interaction in transformation and apoptosis is to induce Myc.
Conclusions
Together, our data support a model in which the binding of E1A to p400 promotes the formation of a Myc–p400 complex at Myc-target gene promoters. The increase in interaction of Myc and p400 is associated with stabilization of the Myc protein and an induction of Myc target genes. These functions of E1A are required for its ability to induce the ARF/p53 pathway, promote apoptosis, and drive cellular transformation, revealing that the downstream arm of the E1A–p400 interaction is mediated via Myc. Although it has long been known that E2F functions as the downstream target of the E1A–Rb interaction, the molecular processes downstream of the E1A–p400 connection have remained obscure. Our data indicate that Myc is the ultimate target of this connection. This finding not only provides an explanation for the overlapping biological functions of Myc and E1A, but also reveals an interesting viral strategy for promoting oncogenesis. By targeting and activating both the E2F and Myc transcriptional networks, E1A can provoke a synergistic response in parallel pathways to efficiently couple cell cycle progression, transformation, and apoptosis. Moreover, given that E1A targets cellular pathways relevant to transformation, our data also support an important role for the Myc–p400 connection in human cancer.
Methods
Antibodies.
The antibodies used in this study were: anti (α)-Ad5 E1A antibodies M73 and M58 (17) and sc-430 (Santa Cruz); α-p400 monoclonal antibody RW144 (2); α-caspase-7 (14); α-Myc 9E10 (Oncogene) and N262 (Santa Cruz); α-Actin AC-15 (Sigma); α-GFP PC408 (Novagen); α-FLAG M2 (Sigma); α-p53 CM1 (Novocastra); α-ARF sc-8340 (Santa Cruz); α-HA 12CA5 (Roche); α-tubulin B512 (Sigma); and α-nucleolin (Santa Cruz).
Cells and Adenoviruses.
U2OS, IMR90, HEK293, HO.15.19, and TGR-1 (13) cells and BJ human fibroblasts were grown under standard conditions. WT adenovirus 5 (Ad5), dl520, was obtained from ATCC. Ad5 mutants were obtained from Arnold Berk (University of California, Los Angeles) (18) and Philip Branton (McGill University, Quebec, Canada) (19). Adenoviruses were propagated in 293 cells and purified by CsCl equilibrium centrifugation. For adenoviral infections, U2OS cells were incubated with virus at a multiplicity of infection of 15 plaque-forming units per cell, for 1 h at 37°C with intermittent rocking.
Protein Assays.
Myc protein turnover was measured by adding CHX (100 μg/ml) to cells, collecting protein samples at the indicated time points, and assaying relevant protein levels by WB. Myc and p53 conjugates were detected by using the His-tagged Ub method after transfection of U2OS cells with pMT107 (20), pCGN-Myc (8), pCGN-p53 (unpublished work), and pCMV-p400 (3) as indicated.
cDNA measurement.
Where appropriate, total cellular RNA was harvested with TRIzol (Invitrogen) and reverse-transcribed with the TaqMan kit (Applied Biosystems), and cDNA levels from the indicated genes were quantified by using the SYBR Green PCR Master Mix (Applied Biosystems) in conjunction with a MJ Research CFD-3240 Chromo 4 Detector. Transcript levels for target genes were normalized to those of actin. Primer sequences are available on request.
RNAi.
Duplex pools of siGenome RNA against Myc and nontargetting control sequences (Dharmacon) were transiently transfected into IMR90 cells via Oligofectamine (Invitrogen). Knockdown of Myc RNA was at least 50% (data not shown).
Cell Viability.
To assay the ability of Myc to rescue the apoptotic defect of the E1A Δ26–35 mutant, IMR90 cells were stably transduced with pLPC, pLPC E1A (21), pLPC E1A Δ26–35 (4), pLPC ΔCR2 (4), or pBabe Hygro HAM Myc (22) by retroviral infection. The resulting cell lines were plated into 12-well dishes at a density of 1 × 105 cells per well. Twenty-four hours later, cells were treated with adriamycin for 24 h. Adherent and nonadherent cells were then pooled and analyzed for viability by trypan blue exclusion. At least 200 cells were counted for each data point. To assay the ability of E1A to induce apoptosis in Myc−/− cells, HO.15.19, and parental Rat1 cells, TGR-1 (13) were transduced with retroviral expression constructs for E1A, E1A Δ26–35, or Myc, in the indicated combinations. Relative apoptosis was determined by comparing cell death 24 h after treatment with increasing doses of adriamycin. Data presented are the average of three independent experiments.
Anchorage-Independent Growth.
BJ normal human primary foreskin fibroblasts were stably transduced with pBABE-Puro Ha-RasV12, pWZL-Neo E1A, or Hygro-MarXII-Myc in the indicated combinations by retroviral infection and analyzed for anchorage-independent growth in semisolid media as described (16).
ChIP and Re-ChIP Analysis.
ChIP analyses were performed in U2OS cells that had been transfected with either control (pUC119) or pCMV-FLAG-p400 construct by using Fugene 6 (Roche). After 48 h, cells were infected with control, dl520, and dl1102 adenovirus for 8 h. Primary immunoprecipitation was performed by using anti-Myc (N262) antibody; for re-ChIP, a secondary immunoprecipitation using anti-FLAG (M2) antibody was performed. Coprecipitating DNAs after each round were assayed by quantitative PCR using either the B23_C (specific) and B23_M (nonspecific control) amplicons (23). Re-ChIP DNA signals for FLAG-p400 were further normalized to those from cells transfected with the vector control.
Supplementary Material
Acknowledgments.
We thank A. Berk, P. Branton, N. Dantuma (Karolinska Institute, Stockholm, Sweden), and D. Livingston (Dana–Farber Cancer Institute, Boston) for reagents. W.P.T. was a Leukemia and Lymphoma Society of America Scholar. This work was supported by Cold Spring Harbor Laboratory Cancer Center Support Grant CA45508, The Irving Hansen Memorial Foundation, and Public Health Service Grant CA-13106 from the National Cancer Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0802095105/DCSupplemental.
References
- 1.Ben-Israel H, Kleinberger T. Adenovirus and cell cycle control. Front Biosci. 2002;7:1369–1395. doi: 10.2741/ben. [DOI] [PubMed] [Google Scholar]
- 2.Arany Z, Sellers WR, Livingston DM, Eckner R. E1A-associated p300 and CREB-associated CBP belong to a conserved family of coactivators. Cell. 1994;77:799–800. doi: 10.1016/0092-8674(94)90127-9. [DOI] [PubMed] [Google Scholar]
- 3.Fuchs M, et al. The p400 complex is an essential E1A transformation target. Cell. 2001;106:297–307. doi: 10.1016/s0092-8674(01)00450-0. [DOI] [PubMed] [Google Scholar]
- 4.Samuelson AV, et al. p400 is required for E1A to promote apoptosis. J Biol Chem. 2005;280:21915–21923. doi: 10.1074/jbc.M414564200. [DOI] [PubMed] [Google Scholar]
- 5.Lohr K, Hartmann O, Schafer H, Dobbelstein M. Mutual interference of adenovirus infection and myc expression. J Virol. 2003;77:7936–7944. doi: 10.1128/JVI.77.14.7936-7944.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Turnell AS, et al. Regulation of the 26S proteasome by adenovirus E1A. EMBO J. 2000;19:4759–4773. doi: 10.1093/emboj/19.17.4759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baluchamy S, Sankar N, Navaraj A, Moran E, Thimmapaya B. Relationship between E1A binding to cellular proteins, c-myc activation, and S-phase induction. Oncogene. 2007;26:781–787. doi: 10.1038/sj.onc.1209825. [DOI] [PubMed] [Google Scholar]
- 8.Salghetti SE, Kim SY, Tansey WP. Destruction of Myc by ubiquitin-mediated proteolysis: Cancer-associated and transforming mutations stabilize Myc. EMBO J. 1999;18:717–726. doi: 10.1093/emboj/18.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dantuma NP, Lindsten K, Glas R, Jellne M, Masucci MG. Short-lived green fluorescent proteins for quantifying ubiquitin/proteasome-dependent proteolysis in living cells. Nat Biotechnol. 2000;18:538–543. doi: 10.1038/75406. [DOI] [PubMed] [Google Scholar]
- 10.Arabi A, Rustum C, Hallberg E, Wright AP. Accumulation of c-Myc and proteasomes at the nucleoli of cells containing elevated c-Myc protein levels. J Cell Sci. 2003;116:1707–1717. doi: 10.1242/jcs.00370. [DOI] [PubMed] [Google Scholar]
- 11.Deleu L, Shellard S, Alevizopoulos K, Amati B, Land H. Recruitment of TRRAP required for oncogenic transformation by E1A. Oncogene. 2001;20:8270–8275. doi: 10.1038/sj.onc.1205159. [DOI] [PubMed] [Google Scholar]
- 12.Adhikary S, Eilers M. Transcriptional regulation and transformation by Myc proteins. Nat Rev Mol Cell Biol. 2005;6:635–645. doi: 10.1038/nrm1703. [DOI] [PubMed] [Google Scholar]
- 13.Mateyak MK, Obaya AJ, Adachi S, Sedivy JM. Phenotypes of c-Myc-deficient rat fibroblasts isolated by targeted homologous recombination. Cell Growth Differ. 1997;8:1039–1048. [PubMed] [Google Scholar]
- 14.Nahle Z, et al. Direct coupling of the cell cycle and cell death machinery by E2F. Nat Cell Biol. 2002;4:859–864. doi: 10.1038/ncb868. [DOI] [PubMed] [Google Scholar]
- 15.Samuelson AV, Lowe SW. Selective induction of p53 and chemosensitivity in RB-deficient cells by E1A mutants unable to bind the RB-related proteins. Proc Natl Acad Sci USA. 1997;94:12094–12099. doi: 10.1073/pnas.94.22.12094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seger YR, et al. Transformation of normal human cells in the absence of telomerase activation. Cancer Cell. 2002;2:401–413. doi: 10.1016/s1535-6108(02)00183-6. [DOI] [PubMed] [Google Scholar]
- 17.Harlow E, Franza BR, Jr, Schley C. Monoclonal antibodies specific for adenovirus early region 1A proteins: Extensive heterogeneity in early region 1A products. J Virol. 1985;55:533–546. doi: 10.1128/jvi.55.3.533-546.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Osborne TF, Gaynor RB, Berk AJ. The TATA homology and the mRNA 5′ untranslated sequence are not required for expression of essential adenovirus E1A functions. Cell. 1982;29:139–148. doi: 10.1016/0092-8674(82)90098-8. [DOI] [PubMed] [Google Scholar]
- 19.Egan C, et al. Mapping of cellular protein-binding sites on the products of early-region 1A of human adenovirus type 5. Mol Cell Biol. 1988;8:3955–3959. doi: 10.1128/mcb.8.9.3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Treier M, Staszewski LM, Bohmann D. Ubiquitin-dependent c-Jun degradation in vivo is mediated by the delta domain. Cell. 1994;78:787–798. doi: 10.1016/s0092-8674(94)90502-9. [DOI] [PubMed] [Google Scholar]
- 21.McCurrach ME, Connor TM, Knudson CM, Korsmeyer SJ, Lowe SW. Bax deficiency promotes drug resistance and oncogenic transformation by attenuating p53-dependent apoptosis. Proc Natl Acad Sci USA. 1997;94:2345–2349. doi: 10.1073/pnas.94.6.2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herbst A, Tansey WP. HAM: A new epitope tag for in vivo protein labeling. Mol Biol Rep. 2000;27:203–208. doi: 10.1023/a:1011008018565. [DOI] [PubMed] [Google Scholar]
- 23.Haggerty TJ, Zeller KI, Osthus RC, Wonsey DR, Dang CV. A strategy for identifying transcription factor binding sites reveals two classes of genomic c-Myc target sites. Proc Natl Acad Sci USA. 2003;100:5313–5318. doi: 10.1073/pnas.0931346100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.