Abstract
Histopathology archives of well-annotated formalin-fixed, paraffin-embedded (FFPE) tissue specimens are valuable resources for retrospective studies of human diseases. Since recovery of quality intact mRNA compatible with molecular techniques is often difficult due to degradation, we evaluated microRNA (miRNA), a novel class of small RNA molecules with growing therapeutic and diagnostic potential, as an alternative analyte for gene expression studies of FFPE samples. Analyzing total RNA yield, miRNA recovery, and robustness of real-time polymerase chain reaction for miRNA, mRNA, and rRNA species, we compared the performance of commercially available RNA isolation kits and identified a preferred methodology. We further implemented modifications to increase tissue throughput and incorporate a quantitative Armored RNA process control to monitor RNA recovery efficiency. The optimized process was tested for reproducibility as well as interoperator and interday variability, and was validated with a set of 30 clinical samples. In addition, we demonstrated that, independent of FFPE block age and RNA quality, miRNAs generate quantitative reverse transcription-polymerase chain reaction signals that are more robust and better correlate with expression levels from frozen reference samples compared with longer mRNAs. Our broad study, including a total of 272 independent RNA isolations from 17 tissue types and 65 FFPE blocks up to 12 years old, indicates that miRNAs are not only suitable but are also likely superior analytes for the molecular characterization of compromised archived clinical specimens.
Since the initial development of standardized methodology for fixation and processing of human tissue samples at the turn of the last century, anatomical pathology practices around the world have been preserving biopsies and other pathological specimens as formalin-fixed, paraffin-embedded (FFPE) blocks. This process has resulted in a widely available and rich archive of well characterized tissue specimens annotated with patient data, which offer a valuable source of information for comparative genomics as well as for biomarker discovery studies.1 With the advent of high-content, high-throughput molecular genetic techniques such as quantitative reverse transcription-polymerase chain reaction (qRT-PCR) and microarrays, there has been growing interest in mining these archival collections as a source of biological data. While the value of retrospective pathological analysis of archived tissues has been extensively validated, until very recently these samples have not been considered reliable sources of mRNA for gene expression studies due to the difficulty in obtaining intact mRNA from these samples. Therefore optimization of the recovery of quality RNA from FFPE tissues is of particular interest to many research laboratories.2,3
The procedure for preserving and archiving tissues involves fixation of the tissue in formalin (10% formaldehyde) or ethanol-based preservatives followed by processing by dehydration with graded ethanol solutions to enhance the infusion of the material with paraffin. While formalin fixation helps preservation of cellular proteins and conserves the tissue architecture, it significantly reduces the recovery and quality of RNA. Extensive cross-linking of RNA with proteins during fixation renders RNA more resistant to extraction. Enzyme degradation, which occurs before and during the fixation process, as well as chemical degradation, results in decreased yield and integrity of RNA. Finally, formalin is responsible for forming mono-methylol adducts with bases of nucleic acids, in particular with adenine.4,5 This covalent modification reduces the efficiency of reverse transcription in qRT-PCR and negatively affects the performance of RNA samples in other downstream applications. Recently, many research groups have attempted to overcome these limitations. The effects of new fixatives on histological properties of tissues as well as on the quality and yield of RNA have been tested.6,7,8,9 Methods that do not require RNA extraction, such as in situ hybridization, have also been evaluated as a novel approach to gene expression analysis in FFPE tissues,10,11,12 but they are not amenable to high-content, high-throughput analyses. To this day, no alternative fixative has replaced formalin fixation as a preservation method in routine clinical use.
In the past seven years there has been an explosion of interest in miRNAs (microRNAs) with many hundreds of publications describing the fundamental role these regulatory biomolecules play in processes as diverse as early development, cell proliferation, differentiation, apoptosis, and oncogenesis.13,14,15,16,17 Due to their small size (19 to 23 nucleotides), miRNAs may be less prone to degradation and modification, and therefore their analysis in FFPE specimens likely provides a more accurate replication of what would be observed in fresh tissue than that of mRNA species. Although several RNA isolation protocols or commercial kits have been optimized for miRNA recovery, no detailed analysis of their relative performance has been performed. In this study we compared different commercially available isolation procedures by evaluating the yield and quality of isolated total RNA as well as the detection efficiency of miRNA, mRNA, and rRNA species. We identified and validated a robust protocol using multiple tissue types and FFPE blocks with ages ranging from 1 to 12 years. We also introduced new procedural improvements to facilitate the increase in tissue throughput as well as to enable better control over the extraction process. Our results suggest that with the appropriate RNA isolation protocol and controls, miRNAs are excellent candidates for biomarker discovery studies using archived clinical specimens.
Materials and Methods
Tissue Samples
Samples derived from human patients were acquired from commercial suppliers by Asuragen's Tissue Procurement Group in compliance with the regulations as outlined in Title 45 of the Code of Federal Regulations, Part 46 (45 CFR 46) and other regulatory guidance. All samples used in this study were collected as part of standard clinical care and were considered to be remnant materials unnecessary for patient treatment. Patient identifiers were thoroughly removed from all samples before distribution to Asuragen. Samples used in the comparison between flash-frozen and FFPE tissues included cervical, breast, and gall bladder from two or three donors for each tissue. Half of each tissue was flash-frozen and half was formalin-fixed and embedded into paraffin according to a standard fixation procedure (<60 minutes elapsed time between surgery and immersion in 10% neutral buffered formalin for 24 hours). Older FFPE tissue blocks, aged from 3 to 12 years, also interrogated in this study included breast cancer, lung cancer, normal kidney, normal cervix, testicular cancer, stomach cancer, uterine cancer, lung normal adjacent tissue (NAT), prostate NAT, spleen, and liver NAT.
RNA Isolation
For the initial evaluation of isolation kits, total RNA from two FFPE tissues blocks, breast tumor and lung tumor, was isolated using five commercially available kits, RNeasy FFPE Kit (Qiagen, Valencia, CA), Absolutely RNA FFPE Kit (Stratagene, La Jolla, CA), High Pure FFPE RNA Micro Kit (Roche, Indianapolis, IN), PureLink FFPE RNA Isolation Kit (Invitrogen, Carlsbad, CA) and RecoverAll Total Nucleic Acid Isolation Kit (Ambion, Austin, TX), according to the respective manufacturers' instructions. In the case of the RNeasy FFPE Kit, a supplementary protocol for “co-purification” of total RNA and miRNA from FFPE tissue sections using the RNeasy FFPE Kit was used. Total RNA from a subset of FFPE blocks, including breast cancer, lung cancer, lung NAT, prostate NAT, spleen, and liver NAT was isolated using RecoverAll and RNeasy kits. Total RNA from remaining tissue blocks (normal kidney, normal cervix, testicular cancer, stomach cancer, and uterine cancer) was isolated using RecoverAll protocol only. To accommodate higher FFPE tissue input, the later protocol was modified based on results of the following experiment. Two prostate cancer FFPE blocks (1 year old), differing in size of cross-sectional tissue area (<75 mm2 or >150 mm2), were cut into 20-μm slices to accommodate four, eight, 12, and 16-slice isolations in duplicate. All slices were placed in a 15-ml conical tube and deparaffinized together (six 10-ml washes with 100% xylene, followed by three 10-ml washes with 100% ethanol, and dried in a Speedvac at 30°C for ∼10 minutes). The dry tissue was resuspended in 4.8 ml of proteinase K buffer and homogenized using a PRO 250 tissue homogenizer (PROScientific, Oxford, CT). The tissue homogenate was aliquoted in volumes corresponding to four-, eight-, 12-, and 16-slice proteinase K tissue digestion (400, 800, 1200, and 1600 μl). On addition of customized proteinase K volume to each tube, the proteinase K digestion and remaining steps were performed as per the manufacturer's protocol.
Total RNA from frozen tissues was extracted using mirVana miRNA Isolation Kit (Ambion) following the manufacturer's protocol. Concentration and purity of the total RNA samples were measured using the NanoDrop ND-1000 Spectrophotometer (NanoDrop Technologies Inc., Wilmington, DE). RNA integrity was assessed with an Agilent 2100 Bioanalyzer and the RNA 6000 LabChip kit (Agilent Technologies, Palo Alto, CA).
Real-Time Quantitative RT-PCR
The recovery of miRNA and mRNA species was estimated via quantification of relative expression levels of miR-24, miR-103, miR-191, 18S, and hGUSB. qRT-PCR experiments were performed using 10 ng total RNA input and TaqMan primer/probe sets (TaqMan Gene Expression Assays, Applied Biosystems, Foster City, CA). For miRNA amplification, a two-step qRT-PCR was performed as follows: RT in duplicate (16°C, 30 minutes; 42°C, 30 minutes; 85°C, 5 minutes, then hold at 4°C); PCR in duplicate from each RT (95°C, 1 minute; 95°C, 15 seconds followed by 60°C, 30 seconds and cycled 40 times). For mRNA amplification, a one-step qRT-PCR was performed in duplicate under the following conditions: 42°C, 15 minutes; 95°C, 2 minutes; then 95°C, 15 seconds followed by 60°C, 45 seconds and cycled 40 times. PCR amplifications were performed on a 7900HT Fast real-time PCR system and data analyzed with the 7900HT Fast system SDS software v2.3 (Applied Biosystems).
Implementation of an Internal Isolation Process Control
A total of 1010 copies of Armored RNA Quant (ARQ) (Asuragen, Austin, TX) containing a non-human RNA sequence was spiked in during RNA isolation from one or four 20-μm slices of FFPE tissue, either at the proteinase K digestion step or at the RNA elution step. One-step qRT-PCR was performed using ARQ-specific primers and probe to quantify the percentage of ARQ recovery. Each qRT-PCR reaction was run in duplicate, including a positive control ARQ heat-lysed for 3 minutes at 95°C in nuclease-free water and a no template control. For extraction from 16 slices (20-μm thickness) of FFPE lung tumor and NAT blocks, ARQ was spiked at the proteinase K digestion step and recovery was assessed as described above.
Results
Method Comparison for RNA Extraction from FFPE Tissues
As a first step toward identifying the most suitable procedure for RNA isolation from FFPE tissues, we isolated total RNA in duplicate from two tissue types, lung cancer and breast cancer, using five different commercially available isolation procedures: Absolutely RNA FFPE Kit, High Pure FFPE RNA Micro Kit, PureLink FFPE RNA Isolation Kit, RecoverAll Total Nucleic Acid Isolation Kit, and RNeasy FFPE Kit. This comparison was based on criteria such as yield of total RNA and efficient recovery of selected miRNA and mRNA species (data not shown). Three kits showed two- to threefold lower total RNA yield and five- to 20-fold lower miRNA qRT-PCR signals at equal RNA mass input, and were not further evaluated. The two RNA isolation kits that demonstrated superior performance, RecoverAll and RNeasy, were subjected to a more comprehensive characterization study.
For a thorough comparison of the two isolation kits, total RNA was isolated from 13 FFPE blocks that ranged in age from 1 to 12 years and comprised seven distinct tissues: normal breast, normal cervix, normal gall bladder, “normal” lung tissue from lung cancer patients (normal adjacent tissue or NAT), prostate NAT, liver NAT, and normal spleen. The comparison of average RNA recovery from triplicate isolations revealed higher yields in samples isolated with RecoverAll by an average of 1.7-fold (Table 1). Eleven of the 13 FFPE blocks analyzed (55% to 98% of the samples with 95% confidence) generated lower RNA yields with the RNeasy protocol. In addition, within each FFPE block, the final RNA amount recovered was more consistent when RecoverAll was used, with %CV for triplicate isolations ranging from 5% to 28%, versus 10% to 43% for isolations performed with RNeasy. Agilent Bioanalyzer electropherograms showed roughly similar RNA integrity for both kits (data not shown). However, RNA purity was greater with RecoverAll, as demonstrated by an average A260/280 ratio of 1.98 for RecoverAll isolations versus 1.68 for RNeasy isolations (P = 0.0003) (Table 1).
Table 1.
Summary of Triplicate RNA Isolations from 13 FFPE Tissue Blocks Using the RecoverAll or RNeasy Procedure
| RecoverAll |
RNeasy |
|||||||
|---|---|---|---|---|---|---|---|---|
| Avg RNA yield (μg) |
A260/A280 |
Avg RNA yield (μg) |
A260/A280 |
|||||
| Tissue | 1 × 20 μm | CV (%) | Avg ratio | CV (%) | 2 × 10 μm | CV (%) | Avg ratio | CV (%) |
| Breast-1 | 0.15 | 11.0 | 2.01 | 16.4 | 0.09 | 43.1 | 1.54 | 5.7 |
| Breast-2 | 0.23 | 5.1 | 2.01 | 6.2 | 0.16 | 39.0 | 1.67 | 10.1 |
| Breast-3 | 0.18 | 25.5 | 2.08 | 8.7 | 0.11 | 42.6 | 1.53 | 3.9 |
| Cervix-1 | 0.69 | 18.1 | 2.06 | 6.3 | 1.61 | 9.8 | 2.06 | 6.5 |
| Cervix-2 | 0.63 | 14.0 | 2.03 | 2.2 | 0.39 | 14.4 | 1.50 | 6.3 |
| Cervix-3 | 0.65 | 28.2 | 1.96 | 7.7 | 0.47 | 10.7 | 1.46 | 3.5 |
| Gall bladder-1 | 0.34 | 14.1 | 1.94 | 7.7 | 0.11 | 26.6 | 1.46 | 5.0 |
| Gall bladder-2 | 0.31 | 7.4 | 1.88 | 22.0 | 0.44 | 11.3 | 1.41 | 3.9 |
| Lung NAT-1 | 2.63 | 27.9 | 1.96 | 0.5 | 1.95 | 16.0 | 1.88 | 1.3 |
| Lung NAT-2 | 1.16 | 8.9 | 1.98 | 3.1 | 0.84 | 33.1 | 1.72 | 6.1 |
| Prostate NAT | 2.44 | 14.0 | 1.89 | 0.6 | 1.47 | 17.0 | 1.72 | 1.2 |
| Spleen | 4.42 | 12.0 | 1.99 | 0.9 | 3.42 | 26.9 | 1.93 | 0.9 |
| Liver NAT-1 | 5.76 | 17.2 | 1.93 | 2.7 | 3.37 | 36.5 | 1.93 | 1.4 |
Quantitative Gene Expression Analyses
To evaluate the quality and efficiency of miRNA and mRNA extraction from FFPE tissues we next interrogated expression levels of miR-24, miR-191, miR-103, 18S rRNA, and hGUSB mRNA by qRT-PCR in a representative set of FFPE RNA samples (two lung NAT, one prostate NAT, one liver NAT, and one spleen) using TaqMan primers and probes and equal input of total RNA for each sample (10 ng). miR-24 and miR-103 were detected at higher levels in the FFPE samples isolated with RecoverAll, 2.2-fold (P = 0.06) and 2.1-fold (P = 0.07) on average, respectively, while detection of miR-191 was comparable for both kits (Figure 1). The performance difference between kits was more striking for larger RNA species, although P values were not significant due to high amplification variability between tissue types. The 18S and the hGUSB were detected 11.7-fold (P = 0.2) and 2.4-fold (P = 0.3) more abundantly in RNA samples isolated with RecoverAll. No amplification signals were obtained when reverse transcriptase or RNA was omitted (data not shown).
Figure 1.
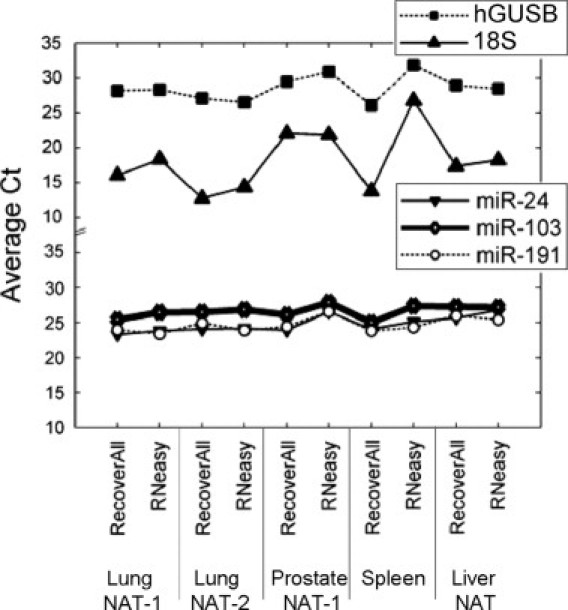
qRT-PCR analysis of RNA samples isolated with the RecoverAll or RNeasy kit. Total RNA was extracted in triplicate from five different FFPE tissue blocks using the indicated method. Each RNA sample was analyzed in duplicate using qRT-PCR assays specific for miR-24, -103, -191, 18S rRNA, or hGUSB mRNA.
Comparison with Matched Frozen Tissues
To further evaluate each method we also compared RNA samples isolated from multiple breast, cervix, and gall bladder blocks, with RNA samples isolated from the corresponding paired flash-frozen tissues using a method optimized for small RNA recovery (see Materials and Methods). As expected, the quality of RNA isolated from frozen tissues was high, with distinct 18S and 28S rRNA bands, while RNA isolated from fixed tissues displayed a low molecular weight distribution, with an average size lower than 100 nucleotides, regardless of the isolation kit used (see Supplemental Figure S1 at http://jmd.amjpathol.org).
Analysis of miR-24, miR-103, and miR-191 expression levels by qRT-PCR showed a significant difference between frozen and FFPE samples with the average cycle threshold (Ct) notably higher in RNA samples isolated from FFPE specimens (Figure 2A; P = 2 × 10−4 for RNeasy and P = 5.5 × 10−7 for RecoverAll). miRNA detection in cervical FFPE samples more closely matched the reference levels from frozen samples when the RNeasy kit was used (P = 0.003; Figure 2C). In contrast, the use of RecoverAll resulted in better miRNA detection in total RNA samples from breast and gall bladder tissues (P = 8.0 × 10−6). The ΔCt between frozen and FFPE samples was on average 0.65 Ct lower (1.6-fold higher miRNA representation) in RecoverAll samples relative to RNeasy samples. For longer RNA species, detection of 18S and hGUSB targets by qRT-PCR in RNA samples from FFPE blocks relative to paired reference frozen tissues were improved on average by 2.45 Ct or 5.5-fold (P = 0.05) with RecoverAll relative to RNeasy (Figure 2, B and C).
Figure 2.
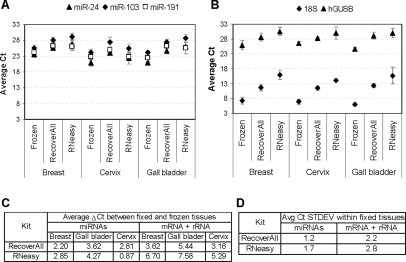
Comparison between FFPE and matching frozen samples. A: Average expression levels for miR-24, -103, and -191 in RNA samples isolated from FFPE and matched frozen breast (three specimens), cervix (three specimens), and gall bladder (two specimens) tissues. Each qRT-PCR reaction was performed in duplicate. B: Same as A for 18S rRNA and hGUSB mRNA. C: Average differential expression (ΔCt) between frozen and matching FFPE samples for miRNAs or mRNA and rRNA species. D: Average standard deviation of qRT-PCR signal (Ct) within the 13 FFPE tissues blocks described in Table 1 for miRNA or mRNA and rRNA species.
Finally, we examined the overall variability in RNA amplification efficiency within the 13 different FFPE blocks tested so far (Figure 2D). We observed that RecoverAll isolation procedure provided a more consistent miRNA, mRNA, and rRNA qRT-PCR signal across all FFPE blocks and tissue types, with a standard deviation on average 1.5-fold lower than with RNeasy. Based on more robust and overall superior performance of the kit we selected RecoverAll as our preferred RNA isolation method and used this kit for the remainder of the study.
Qualification of the RNA Extraction Method
With the recommended RecoverAll protocol (isolation from a maximum of four 20-μm FFPE tissue slices), some tissue types with large cross-sectional area, such as spleen and liver NAT, yielded higher total RNA amounts than other specimens with smaller cross-sectional area, such as normal breast and skin (Table 2). To enable isolation of RNA from FFPE tissue blocks with various cross-sectional areas in quantities sufficient for multiple expression analyses and archiving of the purified RNA, we first determined the maximum number of FFPE slices that could be processed in a single tube. Isolations were performed in duplicate using 4, 8, 12, or 16 slices of FFPE tissue (20-μm thickness) from two prostate cancer FFPE blocks that differed in their tissue cross-sectional area (see Materials and Methods). For the tissue block with a smaller cross-sectional area (<75 mm2) we observed an almost linear increase in RNA recovery for up to 16 slices by proportionately scaling up the reagent volumes (Figure 3). For the prostate cancer block with a larger area (>150 mm2), not all of the tissue was digested in tubes containing more than four 20-μm slices resulting in yields that were lower than expected (data not shown). However, by increasing the amount of proteinase K added to the digestion, we improved the yields by 40% and rescued the linearity of recovery up to 12 slices. For blocks with both large and small cross-sectional areas we were able to recover over 30 μg of purified total RNA in a single isolation (Figure 3).
Table 2.
Summary of Total RNA Yields Recovered from 17 Different Tissue Types Representing a Total of 65 FFPE Blocks and 272 Independent RNA Isolations Using the RecoverAll Procedure
| FFPE tissue | No. of isolations | No. of blocks | Block age (yr) | Avg RNA yield (μg) per 20-μm slice | CV (%) |
|---|---|---|---|---|---|
| B-cell lymphoma | 6 | 2 | 1 | 2.72 | 32.4 |
| Breast | 9 | 3 | 1 | 0.19 | 21.4 |
| Cervix | 20 | 4 | 1–3 | 0.49 | 46.9 |
| Colon | 5 | 2 | 1 | 2.55 | 32.3 |
| Colon cancer | 4 | 1 | 3 | 1.75 | 22.2 |
| Gall bladder | 6 | 2 | 1 | 0.33 | 6.1 |
| Kidney | 12 | 1 | 12 | 0.97 | 6.1 |
| Liver NAT | 8 | 1 | 12 | 3.8 | 72.5 |
| Lung NAT | 70 | 17 | 1–11 | 1.31 | 53.4 |
| Lung cancer | 53 | 15 | 1–11 | 2.71 | 70.1 |
| Myometrium | 28 | 9 | 1–11 | 1.82 | 44.5 |
| Prostate NAT | 3 | 1 | 8 | 1.67 | 13.9 |
| Skin | 6 | 3 | 1–3 | 0.21 | 61.9 |
| Spleen | 6 | 1 | 8 | 3.11 | 59.3 |
| Stomach cancer | 12 | 1 | 9 | 1.28 | 11.5 |
| Testicular cancer | 12 | 1 | 4 | 1.67 | 4.0 |
| Uterine cancer | 12 | 1 | 10 | 1.11 | 5.2 |
Figure 3.
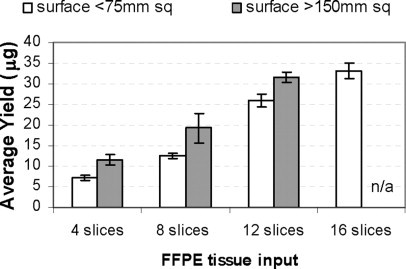
Scale-up of the RecoverAll procedure. RNA isolation was performed with four, eight, 12, or 16 FFPE tissue slices (20-μm) from two prostate cancer FFPE blocks that differed in tissue cross-sectional area. For the block with a tissue area >150 mm2, twice the recommended amount of proteinase K was used.
To determine the reproducibility of this method, we performed repeated isolations using four 20-μm slices from two different FFPE blocks: liver NAT (five independent isolations) and spleen (three independent isolations). The purified RNA samples were compared in terms of yield, purity, miRNA, rRNA, and mRNA amplification by qRT-PCR. The variation in RNA recovery from individual blocks was less than 21% for liver NAT and less than 5% for spleen (Table 3). The A260/280 ratio was highly reproducible, ranging from 1.79 to 1.84 for liver NAT and from 1.91 to 1.94 for spleen. The quantification of miR-24, miR-103, miR-191, 18S, and hGUSB panel by qRT-PCR was also highly reproducible with %CV across all samples less than 2.8% (Table 3).
Table 3.
Summary of Method Variability Data
| RNA isolation |
Target amplification |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Avg yield |
A260/280 |
miR-24 |
miR-103 |
miR-191 |
18S |
hGUSB |
||||||||
| Tissue | μg | CV (%) | Avg ratio | CV (%) | Avg Ct | CV (%) | Avg Ct | CV (%) | Avg Ct | CV (%) | Avg Ct | CV (%) | Avg Ct | CV (%) |
| Liver NAT | 7.42 | 20.7 | 1.80 | 1.1 | 25.52 | 2.4 | 27.58 | 1.4 | 26.11 | 2.7 | 15.93 | 2.4 | 29.05 | 1.6 |
| Spleen | 7.23 | 4.5 | 1.92 | 1.1 | 23.86 | 2.8 | 25.61 | 2.1 | 24.05 | 2.4 | 12.91 | 2.3 | 25.92 | 1.2 |
Five and three independent RNA isolations using four 20-μm slices from a liver NAT and spleen blocks, respectively, were performed by the same operator on the same day using the RecoverAll procedure. qRT-PCR reactions were performed at least in duplicate for each RNA target.
We next investigated interoperator variability with respect to RNA yield as well as extraction efficiency of the miRNA, rRNA, and mRNA fractions. Total RNA from five different FFPE tissue types ranging from 3 to 12 years old was extracted in duplicate by four different operators on the same day (Table 4). The %CV between operators ranged from 7.7% to 25% for yield, and 8.6% to 21.2% for A260/280 ratio. RNA quantification by qRT-PCR was very reproducible with %CV ranging from 0.2% to 6.4% and no significant difference among miRNA, mRNA, and rRNA species. A clear trend was observed between tissue types with cervix generating the highest %CV, while uterine cancer replicates were very consistent in terms of RNA yield, quality, and amplification efficiency. Two operators also performed duplicate RNA isolation from the five different tissues on a second day. On average, day-to-day variability was 20% for RNA yield and %CV for qRT-PCR amplification was lower than 2% for all interrogated targets (see Supplemental Table S1 at http://jmd.amjpathol.org).
Table 4.
Summary of Interoperator Variability Data
| RNA isolation |
Target amplification |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Av yield |
A260/A280 |
miR-24 |
miR-103 |
miR-191 |
18S |
hGUSB |
||||||||
| FFPE tissue | μg | CV (%) | Avg ratio | CV (%) | Avg Ct | CV (%) | Avg Ct | CV (%) | Avg Ct | CV (%) | Avg Ct | CV (%) | Avg Ct | CV (%) |
| Cervix | 0.6 | 22.7 | 2.20 | 21.2 | 22.68 | 2.6 | 28.18 | 2.3 | 27.00 | 2.2 | 17.74 | 2.8 | 30.80 | 3.7 |
| Kidney | 1.9 | 23.5 | 2.16 | 6.6 | 23.25 | 6.4 | 26.80 | 1.7 | 26.23 | 0.9 | 17.09 | 1.7 | 27.11 | 1.4 |
| Uterine cancer | 2.3 | 7.7 | 2.13 | 8.6 | 23.67 | 0.4 | 26.21 | 0.7 | 24.81 | 0.7 | 17.99 | 0.5 | 26.40 | 0.2 |
| Testicular cancer | 3.3 | 8.3 | 2.24 | 9.2 | 22.65 | 2.2 | 27.65 | 0.6 | 26.19 | 1.2 | 14.53 | 0.4 | 26.66 | 1.3 |
| Stomach cancer | 2.3 | 25.0 | 2.11 | 12.9 | 22.79 | 1.5 | 26.29 | 1.8 | 26.22 | 0.6 | 17.45 | 2.0 | 26.16 | 0.6 |
Duplicate RNA isolations using two 20-μm slices from five different FFPE tissue blocks were performed by four different operators using the RecoverAll procedure. qRT-PCR reactions were performed at least in duplicate for each RNA target.
Implementation of a RNA Isolation Process Control
The inherent variability in FFPE block size, tissue content, and RNA quality make it difficult to determine whether low RNA yield and/or low RNA detection signal are due to poor RNA isolation processing (process failure) or poor tissue/RNA initial quality (sample failure). To better control the RNA isolation process we evaluated the use of a precisely quantified Armored RNA Quant as a quality control check point. Armored RNA is formulated to resist degradation by encapsulating RNA in a bacteriophage protein coat.18 The RNA component is released by most RNA isolation methods or by heat denaturation and thus serves as an excellent isolation control. Preliminary qRT-PCR experiments showed that ARQ molecules spiked at the beginning of the FFPE RNA isolation procedure, ie, during the proteinase K digestion step, are consistently and efficiently recovered and detected regardless of the number of FFPE tissue slices processed (see Supplemental Figure S2 at http://jmd.amjpathol.org). Thus, ARQ can be used as an external process control to rule out putative process failure and identify those FFPE samples with intrinsic poor RNA quality.
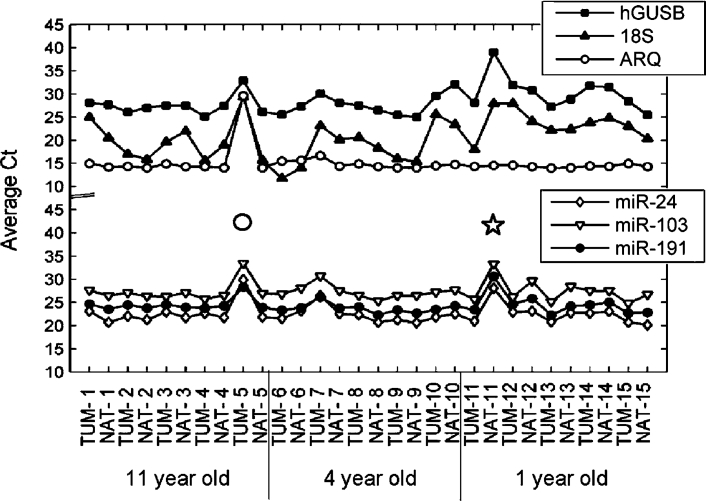
To validate our modified protocol, we tested the performance on 15 pairs of matched lung non-small cell carcinoma and NAT (normal adjacent tissue) FFPE blocks that were 1, 4, or 11 years old. For each block, 16 20-μm tissue slices were processed in a single tube in the presence of ARQ spiked in at the proteinase K digestion step. On average, 28.5 μg of total RNA was recovered per isolation with yield ranging from 0.36 to 5.9 μg per 20-μm slice and A260/280 ratio ranging from 1.52 to 2.02 (see Supplemental Table S2 at http://jmd.amjpathol.org). Expression analysis on the 30 samples using equal RNA mass input confirmed that qRT-PCR detection is more robust and less variable for miRNA than for longer mRNA or rRNA species (Figure 4 and Supplemental Table S2 at http://jmd.amjpathol.org). However, an unexpected low detection signal was observed for all endogenous RNA species in two samples, TUM-5 and NAT-11. Further analysis of ARQ recovery at constant RNA volume input (Figure 4) identified NAT-11 as a sample failure (expected ARQ signal, low endogenous signal) and TUM-5 as a process failure (low ARQ and endogenous signals). Together, these data demonstrate that with the appropriate endogenous expression controls, external process controls, and RNA isolation methods, robust extraction and accurate expression analyses can be performed on miRNAs from archival clinical specimens, regardless of the age of the FFPE block.
Figure 4.
Armored RNA Quant as a process control. RNA isolation was performed using 16 20-μm slices from 15 pairs of matched FFPE lung tumor and NAT specimens with 1010 copies of ARQ spiked at the proteinase K digestion step. qRT-PCR were performed in duplicate using 2 μl of purified RNA for ARQ and 10 ng of purified RNA for miR-24, -191, -103, hGUSB mRNA, and 18S rRNA. Samples with abnormally high Ct for endogenous RNAs and ARQ or for endogenous RNAs only are indicated by a circle and a star, respectively.
Discussion
Clinical practices worldwide have created valuable archives of well characterized FFPE pathological specimens. However, these preservation methods were initially optimized for histological and immunochemical evaluation of specimens instead of RNA integrity. Total RNA extracted from these specimens has generally been considered unsuitable for studying gene expression patterns using modern molecular techniques, such as quantitative RT-PCR or microarrays. Therefore, there is a considerable interest in the scientific community in the development of new fixation methods, compatible molecular detection techniques, or improved nucleic acids recovery protocols. To the best of our knowledge, this study is the first report on the performance of different isolation methods with respect to the recovery of mRNA, rRNA, and miRNA from FFPE tissue blocks. We implemented and validated a process allowing robust and reproducible isolation of total RNA of sufficient quality and quantity to support gene expression profiling studies. Based on a thorough analysis of 17 FFPE tissue types, we conclude that miRNA recovery and expression analysis are more robust and accurate than that of mRNAs. These improvements will facilitate retrospective molecular analysis and characterization of archived clinical samples using miRNA analytes.
By comparing the performance of commercially available RNA isolation kits, we have identified a preferred methodology, the RecoverAll kit, for extraction of total RNA from archived fixed tissues. This protocol allowed robust and reproducible recovery of approximately twofold higher yield of total RNA, which included the small RNA fraction. It is well documented that RNA extracted from FFPE samples is often of poor quality.19,20 As expected, the integrity of RNA samples isolated here was compromised, with a lack of distinct 18S and 28S rRNA peaks and an average RNA size lower than 100 nucleotides in most cases (see Supplemental Figure S1 at http://jmd.amjpathol.org). The variability from repeated isolations from the same FFPE block was within 20% for yield and within 3% for qRT-PCR signal (Table 3). Noteworthy is that the yield variation most likely reflected changes of tissue shape within a given block during sectioning rather than lack of process robustness. When blocks of similar size were used, inter-run as well as interoperator variability was well below 10% (Tables 3 and 4). Similarly, the yield variability observed between the 15 tumor/NAT pairs (0.36 to 5.89 μg per 20-μm slice; see Supplemental Table S2 at http://jmd.amjpathol.org) could be linked to the disease state. In fact, the NAT tissues yielded on average 2.2-fold less RNA per slice than the matched tumor tissues. Also, the best quality RNA and the highest yields were obtained from the oldest FFPE blocks that were fixed in 1996. This observation could reflect differences in postsurgery tissue handling techniques and/or fixation protocol, factors as critical as the choice of an appropriate RNA isolation method for recovery of large amounts of good quality RNA from FFPE specimens.21
Significant performance differences among five commercially available kits were identified in our study. Although the Absolutely RNA, High Pure RNA, and Pure Link kits do not claim to be optimized for miRNA isolation, the two- to threefold lower yields of total RNA obtained with these kits cannot be attributed only to the lack of miRNA fraction recovery. Possible explanations for the kit-to-kit variation in total RNA yield and recovery of different RNA fractions are the conditions recommended for the proteinase K digestion and/or the binding/washing steps. For example, we noted that the top two best performing kits, RecoverAll and RNeasy, use the highest ethanol concentration for binding the sample onto small filter cartridges provided with the kits (55 and 70%, respectively). In addition, both protocols recommend identical amounts of proteinase K enzyme and perform this reaction at similar temperature (200 μg per reaction, 50°C or 55°C for RecoverAll and RNeasy, respectively). However, the tissue incubation time with the proteinase K enzyme is considerably shorter in the RNeasy protocol, 15 minutes versus 3 hours for RecoverAll. It is therefore possible that the digestion process in the RNeasy protocol is less efficient, leaving RNA trapped in undigested tissue matter and thus affecting the final total yield. Additionally, as smaller RNA species might be released from protein cross-links more easily than longer RNA species, the shorter proteinase K digestion procedure could generate total RNA whose composition is biased toward the small RNA fraction, ie, miRNAs and short, partially degraded mRNA and rRNA fragments. Indeed, we observed the largest performance differences between the two kits for the longer RNA species, mRNA and rRNA, with detection signals two- to 12-fold lower for RNeasy at equal input of total RNA (Figures 1 and 2).
Previously it has been shown that RNA samples derived from FFPE tissues can serve as effective templates for qRT-PCR and that these samples can be used to correctly differentiate diseased tissue from non-diseased tissue.19,22,23,24 Here we show that nanogram quantities of RNA from FFPE tissue blocks up to 12 years of age are sufficient for gene expression analysis using TaqMan assays for miRNA, mRNA, and rRNA targets. To ensure that the recovery of different RNA species was adequately assessed it was crucial to select appropriate RNA targets for interrogation by qRT-PCR. 18S rRNA and hGUSB mRNAs are constitutively expressed across various tissues and have been commonly used to calibrate measurements of gene expression.25 miR-24, miR-103, and miR-191 were chosen because they are relatively stably expressed across many tissue types and represent high, medium, and lower abundance miRNA species.26 The RT-PCR amplification results reported here demonstrate that miRNA targets are detected at levels that better match expression levels from reference frozen tissue than mRNA targets. In fact, the average representation of the three miRNA panels in FFPE samples is approximately 2.3-fold higher and closer to frozen samples than that of 18S and hGUSB, but can be as much as 3.5-fold higher depending on tissue type (Figure 2C and Supplemental Table S3 at http://jmd.amjpathol.org). This observation supports the notion that miRNAs may be more stable or more easily recovered than mRNAs and therefore are likely better analytes for molecular characterization of clinical samples.27,28
In addition to variations resulting from isolation procedure, tissue type, block preparation, and storage, other factors can affect the robustness of RNA isolation from archived fixed samples. The carryover of contaminants or process failures can strongly reduce or introduce bias in the detection of specific RNA targets. We therefore further modified the RecoverAll protocol to address these issues. We adapted the procedure to accommodate up to 16 slices of FFPE tissue in single tube isolation without the need to repeat the time-consuming process of deparaffinization and proteinase K digestion. This is particularly beneficial for RNA extraction from tissues that are known to generate low RNA yield or from FFPE blocks with very small tissue cross-sectional areas that may require several independent isolations per specimen to recover enough material for expression analysis. We also implemented an Armored RNA Quant spike-in control into our isolation protocol to monitor for potential failure of the isolation process (Figure 4). Armored RNA is currently widely used as a ribonuclease resistant control for viral testing in various clinical specimens.29,30,31 Furthermore, Armored RNA can be precisely quantified in comparison to National Institute of Standards and Technology standards. Our results suggest that the technology is also useful for RNA isolation from FFPE samples and may have broader applications such as oncology testing, biomarker discovery, or clinical signature validation. Incorporation of the Armored RNA process control could indeed enable selection early in the process of the RNA samples with acceptable quality and yield for downstream molecular studies using often costly gene expression analysis platforms.
Overall, we analyzed 17 different tissue types during the course of this study, including normal, tumor and normal adjacent tissues. For other purposes, we also isolated and characterized total RNA from B-cell lymphoma, colon tumor, myometrium, and skin FFPE tissue blocks using the same RecoverAll procedure (data not shown). In total we have performed over 272 RNA isolations from 65 different tissue blocks and successfully recovered enough total RNA compatible with downstream qRT-PCR analysis. On average, we obtained 1.6 μg of total RNA per 20-μm FFPE slice, with only three tissues (breast, gall bladder, and skin) yielding less than 0.5 μg per slice (Table 2). Forty-eight total RNA samples from liver NAT, kidney, spleen, stomach cancer, testicular cancer, or uterine cancer tissue blocks also generated robust expression signals when labeled and hybridized onto miRNA microarrays (data not shown). In an independent study, we demonstrate that miRNA expression profiling in FFPE samples is in fact robust and reasonably accurate relative to frozen samples.32 miRNAs are therefore legitimate targets for biomarker discovery in compromised clinical samples such as FFPE tissues. In the long term, the ability to retrospectively interrogate well-annotated archived samples collected as part of standard clinical care should facilitate the development of novel miRNA-based diagnostic and/or therapeutic applications.
Acknowledgements
We thank Gary Latham for technical assistance and Bernard Andruss for critical review of the manuscript.
Footnotes
Supported in part by National Cancer Institute grant 1R43CA118785.
Supplemental material for this article can be found on http://jmd.amjpathol.org.
Supplementary data
References
- 1.Lewis F, Maughan NJ, Smith V, Hillan K, Quirke P. Unlocking the archive–gene expression in paraffin-embedded tissue. J Pathol. 2001;195:66–71. doi: 10.1002/1096-9896(200109)195:1<66::AID-PATH921>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 2.Coombs NJ, Gough AC, Primrose JN. Optimisation of DNA and RNA extraction from archival formalin-fixed tissue. Nucleic Acids Res. 1999;27:e12. doi: 10.1093/nar/27.16.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chung JY, Braunschweig T, Hewitt SM. Optimization of recovery of RNA from formalin-fixed, paraffin-embedded tissue. Diagn Mol Pathol. 2006;15:229–236. doi: 10.1097/01.pdm.0000213468.91139.2d. [DOI] [PubMed] [Google Scholar]
- 4.Masuda N, Ohnishi T, Kawamoto S, Monden M, Okubo K. Analysis of chemical modification of RNA from formalin-fixed samples and optimization of molecular biology applications for such samples. Nucleic Acids Res. 1999;27:4436–4443. doi: 10.1093/nar/27.22.4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Korga A, Wilkolaska K, Korobowicz E. Difficulties in using archival paraffin-embedded tissues for RNA expression analysis. Postepy Hig Med Dosw (online) 2007;61:151–155. [PubMed] [Google Scholar]
- 6.Benchekroun M, DeGraw J, Gao J, Sun L, von Boguslawsky K, Leminen A, Andersson LC, Heiskala M. Impact of fixative on recovery of mRNA from paraffin-embedded tissue. Diagn Mol Pathol. 2004;13:116–125. doi: 10.1097/00019606-200406000-00008. [DOI] [PubMed] [Google Scholar]
- 7.Stanta G, Mucelli SP, Petrera F, Bonin S, Bussolati G. A novel fixative improves opportunities of nucleic acids and proteomic analysis in human archive's tissues. Diagn Mol Pathol. 2006;15:115–123. doi: 10.1097/00019606-200606000-00009. [DOI] [PubMed] [Google Scholar]
- 8.Gillespie JW, Best CJ, Bichsel VE, Cole KA, Greenhut SF, Hewitt SM, Ahram M, Gathright YB, Merino MJ, Strausberg RL, Epstein JI, Hamilton SR, Gannot G, Baibakova GV, Calvert VS, Flaig MJ, Chuaqui RF, Herring JC, Pfeifer J, Petricoin EF, Linehan WM, Duray PH, Bova GS, Emmert-Buck MR. Evaluation of non-formalin tissue fixation for molecular profiling studies. Am J Pathol. 2002;160:449–457. doi: 10.1016/S0002-9440(10)64864-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paska C, Bogi K, Szilak L, Tokes A, Szabo E, Sziller I, Rigo J, Jr, Sobel G, Szabo I, Kaposi-Novak P, Kiss A, Schaff Z. Effect of formalin, acetone, and RNAlater fixatives on tissue preservation and different size amplicons by real-time PCR from paraffin-embedded tissue. Diagn Mol Pathol. 2004;13:234–240. doi: 10.1097/01.pdm.0000134778.37729.9f. [DOI] [PubMed] [Google Scholar]
- 10.Henke RT, Maitra A, Paik S, Wellstein A. Gene expression analysis in sections and tissue microarrays of archival tissues by mRNA in situ hybridization. Histol Histopathol. 2005;20:225–237. doi: 10.14670/HH-20.225. [DOI] [PubMed] [Google Scholar]
- 11.Henke RT, Eun Kim S, Maitra A, Paik S, Wellstein A. Expression analysis of mRNA in formalin-fixed, paraffin-embedded archival tissues by mRNA in situ hybridization. Methods. 2006;38:253–262. doi: 10.1016/j.ymeth.2005.11.013. [DOI] [PubMed] [Google Scholar]
- 12.To MD, Done SJ, Redston M, Andrulis IL. Analysis of mRNA from microdissected frozen tissue sections without RNA isolation. Am J Pathol. 1998;153:47–51. doi: 10.1016/S0002-9440(10)65544-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 14.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 15.Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, Iorio MV, Visone R, Sever NI, Fabbri M, Iuliano R, Palumbo T, Pichiorri F, Roldo C, Garzon R, Sevignani C, Rassenti L, Alder H, Volinia S, Liu CG, Kipps TJ, Negrini M, Croce CM. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 16.Esquela-Kerscher A, Slack FJ. Oncomirs—microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 17.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC, Croce CM. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pasloske BL, Walkerpeach CR, Obermoeller RD, Winkler M, DuBois DB. Armored RNA technology for production of ribonuclease-resistant viral RNA controls and standards. J Clin Microbiol. 1998;36:3590–3594. doi: 10.1128/jcm.36.12.3590-3594.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Godfrey TE, Kim SH, Chavira M, Ruff DW, Warren RS, Gray JW, Jensen RH. Quantitative mRNA expression analysis from formalin-fixed, paraffin-embedded tissues using 5′ nuclease quantitative reverse transcription-polymerase chain reaction. J Mol Diagn. 2000;2:84–91. doi: 10.1016/S1525-1578(10)60621-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cronin M, Pho M, Dutta D, Stephans JC, Shak S, Kiefer MC, Esteban JM, Baker JB. Measurement of gene expression in archival paraffin-embedded tissues: development and performance of a 92-gene reverse transcriptase-polymerase chain reaction assay. Am J Pathol. 2004;164:35–42. doi: 10.1016/S0002-9440(10)63093-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lisowski AR, English ML, Opsahl AC, Bunch RT, Blomme EA. Effect of the storage period of paraffin sections on the detection of mRNAs by in situ hybridization. J Histochem Cytochem. 2001;49:927–928. doi: 10.1177/002215540104900716. [DOI] [PubMed] [Google Scholar]
- 22.Abrahamsen HN, Steiniche T, Nexo E, Hamilton-Dutoit SJ, Sorensen BS. Towards quantitative mRNA analysis in paraffin-embedded tissues using real-time reverse transcriptase-polymerase chain reaction: a methodological study on lymph nodes from melanoma patients. J Mol Diagn. 2003;5:34–41. doi: 10.1016/S1525-1578(10)60449-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Finke J, Fritzen R, Ternes P, Lange W, Dolken G. An improved strategy and a useful housekeeping gene for RNA analysis from formalin-fixed, paraffin-embedded tissues by PCR. Biotechniques. 1993;14:448–453. [PubMed] [Google Scholar]
- 24.Specht K, Richter T, Muller U, Walch A, Werner M, Hofler H. Quantitative gene expression analysis in microdissected archival formalin-fixed and paraffin-embedded tumor tissue. Am J Pathol. 2001;158:419–429. doi: 10.1016/S0002-9440(10)63985-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aerts JL, Gonzales MI, Topalian SL. Selection of appropriate control genes to assess expression of tumor antigens using real-time RT-PCR. Biotechniques. 2004;36:84–91. doi: 10.2144/04361ST04. [DOI] [PubMed] [Google Scholar]
- 26.Peltier HJ, Latham GJ. Normalization of microRNA expression levels in quantitative RT-PCR assays: identification of suitable reference RNA targets in normal and cancerous human solid tissues. RNA. 2008 doi: 10.1261/rna.939908. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hampton T. MicroRNAs move into cancer research. JAMA. 2005;294:411–412. doi: 10.1001/jama.294.4.411. [DOI] [PubMed] [Google Scholar]
- 28.Garber K. No miR hype: microRNA's cancer role expands. J Natl Cancer Inst. 2006;98:885–887. doi: 10.1093/jnci/djj286. [DOI] [PubMed] [Google Scholar]
- 29.WalkerPeach CR, Winkler M, DuBois DB, Pasloske BL. Ribonuclease-resistant RNA controls (Armored RNA) for reverse transcription-PCR, branched DNA, and genotyping assays for hepatitis C virus. Clin Chem. 1999;45:2079–2085. [PubMed] [Google Scholar]
- 30.Beld M, Minnaar R, Weel J, Sol C, Damen M, van der Avoort H, Wertheim-van Dillen P, van Breda A, Boom R. Highly sensitive assay for detection of enterovirus in clinical specimens by reverse transcription-PCR with an armored RNA internal control. J Clin Microbiol. 2004;42:3059–3064. doi: 10.1128/JCM.42.7.3059-3064.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Drosten C, Seifried E, Roth WK. TaqMan 5′-nuclease human immunodeficiency virus type 1 PCR assay with phage-packaged competitive internal control for high-throughput blood donor screening. J Clin Microbiol. 2001;39:4302–4308. doi: 10.1128/JCM.39.12.4302-4308.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Szafranska AE, Davison T, Shingara J, Doleshal M, Riggenbach JA, Morrison CD, Jewell S, Labourier E. Accurate molecular characterization of formalin-fixed paraffin-embedded tissues by microRNA expression profiling. J Mol Diagn. 2008 doi: 10.2353/jmoldx.2008.080018. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.