Abstract
The Long-Evans Cinnamon (LEC) rat strain (Atp7b m/m), which accumulates copper in the liver due to mutations in the Atp7b gene, is a useful model for investigating the relationship between oxidative stress and hepatocarcinogenesis. To determine the effect of this mutation on oxidative stress marker genes, we performed oligonucleotide array analysis (Affymetrix), and compared the results in Atp7b m/m rats with those of a sibling line with the Atp7b w/w genotype. We focused our studies on the expression of the aldo-keto reductase 1 family B7 (AKR1B7)-like protein gene, since this gene codes for reductase enzymes involved in the detoxification of oxidizing compounds (e.g., aldehydes) and was differentially expressed in Atp7b m/m and Atp7b w/w rat liver. Akr1B7 mRNA expression was significantly increased in comparison with the expression of 4 other known oxidative stress responsive genes, haem-oxygenase-1 (HO-1), thioredoxin (Trx), aldehyde reductase (AKR1A1), and glucose-6-phosphate dehydrogenase (G6PDH). By searching binding motifs, five nuclear factor kappa B (NF-κB) binding sites were located in the 5′-upstream region of the Akr1b7 gene. Transient co-transfection with both I-κBα and the Akr1b7 6 kb promoter (p6.0-AKR-Luc) inhibited luciferase activity of p6.0-AKR-Luc in HepG2 cells. Cuprous ion however did not affect the transcription activity induced by p6.0-AKR-Luc. Gel-shift assay showed that the DNA binding activity of NF-κB increased in the livers of LEC rats, suggesting that the oxidative stress is mediated through NF-κB. The results indicate conclusively that in LEC rat liver, Akr1b7 might be up-regulated by oxidative stress mediated through NF-κB, but not that mediated directly by copper.
Keywords: AKR1B7, oxidative stress, oligonucleotide array, real-time PCR, Long Evans Cinnamon rat
Introduction
Hepatocellular carcinoma (HCC) is the fifth most common cancer worldwide and one of the most deadly cancers with approximately 600,000 yearly deaths (1). Hepatocarcinogenesis is a multi-step process and cellular oxidative stress is one of the factors responsible for the propagation of liver diseases, such as hepatitis, cirrhosis, and hepatoma (2-4). Although much is known about the cellular pathogenesis and etiological agents leading to HCC, the molecular events are not well understood. Recent advances in genomics and proteomics have had the potential to identify characteristics of HCC that may be related to prognosis or to etiology, and these may be useful in HCC screening or diagnosis. Here the Long-Evans Cinnamon (LEC) rat model provides a unique opportunity to unveil the early molecular event associated with oxidative stress-related hepatocarcinogenesis.
The Long-Evans Cinnamon (LEC) rat strain, which accumulates copper in the liver due to mutations in the Atp7b gene, has been used as a model to investigate the relationship between oxidative stress and hepatocarcinogenesis (5-11). The hepatic levels of oxidative stress indicators in LEC rats were found to be elevated during the onset of fulminant hepatitis between 16 and 24 weeks of age, while the levels of antioxidants such as ascorbate and ubiquinol were concomitantly decreased. Oxidative stress levels in LEC rats were highest at the onset of hepatitis, which occurred when the rats were around 20 weeks of age (Fig. 1.) The oxidative stress levels decreased from peak levels but still remained elevated during the subsequent phases of chronic hepatitis and hepatocellular carcinoma in LEC rats. High oxidative stress levels in the liver of LEC rats at the hepatitis stage may be involved in the subsequent development of liver cancer. Therefore, the LEC rat model can be considered to be useful for exploring oxidative stress-responsive genes associated with the development of neoplastic lesions.
Figure 1.
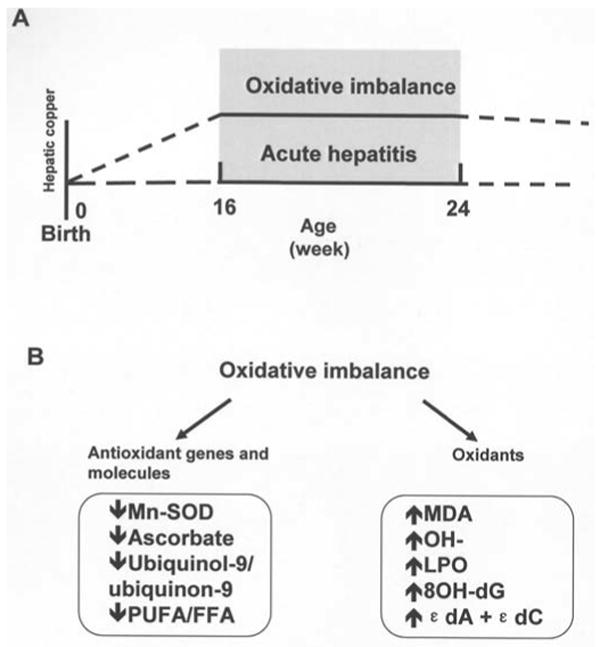
A, a high level of oxidative status persisted in livers of 16- to 24-week-old LEC rats due to copper accumulation. B, oxidative imbalance may cause a decrease in antioxidant biomarkers and an increase in oxidant levels. Studies on these biomarkers have been published before by our group and by others. Hydroxyl free radical (OH•) (5), LPO (6), etheno adducts (sum of dA + dC) (8), 8-hydroxy-deoxyguanosin (8-OHdG) (7), and malondialdehyde (MDA) (8) are products of oxidation. Ascorbate, superoxide dismutase, polyunsaturated fatty acid: free acid ratio (PUFA/FFA), and ubiquinol-9 (9) are antioxidants. In LEC rats at the age of 16 to 24 weeks old, the levels of oxidative products including MDA, OH•, 8-OHdG and εdA + εdB, were significantly higher than those of age-matched healthy LEA rats (controls); however, the reductant ability, including that of Mn-SOD, ascorbate, ubiquinol-9/ubiquinon-9 and PUFA/FFA was decreased significantly when compared with LEA rats, which indicated that the redox status was unbalanced and that oxidative stress levels peaked in LEC rats during this period.
Thus, the present study examined the gene expression patterns associated with oxidative stress by utilizing an oligonucleotide array containing more than 8732 expressed sequence tag (EST) clusters. The difference in gene expression between Atp7b m/m and Atp7b w/w rats was analyzed. Based on the gene array results, candidate genes were further evaluated by RT-PCR and real-time PCR. We found that the gene whose expression was most altered in response to oxidative stress was Akr1B7, and we describe the expression and transcriptional regulation of this gene in the present study.
Materials and methods
Chemicals and animals
Taqman reverse transcription reagents and Taqman 1000 Rxn Gold/Buffer A were provided by Applied Biosystems Roche (Branchburg, New Jersey, USA). RNeasy mini kit was from Qiagen (Tokyo, Japan). GeneChip rat genome RG-U34B set was obtained from Affymetrix (Santa Clara, CA, USA). Atp7b w/w (wild/wild) and Atp7b m/m (mutant/mutant) in siblings of LEC x (F344xLEC)F1 backcrossed rats (20 weeks old) were used for oligonucleotide array analysis to minimize background noise due to factors such as strain differences. Atp7b m/m rats have the same biological features as LEC rats with respect to hepatic copper accumulation and the onset of fulminant hepatitis (10). For quantitative determination of gene expression, to verify the array data, 24- to 48-week-old male LEC rats with the Atp7b m/m genotype and healthy LEA rats with the Atp7b w/w genotype were used. The livers were dissected, snap-frozen in liquid nitrogen, and stored at -80°C until use.
Oligonucleotide array and RT-PCR
RNA from 3 rats with either the Atp7b w/w or Atp7b m/m genotype (age, 20 weeks) was pooled. Total RNA was extracted from frozen livers using an RNeasy kit and DNase kit according to the manufacturer's protocols (Qiagen, Chatsworth, CA). The GeneChip rat genome RG-U34B set (Affymetrix, Santa Clara, CA) was used according to the protocol provided. Data were analyzed using GeneChip expression analysis software. Expression changes were reflected in the differences in signal intensity from array hybridization. A two-fold or greater change in signal intensity was considered a significant change in expression.
For RT-PCR, Taqman reverse transcription reagents and Taqman 1000 Rxn Gold/Buffer A were used for cDNA synthesis and PCR, respectively according to the manufacturer's protocols. All signals were normalized against the glyceraldehyde-3-phosphate dehydrogenase (G3PDH) gene, which was amplified from the same dilution series. Primers used for RT-PCR were as follows: Akr1b7, 5′-TGAGAGTGAGGTGGGAGAA-3′ and 5′-TCCTCGTGGAAAGGAAAGTCC-3′; glucose-6-phosphate dehydrogenase (G6PDH), 5′-ATGGCCTTCTACCCGAAGACA-3′ and 5′-TCATGTGGCTGTTGAGGTGCT-3′; haem-oxygenase 1 (HO-1), 5′-GTGTCCAGGGAAGGCTTTAAGC-3′ and 5′-TGCTGTGTGGCTGGTGTGTAAG-3′; Thioredoxin (Trx), 5′-GCCAAAATGGTGAAGCTGATC-3′ and 5′-AACATCCTGGCAGTCATCCAC-3′; aldehyde reductase (Akr1a1), 5′-AGATGCCTCTGATTGGTCTGG-3′ and 5′-TGGAGGTCACAAACAGCTCCT-3′.
Taqman real-time PCR method (12-13) cDNA synthesis
cDNA was synthesized from total RNA (1 μg) in a final volume of 40 μl using Taqman reverse transcription reagents according to the manufacturer's protocol.
Primers and probes
Primers and probes that recognized rat cDNA sequences for Akr1b7 and G3PDH were designed using Primer Express software (PE Applied Biosystems, Foster City, CA, USA). G3PDH was used as an endogenous control gene. The primers and probes used were as follows: Akr1b7, 5′-GAAGCAGATTTTAGCCACGTCTCT-3′, 5′-GAGTCTGCACCTTGATG-3′ and probe 5′-CCTGGATGTAGGCTACCATTTGGCCA-3′; G3PDH, 5′-GGCTGCCTTCTCTTGTGACAA-3′, 5′-CCGTGGGTAGAGTCATACTGG-3′ and probe 5′-TGCCATCAACGACCCCTTCATTGAC-3′. All reactions were performed with an ABI Prism 7000 sequence detection system (PE Applied Biosystems). Each sample was amplified in triplicate. On the same plate, samples for standard curves for the target genes as well as a negative control (NTC) sample prepared without cDNA were included. The standard curves for the Akr1b7 target gene and G3PDH were constructed with 10-fold serial dilutions of the target gene cDNA. The target messages in unknown samples were quantified according to the copy numbers by using the standard curves, and then normalized on the basis of G3PDH expression and expressed as copy number per 1000 copies of G3PDH.
Cloning of the 5′ regulatory region of the rat Akr1b7 gene and construction of plasmids
The rat Akr1b7 6.0-kb 5′ region (between nt -15 and -6000 from the transcription initiation site) was cloned by PCR with Advantage 2 polymerase mix (Clontech) in the presence of 1 μg of normal rat fibroblast NRF-49F genomic DNA as a template and 100 ng each of forward primer (5′-AGGAGACAGACTCACATAGCCCAATG-3′) and reverse primer (5′- GTTTGTGGACCTAAATAACGCAGTG-3′). The PCR-amplified 6.0-kb DNA was first cloned into pGEM-T vector (Promega), and then digested with ApaI, followed by filling in with the large fragment of DNA polymerase I and SacI digestion. The resultant 6.0-kb Akr1b7 DNA was cloned into SacI- and SmaI-digested pGL3 basic luciferase vector (Promega). The ARE-positive and -negative control reporters were derived from luciferase plasmids containing the 4.5 kb and 4.4 kb of the human ferritin H 5′ region, respectively (14). ARE (antioxidant response element)-positive and -negative control reporters, hFHARE (+)-Luc and hFHARE(-)-Luc, were constructed from the 5′-upstream region of human ferritin H. Internal control reporter Renilla luciferase pRL-TK plasmid (the TK promoter region linked to the cDNA encoding Renilla luciferase) was purchased from Promega (Madison, WI), and I-κB expression plasmid pCMV-I-κBα was from BD Biosciences Clontech, Palo Alto, CA. All plasmids were purified by column chromatography (QIAfilter Maxi column, Qiagen, Santa Cruz, CA). The sequence of all plasmids was verified by automated sequencing.
Cell culture, transient transfection and luciferase assay
Human liver hepatocellular carcinoma HepG2 cells (ATCC#HB-8065) were maintained in Eagle's minimum essential medium supplemented with 100 units/ml penicillin, 10 μg/ml streptomycin, and 10% fetal bovine serum at 37°C under 5% CO2. The transfections were carried out with Qiagen effectene transfection reagent (Qiagen, Valencia, CA) according to the manufacturer's instructions. To investigate the reporter activities in HepG2 cells, the total DNA transfected in each experiment was kept constant with reporter constructs (p6.0-AKR-Luc, Luc, hFHARE(+)-Luc, or hFHARE(-)-Luc; 100 ng per well), internal control Renilla reporter pRL-TK plasmid (2 ng per well), I-κB expression plasmid, or the empty vector pCMV to make the total amount of 152 ng. Prior to transfections, cells were plated in 24-well plates and grown overnight. The cells were transfected for 24 h, then washed, reincubated with fresh medium, and cultured for another 24 h. In the CuSO4 and tBHQ treatment experiments, the cells were treated with or without CuSO4 or tBHQ for 24 h at various concentrations. Cells were collected 24 h after transfection and the firefly and Renilla luciferase activities were measured with a dual-luciferase reporter assay system (Promega) on a Fluoroskan Ascent FL fluorometer (Labsystems, Franklin, MA). The firefly luciferase reporter activities were normalized by Renilla luciferase activities and expressed in relative light units (RLUs). The RLU of the p6.0-AKR-Luc or hFHARE(+)-Luc was set as 100%. The data were the mean ± SD from a minimum of 3 independent experiments with duplicates for each experiment and presented as RLU or percentage of activity.
Northern blot analysis
Total RNA (10 ug) was electrophoresed on a 1.5% denaturing agarose gel and then transferred to nylon membranes (Gibco BRL) and fixed by UV irradiation. The membrane was subsequently hybridized to an appropriate 32P-labeled synthesized antisense oligonucleotide as probes (15). The Northern blot signals were visualized using a Storm 860 (Amersham Biosciences) scanner system.
Gel mobility shift assay
Preparation of nuclear extracts from liver was performed as described previously. Nuclear extracts containing 10 ug protein were incubated with 50 pg of 5′-end-labeled, double-stranded oligonucleotide (upper strand, 5′-AGTTGAGGGGACTTTCCCAGGC-3′) for NF-κB, and subjected to a gel shift assay on a native 4% polyacrylamide gel, as described previously (16-17).
Immunohistochemistry for glutathione S-transferase placental form (GST-P)-positive cells
GST-P immunostaining was performed as previously reported (11). Briefly, sections were treated sequentially with rabbit anti-GST-P antibody (1;2000), biotin-labeled goat anti-rabbit IgG and the ABC staining system and then counterstained with methyl green. The numbers of GST-P-positive cells were counted manually over the entire portion of the cut surface of livers from 24-week-old LEC and LEA rats.
Statistical analysis
ANOVA with subsequent post hoc tests were used where appropriate. All values are expressed as the mean ± SD for 6 animals. Differences were considered significant at p<0.05.
Results
Oxidative stress in livers of LEC rats
Biochemical and pathological changes were measured to determine hepatic oxidative status in LEC rats. Fig. 2A shows Northern blots of the mRNAs for metallothionein (MT)-I, MT-II, and hsp73 mRNAs. The levels of both MT-I and MT-II mRNA in LEC rat liver were approximately 40-fold higher than those observed in age-matched LEA rat liver. Hsp73 (hsc70) is a member of the constitutively expressed heat shock proteins and is significantly induced by various stressors (e.g., heat-shock, heavy metals, oxidation). The hepatic levels of hsp73 mRNA in LEC rats were also significantly increased (∼3-fold) as compared with LEA rats.
Figure 2.
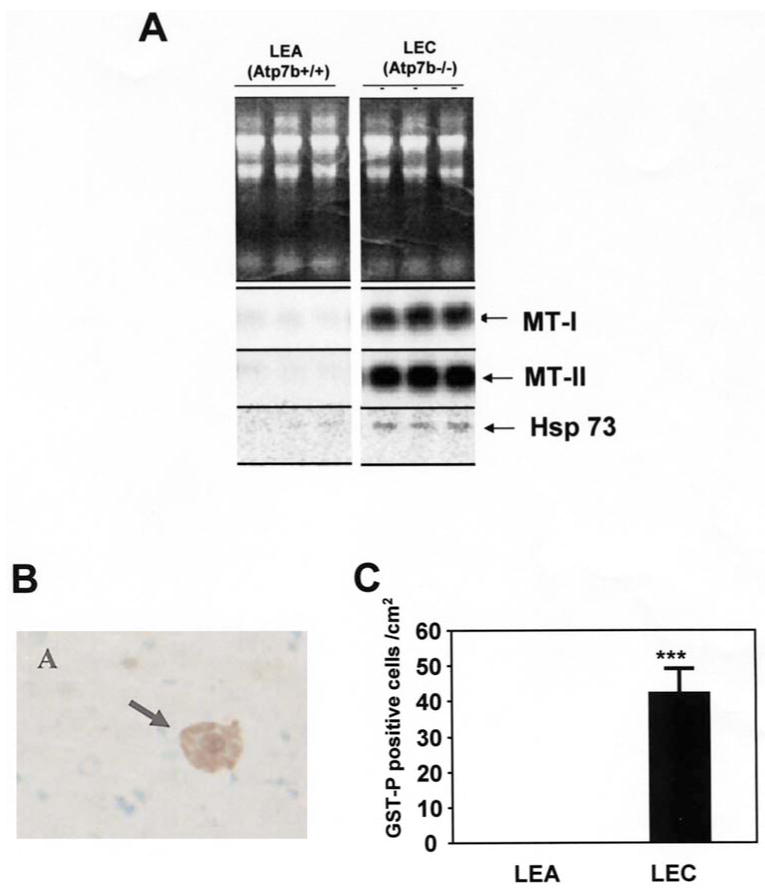
Biochemical and biophysical responses in LEC rat liver with the development of hepatitis. A, Northern blots of the mRNAs for metallothionein (MT)-I, -II, and hsp73 mRNAs. B, a typical picture of GST-P+ cells in the liver. GST-P+ cells were detected by immunohistochemistry using anti-GST-P antibody. C, the number of GST-P+ cells per cm2 in the livers of LEA and LEC rats aged 24 weeks of age. Data are expressed as mean ± SD of 7 rats. Student's t-test was performed for comparison of the number of GST-P+ cells per cm2. ***Significant difference at p<0.001 when compared with age-matched LEA rat group.
Fig. 2B shows the number of GST-P-positive cells detected by immunohistochemistry in the livers of LEC and LEA rats at 24 weeks of age. GST-P-positive (GST-P+) cells were first observed at 20 weeks of age and reached a peak at 24 weeks of age. No GST-P+ cells were observed in livers of LEA rats at 24 weeks of age.
These observations regarding MT-I, MT-II, hsp73, and GST-P indicate that the livers of LEC rats at 24 weeks of age underwent high levels of oxidative stress.
Microarray analysis of LEC rat liver
In order to profile oxidative-responsive genes, RNA from the livers of LEC rats with hepatitis and age-matched wild-type rats was subjected to microarray analysis. Forty-five transcripts were up- or down-regulated in Atp7b m/m rat liver as compared with ATP7b w/w control rats. The differentially expressed genes were mainly related to oxidative stress, growth regulation, and transcriptional regulation. To verify the changes in gene expression observed in the array analysis, mRNA expression of these transcripts was further evaluated by RT-PCR. The relative expression detected by RT-PCR showed only ∼38% concordance with the relative expression determined by gene array analysis. Nine transcripts showed higher mRNA expression in Atp7b m/m rat liver, whereas 8 transcripts showed lower expression as compared with Atp7b w/w rats (Table I). Known genes out of the 9 genes that were up-regulated were TRADD, Rat cofactor required for Sp1 transcriptional activation, subunit 6(CRSP6), 20s proteasome non-ATPase subunit mouse homologue, Tclone15/calcyclin, and human homologue MBD2 and MVDP/AKR1B7. Known genes out of the 8 down-regulated genes were CL-6, glucose-6-phosphatase catalytic subunit, and alternative transcript of mouse homologue growth hormone receptor. Out of the 17 up- and down-regulated genes, only akr1b7 is associated with oxidation-reduction. Therefore, we next focused on transcriptional regulation of Akr1b7 by oxidative stress in the livers of LEC rats.
Table I.
List of genes confirmed by semi-quantitative RT-PCR.
| Name of probe set | Fold changea | Description |
|---|---|---|
| Up-regulated genes | ||
| rc-AA946443 | 12.1 | Npdc1: Neural proliferation, differentiation and control, 1 |
| rc-AA851204 | 11.5 | Rat TNF receptor-1 associated protein (TRADD) mRNA, 3′ end of cds |
| rc-AI010116 | 10.5 | Rat cofactor required for Spl transcriptional activation, subunit 6(CRSP6) mRNA |
| rc-AA925183 | 8.8 | Mouse homologue mRNA for 20 S proteasome non-ATPase subunit |
| rc-AI008106 | 8.1 | Rat T clonel5 mRNA. mRNA for rat calcyclin |
| rc-AA850378 | 5.9 | Human homologue methyl-CpG binding domain protein2 (MBD2) mRNA |
| rc-AI009350 | 4.8 | Abhydrolase domain containing 4 (predicted) Rn. 1831 |
| rc-AA925864 | 4.4 | Rat aldose-reductase-like protein MVDP/AKR1B7 mRNA, complete cds |
| rc-AA899498 | 2.6 | Catechol-O-methyltransferase domain containing 1 (predicted) |
| Down-regulated genes | ||
| rc-AI029795 | -7.7 | Rat growth response protein (CL-6), complete cds |
| rc-AA924301 | -6.2 | ATP-binding cassette, sub-family C (CFTR/MRP), member 2 (Abcc2) |
| rc-AI964628 | -6.1 | Rat mRNA for glucose-6-phosphatase catalytic subunit, complete cds |
| rc-AI044453 | -4.9 | Strongly similar to XP_544438.2 Predicted: similar to 1810010N17Rik protein |
| rc-AI029455 | -4.2 | Ttpa: tocopherol (alpha) transfer protein |
| rc-AI010568 | -4.2 | Ghr: Growth hormone receptor |
| rc-AI045040 | -3.9 | Dgat2: Diacylglycerol O-acyltransferase homolog 2 (mouse) |
| rc-AI045066 | -3.5 | Human homologue histidine-rich glycoprotein (HRG) mRNA |
Fold change column indicates results from the oligonucleotide array.
Analysis of the correlation between akr1b7 and oxidative stress level
To further characterize the gene encoding AKR1B7 on the array, we detected alterations in its expression in the livers of LEC rats under different levels of oxidative stress. Another 4 known oxidative stress-responsive genes, including G6PDH (18), thioredoxin (Trx) (19-20), HO-1 (21-22), and Akr1a1 (23-25) were also analyzed. A summary of the relative expression levels of the genes detected by RT-PCR is shown in Fig. 3. The induction of Akr1b7 in the livers of LEC rats aged 24 and 48 weeks was 486- and 27.5-fold higher, respectively, than that of age-matched LEA rats. In contrast, the expression of well-known oxidative stress genes including G6PDH, Trx, HO-1, and Akr1a1 in LEC rat liver was only 1.2- to 2.3-fold higher than that of age-matched LEA rats. Consistent with the high level of oxidative stress observed, the expression of these 5 genes in LEC rats was higher than that of 24-week-old LEA rats.
Figure 3.
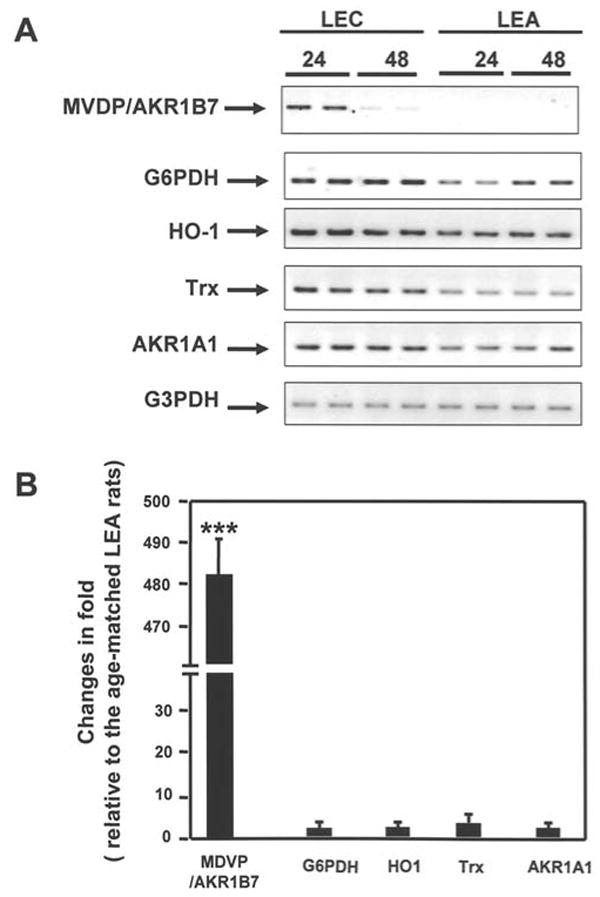
Summary of representative gene expression detected by RT-PCR. A, expression of MVDP/AKR1B7 (Akr1b7), G6PDH, HO-1, thioredoxin (Trx), AKR1A1, and G3PDH in the livers of LEC and LEA rats aged 24 and 48 weeks. B, relative expression of MVDP/AKR1B7, G6PDH, HO-1, TRX, AKR1A1, and G3PDH in the liver of LEC rats as compared with 24-week-old LEA rats. Values are means ± SD for 6 pairs of age-matched rats. ***Significantly different from the corresponding control group (*p<0.001).
Since real-time PCR is a more quantitative, specific, sensitive, and reliable method for detecting the expression of target genes at the RNA level when compared with traditional ‘end-point’ measurements of PCR products, we performed real-time PCR for Akr1b7. The results are shown in Fig. 4. The expression of Akr1b7 in livers of LEC rats aged 24 weeks was 85-fold higher than that of age-matched LEA rats, and 16-fold higher than that of LEC rats aged 48 weeks old.
Figure 4.
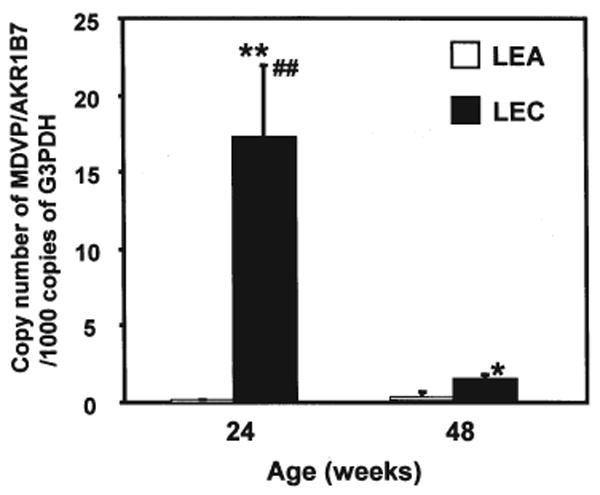
Expression of Akr1b7 quantitatively detected by real-time RT-PCR in livers of LEA and LEC rats at different ages. All values are expressed as mean ± SD of 4 animals. Student's t-test was performed for comparison of Akr1b7 expression. * and **, significant difference at p<0.05, p<0.01 when compared with age-matched LEA rat group. ##Significant difference at p<0.01 when compared with 48-week-old LEC rats.
Transcriptional activity of nuclear factor kappa B (NF-κB)
Several conserved sequence motifs throughout the promoter were found to be correlated with potential sites for the binding of transcription regulatory factors, as determined by computational analysis of ∼6000-bp DNA sequences of the 5′-flanking region of rat Akr1b7 using the TRANSFAC database (Fig. 5A). The key binding domains associated with oxidative stress were identified. The nuclear factor kappa B (NF-κB) sites (26-27) were located at position -389/398, -650/660, -3758/-3769, and -5657/-5667. The ARE/EpRE (28-29) sites were located at -1187/-1203, -2416/-2432, -2709/-2725, and -4226/-4242 (Fig. 5B).
Figure 5.
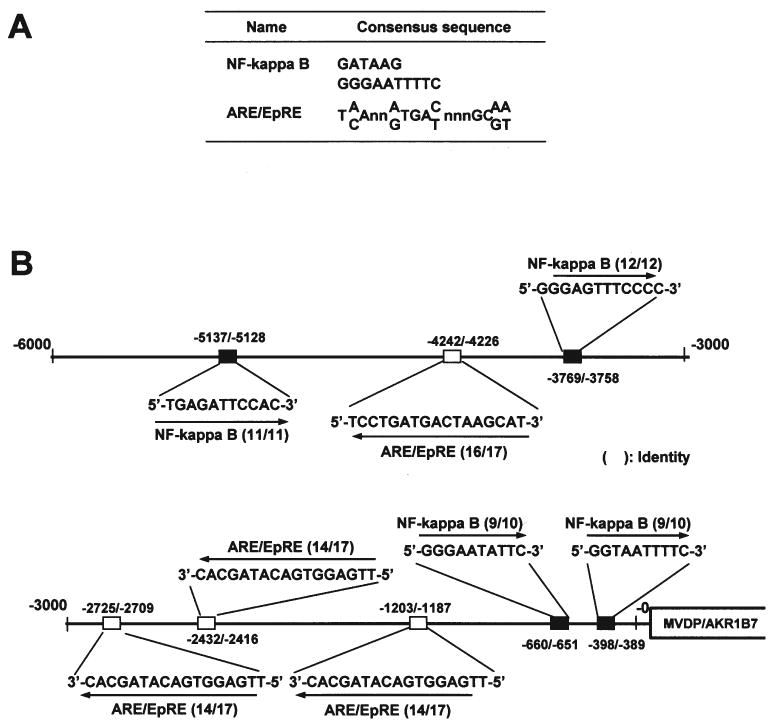
Genome informative analysis of binding motifs of the 5′-upstream regions of Akr1b7 genes. A, typical genes having the NF-κB site or EpRE/ARE sequences. B, distributions of the NF-κB site and EpRE/ARE sequences are located 6 kb upstream of the Akr1b7 gene.
To examine whether NF-κB is induced in livers of LEC rats in response to oxidative stress and serves as a potential mediator of the Akr1b7 gene, we determined the binding activity of NF-κB in hepatic nuclear extracts from both strains by gel mobility shift assay using NF-κB binding consensus oligonucleotide. As shown in Fig. 6, NF-κB binding was detected as two bands, designated upper and lower to indicate slower and faster mobility through the gel, respectively. A faster migrating band was detected in both LEA and LEC rats. However, the marked increase in the amount of a slow migrating complex was observed in LEC rats. Previous studies have shown that the complex of slower and faster mobility observed in the nuclear extract from rat liver is a p65/p50 heterodimer and a p50/p50 homodimer, respectively (30). Since the activation of the p50/p60 heterodimeric form of NF-κB is particularly sensitive to oxidative stress and changes in cellular redox conditions (31-32), its observed up-regulation is consistent with a heightened state of oxidative stress in LEC rat liver.
Figure 6.
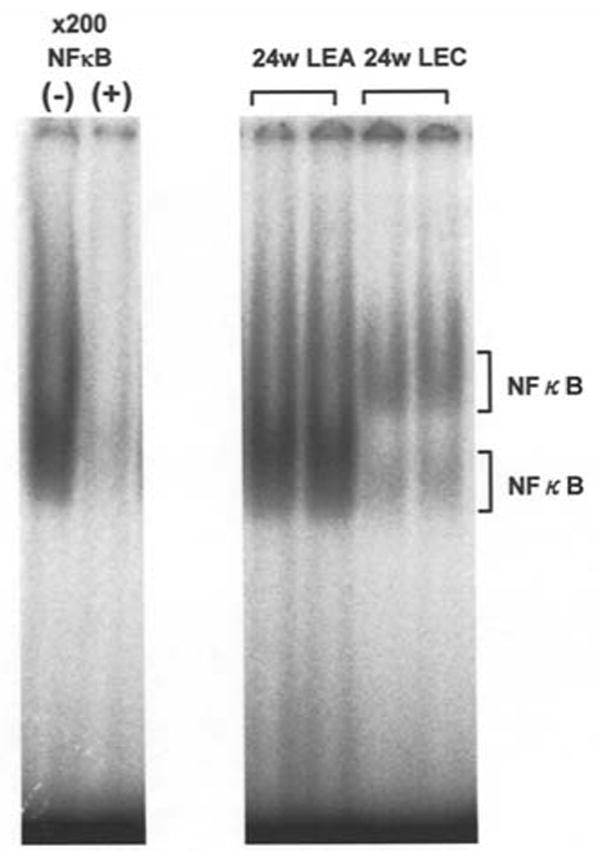
Involvement of transcription factor NF-κB in the transcriptional regulation of the MVDP/AKR1B7 gene. NF-κB is activated by hepatitis onset in LEC rat liver. NF-κB oligonucleotide binding activity in nuclear extracts prepared from the livers of 24-week-old LEA and LEC rats was determined by gel mobility shift assay using 32P-labeled oligonucleotide as described under Materials and methods. The specificity of the NF-κB complex was verified by displacement with excess (250-fold) unlabeled oligonucleotide for NF-κB.
To explore the possibility that NF-κB or cupric ion regulate transcriptional activity of 5′-upstream regions of Akr1b7, we investigated their effects on AKR reporter plasmids in transfection assays with HepG2 cells (Fig. 7). The reporter plasmid p6.0-AKR-Luc contains the upstream 6-kb DNA sequence of Akr1b7. To verify that p6.0-AKR-Luc shows transcriptional activity, reporter activities of the plasmids lacking or with the promoter were determined (Fig. 7A). When I-κB expression plasmid was co-transfected with p6.0-AKR-Luc in HepG2 cells, I-κB repressed its expression dose-dependently (Fig. 7B). In contrast, treatment with copper sulfate did not affect the transcriptional activity of p6.0-AKR-Luc, suggesting that it is unlikely that copper mediates transcriptional activity of the 6-kb sequence upstream of Akr1b7 (Fig. 7C). In contrast with the transcriptional activity of ARE to redox-active agents, the transcriptional activity of p6.0-AKR-Luc was not induced by tBHQ (Fig. 7D). The ARE-reporter gene however was induced by tBHQ treatment (Fig. 7E).
Figure 7.
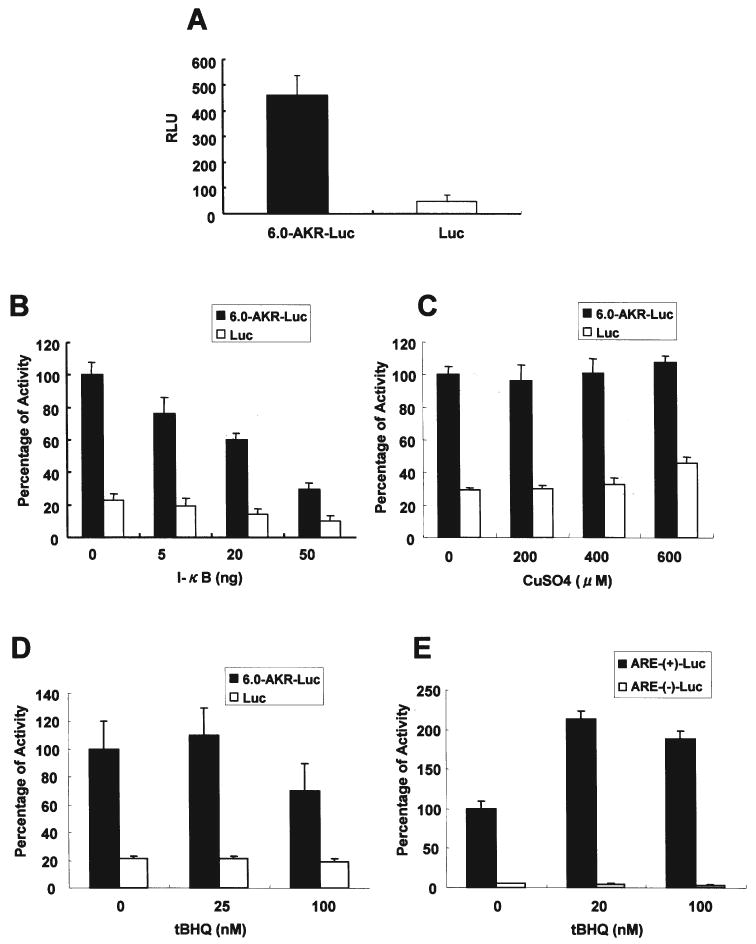
Effects of I-κB and copper on AKR promoter reporter luciferase activity. HepG2 cells were transfected with 6.0-AKR-Luc or Luc (100 ng/well, in a 24-well plate) reporter construct with internal control reporter Renilla luciferase (pRL-TK, 2 ng/well) for 24 h. Cells were collected 24 h after transfection and the firefly and Renilla luciferase activities measured using a Dual-luciferase reporter assay system. Firefly luciferase reporter activities were normalized to Renilla luciferase activities and expressed as relative light units (RLUs). A, AKR promoter reporter 6.0-AKR-Luciferase has activity in HepG2 cells. B, I-κB overexpression represses AKR promoter reporter 6.0-AKR-Luciferase activity in HepG2 cells. HepG2 cells were transfected with 6.0-AKR-Luc or Luc with various amounts of I-κB expression plasmids for 24 h. The RLU of the 6.0-AKR-Luc without I-kB overexpression was set as 100%. C, AKR promoter reporter 6.0-AKR-Luciferase did not respond to CuSO4 in HepG2 cells. The RLU of the 6.0-AKR-Luc without CuSO4 treatment was set as 100%. D, effects of tBHQ on AKR promoter reporter 6.0-AKR-Luciferase in HepG2 cells. The RLU of the 6.0-AKR-Luc without tBHQ treatment was set as 100%. E, effects of tBHQ on ARE(+)promoter reporter ARE(+)-Luciferase in HepG2 cells. The RLU of the ARE(+)-Luc without tBHQ treatment was set as 100%.
Discussion
In our study, high levels of expression of MT (I/II), Hsp73, and other indicators of oxidative stress suggest that LEC rats at approximately 24 weeks of age undergo high oxidative stress levels. Furthermore, the high number of GST-P+ single cells also suggested an increase in the number of preneoplastic cells in LEC rats at around 24 weeks of age. Moore and colleagues (33) reported that single GST-P+ liver cells are a putative marker of initiated hepatocytes that can be useful for analyzing the initiation stage of carcinogenesis (34). Oligonucleotide array analysis and RT-PCR of liver tissue from 24-week old rats identified 17 transcripts that were differentially expressed in Atp7b m/m rats, a genotype generated by backcrossing LEC rats. Of the 17 differentially expressed genes, the expression of Akr1b7 gene was most altered in LEC rats as well as LEA rats.
To compare the effects of oxidative stress on akr1b7 with those on other well-known oxidative stress-related genes, we selected G6PDH, HO1, Trx and Akr1a1, (which are sensitive biomarkers of oxidative-stress) for RT-PCR and measurement of hepatic mRNA expression in 24- and 48-week-old LEC and LEA rats. HO1 plays an important functional role as an endogenous antioxidant (21,22). Trx in conjunction with thioredoxin reductase, is a ubiquitous oxido-reductase system, positioned at the core of cellular thiol redox control and antioxidant defense (19). There is emerging evidence that the Trx system is as important as the glutathione system for cellular redox regulation of oxidative stress (20,35). MT and Trx proteins belong to the same family of thiol-rich compounds (36), and play important roles in the defense against reactive oxygen species (32). G6PDH can be up-regulated by hepatocellular oxidative stress (18), and AKR1A1 is responsible for the reduction of aldehydes, which are the main byproducts of lipid peroxidation (23,24). Interestingly, Akr1b7 was the gene whose expression was up-regulated most in response to oxidative stress in LEC rat liver in the present study.
AKR1B7 encodes an aldose-reductase involved in detoxification processes (37). It is the major enzyme responsible for the reduction of isocaproaldehyde, which is formed from the side-chain cleavage of cholesterol in the first step of murine steroidogenesis (38). Transcriptional regulation of Akr1b7 has been well defined in two of the major organs in which it is expressed. In mouse adrenal cortex, its expression is acutely regulated by ACTH (39), whereas in the mouse vas deferens, a strong androgen-dependent accumulation has been described (40). Lau et al have shown by RNase protection that Akr1b7 is also expressed in mouse kidney, eye, intestine and, to a very low extent, in liver (41). The present study is the first to report that Akr1b7 is induced by copper overload in LEC rat liver.
Lefrançois-Martinez et al have shown that AKR1B7 can detoxify 4-hydroxynonenal (4-HNE), a cytotoxic, β-unsaturated acyl aldehyde (38). This product is a natural result of the lipid peroxidation and cleavage that occurs in response to oxidative stress and aging (42). The previous studies showed that 4-HNE levels increased during the onset of hepatitis and development of hepatic tumors in LEC rats (43). Hence, we hypothesized that AKR1B7 could be one of several enzymes involved in the detoxification of lipid peroxides in the livers of LEC rats.
We also determined the expression of Akr1a1, which is ubiquitously localized and activates trans-dihydrodiols by converting them to reactive and redox-active o-quinones (44); however it was not induced in the livers of LEC rats. Therefore, it is likely that AKR1B7 is primarily responsible for detoxification of specific oxidized molecules such as 4-HNE and etheno adenine, which are produced in LEC rat liver. Further studies should be conducted to confirm this possibility.
Since a search for binding motifs revealed 4 NF-κB binding sites in the 5′-upstream regions of the Akr1b7 gene (Fig. 5), our results suggest that overexpression of the Akr1b7 gene might be regulated partly through the NF-κB pathway. NF-κB is activated in response to a variety of stimuli, most of which are relevant to oxidative stress conditions (e.g., UV light, virus infections, H2O2, heat shock, phorbol esters, cytokines, etc.) (45). Furthermore, these oxidative stimuli cause a preferential increase in the p50/p65 heterodimeric form of NF-κB over the p50/p50 homodimer (31,45). As shown in Fig. 6, the nuclear level of the p50/p60 heterodimeric form of NF-κB was markedly increased in the livers of LEC rats as compared with LEA rats. These results suggest that NF-κB is activated by oxidative stress during the development of chronic hepatitis and hepatocellular carcinoma and may play a significant role in the induction of the Akr1b7 gene in LEC rat liver.
Another interesting observation in this study was that electrophile response element (EpRE), RGCNNN(C/G)TCA (28,46), and NF-κB were located at several sites in the 5′-upstream regions of Akr1b7 (26), as shown in Fig. 5. The information about these response elements suggests that expression of Akr1b7, as well as the GST-P gene, reflects oxidative stress from copper overload, such as that which occurs in the LEC rat model used in the present study, as well as the GST-P gene. Therefore, a transient transfection assay using the 6-kb upstream of Akr1b7 fragment (p6.0 kb-AKR-Luc) was conducted. Since NF-κB is activated constitutively in HepG2 cell lines (47), IκB inhibited luciferase activity induced by p6.0 kb-AKR-luc even in unstimulated HepG2 cells (Fig. 7), indicating that transcriptional activity of Akr1b7 is involved in the NF-κB pathway. In contrast, cuprous ions did not induce the luciferase activity of p6.0 kb-AKR-Luc, suggesting that copper signaling may not directly affect its transcriptional activity. However, several studies demonstrated that cellular damage by copper mediates NF-κB signaling and that the copper chelating agent thiomolibdate suppresses NF-κB activation (48-51). Based on these studies and our present observations from the transfection assays, we speculate that the Akr1b7 gene is mainly induced by NF-κB activation, which is mediated through copper accumulation in the hepatocytes of LEC rats.
In conclusion, oligonucleotide array analysis showed that oxidative stress induced by abnormal accumulation of copper can up- or down-regulate the expression of a battery of genes in LEC rat liver, which are mainly related to oxidative stress, growth regulation, and transcriptional regulation. We characterized gene expression changes that were specifically dependent on the oxidative stress level. Compared with well-known oxidative stress-responsive genes, the Akr1b7 gene was highly expressed in LEC rats as a consequence of high oxidative stress levels, suggesting that it is involved in the defense against oxidants in the response to oxidative stress. Our study also demonstrated that overexpression of the Akr1b7 gene might be regulated partly through the NF-κB pathway.
Acknowledgments
We thank Dr Junzo Yonemoto (National Institute for Environmental Studies) for useful discussions and Ms. Chizu Suzuki and Mr. Kazuhiro Shiizaki for their technical support. This work was supported in part by a grant from the Japan Science and Technology Agency and DK60007 from the National Institutes of Health, USA.
Abbreviations
- AKR1A1
aldo-keto reductase family 1, member A1
- AKR1B7
aldo-keto reductase 1 family B7
- ATP7b
ATPase7b
- EpRE
electrophile response element
- EST
expressed sequence tag
- G6PDH
glucose-6-phosphate dehydrogenase
- GST-P
glutathione S-transferase-placental
- HO-1
haemoxygenase-1
- 8-OHdG
8-hydroxydeoxyguanosin
- IκBα
inhibitory kappa B alpha
- LEC
Long-Evans Cinnamon
- MT
metallothionein
- NF-κB
nuclear factor kappa B
- PUFA
polyunsaturated fatty acid
- Trx
thioredoxin
References
- 1.Llovet JM, Burroughs A, Bruix J. Hepatocellular carcinoma. Lancet. 2003;362:1907–1917. doi: 10.1016/S0140-6736(03)14964-1. [DOI] [PubMed] [Google Scholar]
- 2.Yamamoto Y, Yamashita S, Fujisawa A, Kokura S, Yoshikawa T. Oxidative stress in patients with hepatitis, cirrhosis, and hepatoma evaluated by plasma antioxidants. Biochem Biophys Res Commun. 1998;247:166–170. doi: 10.1006/bbrc.1998.8752. [DOI] [PubMed] [Google Scholar]
- 3.Feitelson MA, Sun B, Satiroglu Tufan NL, Liu J, Pan J, Lian Z. Genetic mechanisms of hepatocarcinogenesis. Oncogene. 2002;21:2593–2604. doi: 10.1038/sj.onc.1205434. [DOI] [PubMed] [Google Scholar]
- 4.Jungst C, Cheng B, Gehrke R, Schmitz V, Nischalke HD, Ramakers J, et al. Oxidative damage is increased in human liver tissue adjacent to hepatocellular carcinoma. Hepatology. 2004;39:1663–1672. doi: 10.1002/hep.20241. [DOI] [PubMed] [Google Scholar]
- 5.Yamamoto H, Watanabe T, Mizuno H, Endo K, Hosokawa T, Kazusaka A, et al. In vivo evidence for accelerated generation of hydroxyl radicals in liver of Long-Evans Cinnamon (LEC) rats with acute hepatitis. Free Radic Biol Med. 2001;30:547–554. doi: 10.1016/s0891-5849(00)00496-2. [DOI] [PubMed] [Google Scholar]
- 6.Yamamoto H, Hirose K, Hayasaki Y, Masuda M, Kazusaka A, Fujita S. Mechanism of enhanced lipid peroxidation in the liver of Long-Evans cinnamon (LEC) rats. Arch Toxicol. 1999;73:457–464. doi: 10.1007/s002040050635. [DOI] [PubMed] [Google Scholar]
- 7.Yamamoto F, Kasai H, Togashi Y, Takeichi N, Hori T, Nishimura S. Elevated level of 8-hydroxydeoxyguanosine in DNA of liver, kidneys, and brain of Long-Evans Cinnamon rats. Jpn J Cancer Res. 1993;84:508–511. doi: 10.1111/j.1349-7006.1993.tb00168.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nair J, Sone H, Nagao M, Barbin A, Bartsch H. Copper-dependent formation of miscoding etheno-DNA adducts in the liver of Long Evans cinnamon (LEC) rats developing hereditary hepatitis and hepatocellular carcinoma. Cancer Res. 1996;56:1267–1271. [PubMed] [Google Scholar]
- 9.Yamamoto Y, Sone H, Yamashita S, Nagata Y, Niikawa H, Hara K, et al. Oxidative stress in LEC rats evaluated by plasma antioxidants and free fatty acids. J Trace Elem Exp Med. 1997;10:129–134. [Google Scholar]
- 10.Sone H, Li YJ, Ishizuka M, Aoki Y, Nagao M. Increased mutant frequency and altered mutation spectrum of the lacI transgene in Wilson disease rats with hepatitis. Cancer Res. 2000;60:5080–5086. [PubMed] [Google Scholar]
- 11.Jia G, Tohyama C, Sone H. DNA damage triggers imbalance of proliferation and apoptosis during development of preneoplastic foci in the liver of Long-Evans Cinnamon rats. Int J Oncol. 2002;21:755–761. [PubMed] [Google Scholar]
- 12.Freeman WM, Walker SJ, Vrana KE. Quantitative RT-PCR: pitfalls and potential. Biotechniques. 1999;26:112–125. doi: 10.2144/99261rv01. [DOI] [PubMed] [Google Scholar]
- 13.Livak KJ, Flood SJ, Marmaro J, Giusti W, Deetz K. Oligonucleotides with fluorescent dyes at opposite ends provide a quenched probe system useful for detecting PCR product and nucleic acid hybridization. PCR Methods Appl. 1995;4:357–362. doi: 10.1101/gr.4.6.357. [DOI] [PubMed] [Google Scholar]
- 14.Tsuji Y. JunD activates transcription of the human ferritin H gene through an antioxidant response element during oxidative stress. Oncogene. 2005;24:7567–7578. doi: 10.1038/sj.onc.1208901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heydari AR, Wu B, Takahashi R, Strong R, Richardson A. Expression of heat shock protein 70 is altered by age and diet at the level of transcription. Mol Cell Biol. 1993;13:2909–2918. doi: 10.1128/mcb.13.5.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takahashi R, Heydari AR, Gutsmann A, Sabia M, Richardson A. The heat shock transcription factor in liver exists in a form that has DNA binding activity but no transcriptional activity. Biochem Biophys Res Commun. 1994;201:552–558. doi: 10.1006/bbrc.1994.1737. [DOI] [PubMed] [Google Scholar]
- 17.Takahashi R, Toyoda E, Aoki Y, Suzuki KT, Goto S. Paradoxical increase of heat-shock response with age in a substrain of F344 rats: comparison between F344/DuCrj and F344/Jcl. Mech Ageing Dev. 2002;123:1605–1615. doi: 10.1016/s0047-6374(02)00096-9. [DOI] [PubMed] [Google Scholar]
- 18.Cramer CT, Cooke S, Ginsberg LC, Kletzien RF, Stapleton SR, Ulrich RG. Upregulation of glucose-6-phosphate dehydrogenase in response to hepatocellular oxidative stress: studies with diquat. J Biochem Toxicol. 1995;10:293–298. doi: 10.1002/jbt.2570100603. [DOI] [PubMed] [Google Scholar]
- 19.Nordberg J, Arner ES. Reactive oxygen species, antioxidants, and the mammalian thioredoxin system. Free Radic Biol Med. 2001;31:1287–1312. doi: 10.1016/s0891-5849(01)00724-9. [DOI] [PubMed] [Google Scholar]
- 20.Tanaka T, Nakamura H, Nishiyama A, Hosoi F, Masutani H, Wada H, et al. Redox regulation by thioredoxin superfamily; protection against oxidative stress and aging. Free Radic Res. 2000;33:851–855. doi: 10.1080/10715760000301361. [DOI] [PubMed] [Google Scholar]
- 21.Ryter SW, Kvam E, Tyrrell RM. Heme oxygenase activity. Current methods and applications. Methods Mol Biol. 2000;99:369–391. doi: 10.1385/1-59259-054-3:369. [DOI] [PubMed] [Google Scholar]
- 22.Elbirt KK, Bonkovsky HL. Heme oxygenase: recent advances in understanding its regulation and role. Proc Assoc Am Physicians. 1999;111:438–447. [PubMed] [Google Scholar]
- 23.Hyndman D, Flynn T. The also-keto reductases and their role in cancer. Vol. 7. New York: Kluwer Academic/Plenum Publishers; 1999. pp. 427–434. [DOI] [PubMed] [Google Scholar]
- 24.Cao D, Fan ST, Chung SS. Identification and characterization of a novel human aldose reductase-like gene. J Biol Chem. 1998;273:11429–11435. doi: 10.1074/jbc.273.19.11429. [DOI] [PubMed] [Google Scholar]
- 25.Srivastava S, Watowich SJ, Petrash JM, Srivastava SK, Bhatnagar A. Structural and kinetic determinants of aldehyde reduction by aldose reductase. Biochemistry. 1999;38:42–54. doi: 10.1021/bi981794l. [DOI] [PubMed] [Google Scholar]
- 26.Zabel U, Schreck R, Baeuerle PA. DNA binding of purified transcription factor NF-kappa B. Affinity, specificity, Zn2+ dependence, and differential half-site recognition. J Biol Chem. 1991;266:252–260. [PubMed] [Google Scholar]
- 27.Lavrovsky Y, Schwartzman ML, Levere RD, Kappas A, Abraham NG. Identification of binding sites for transcription factors NF-kappa B and AP-2 in the promoter region of the human heme oxygenase 1 gene. Proc Natl Acad Sci USA. 1994;91:5987–5991. doi: 10.1073/pnas.91.13.5987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sasaki H, Sato H, Kuriyama-Matsumura K, Sato K, Maebara K, Wang H, et al. Electrophile response element-mediated induction of the cystine/glutamate exchange transporter gene expression. J Biol Chem. 2002;277:44765–44771. doi: 10.1074/jbc.M208704200. [DOI] [PubMed] [Google Scholar]
- 29.Rushmore TH, Pickett CB. Transcriptional regulation of the rat glutathione S-transferase Ya subunit gene. Characterization of a xenobiotic-responsive element controlling inducible expression by phenolic antioxidants. J Biol Chem. 1990;265:14648–14653. [PubMed] [Google Scholar]
- 30.Morales A, Garcia-Ruiz C, Miranda M, Mari M, Colell A, Ardite E. Tumor necrosis factor increases hepatocellular glutathione by transcriptional regulation of the heavy subunit chain of gamma-glutamylcysteine synthetase. J Biol Chem. 1997;272:30371–30379. doi: 10.1074/jbc.272.48.30371. [DOI] [PubMed] [Google Scholar]
- 31.Janssen-Heininger YM, Poynter ME, Baeuerle PA. Recent advances towards understanding redox mechanisms in the activation of nuclear factor kappaB. Free Radic Biol Med. 2000;28:1317–1327. doi: 10.1016/s0891-5849(00)00218-5. [DOI] [PubMed] [Google Scholar]
- 32.Moran LK, Gutteridge JM, Quinlan GJ. Thiols in cellular redox signalling and control. Curr Med Chem. 2001;8:763–772. doi: 10.2174/0929867013372904. [DOI] [PubMed] [Google Scholar]
- 33.Moore MA, Nakagawa K, Satoh K, Ishikawa T, Sato K. Single GST-P positive liver cells - putative initiated hepatocytes. Carcinogenesis. 1987;8:483–486. doi: 10.1093/carcin/8.3.483. [DOI] [PubMed] [Google Scholar]
- 34.Pinkus R, Weiner LM, Daniel V. Role of quinone-mediated generation of hydroxyl radicals in the induction of glutathione S-transferase gene expression. Biochemistry. 1995;34:81–88. doi: 10.1021/bi00001a010. [DOI] [PubMed] [Google Scholar]
- 35.Nishinaka Y, Masutani H, Nakamura H, Yodoi J. Regulatory roles of thioredoxin in oxidative stress-induced cellular responses. Redox Rep. 2001;6:289–295. doi: 10.1179/135100001101536427. [DOI] [PubMed] [Google Scholar]
- 36.Deneke SM. Thiol-based antioxidants. Curr Top Cell Regul. 2000;36:151–180. doi: 10.1016/s0070-2137(01)80007-8. [DOI] [PubMed] [Google Scholar]
- 37.Taragnat C, Berger M, Jean C. Preliminary characterization, androgen-dependence and ontogeny of an abundant protein from mouse vas deferens. J Reprod Fertil. 1988;83:835–842. doi: 10.1530/jrf.0.0830835. [DOI] [PubMed] [Google Scholar]
- 38.Lefrancois-Martinez AM, Tournaire C, Martinez A, Berger M, Daoudal S, Tritsch D, Veyssiere G, Jean C. Product of side-chain cleavage of cholesterol, isocaproaldehyde, is an endogenous specific substrate of mouse vas deferens protein, an aldose reductase-like protein in adrenocortical cells. J Biol Chem. 1999;274:32875–32880. doi: 10.1074/jbc.274.46.32875. [DOI] [PubMed] [Google Scholar]
- 39.Aigueperse C, Martinez A, Lefrancois-Martinez AM, Veyssiere G, Jean CI. Cyclic AMP regulates expression of the gene coding for a mouse vas deferens protein related to the aldo-keto reductase superfamily in human and murine adrenocortical cells. J Endocrinol. 1999;160:147–154. doi: 10.1677/joe.0.1600147. [DOI] [PubMed] [Google Scholar]
- 40.Martinez A, Aigueperse C, Val P, Dussault M, Tournaire C, Berger M, et al. Physiological functions and hormonal regulation of mouse vas deferens protein (AKR1B7) in steroidogenic tissues. Chem Biol Interact. 2001:130–132. 903–917. doi: 10.1016/s0009-2797(00)00244-1. [DOI] [PubMed] [Google Scholar]
- 41.Lau ET, Cao D, Lin C, Chung SK, Chung SS. Tissue-specific expression of two aldose reductase-like genes in mice: abundant expression of mouse vas deferens protein and fibroblast growth factor-regulated protein in the adrenal gland. Biochem J. 1995;312:609–615. doi: 10.1042/bj3120609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Comporti M. Lipid peroxidation and biogenic aldehydes: from the identification of 4-hydroxynonenal to further achievements in biopathology. Free Radic Res. 1998;28:623–635. doi: 10.3109/10715769809065818. [DOI] [PubMed] [Google Scholar]
- 43.Ma Y, Zhang D, Kawabata T, Kiriu T, Toyokuni S, Uchida K, et al. Copper and iron-induced oxidative damage in non-tumor bearing LEC rats. Pathol Int. 1997;47:203–208. doi: 10.1111/j.1440-1827.1997.tb04481.x. [DOI] [PubMed] [Google Scholar]
- 44.Palackal NT, Burczynski ME, Harvey RG, Penning TM. The ubiquitous aldehyde reductase (AKR1A1) oxidizes proximate carcinogen trans-dihydrodiols to o-quinones: potential role in polycyclic aromatic hydrocarbon activation. Biochemistry. 2001;40:10901–10910. doi: 10.1021/bi010872t. [DOI] [PubMed] [Google Scholar]
- 45.Baeuerle PA, Baltimore D. NF-kappa B: ten years after. Cell. 1996;87:13–20. doi: 10.1016/s0092-8674(00)81318-5. [DOI] [PubMed] [Google Scholar]
- 46.Ansell PJ, Espinosa-Nicholas C, Curran EM, Judy BM, Philips BJ, Hannink M, et al. In vitro and in vivo regulation of antioxidant response element-dependent gene expression by estrogens. Endocrinology. 2004;145:311–317. doi: 10.1210/en.2003-0817. [DOI] [PubMed] [Google Scholar]
- 47.Saliou C, Rihn B, Cillard J, Okamoto T, Packer L. Selective inhibition of NF-kappaB activation by the flavonoid hepatoprotector silymarin in HepG2. Evidence for different activating pathways. FEBS Lett. 1998;440:8–12. doi: 10.1016/s0014-5793(98)01409-4. [DOI] [PubMed] [Google Scholar]
- 48.Song YS, Lee YS, Chan PH. Oxidative stress transiently decreases the IKK complex (IKKalpha, beta, and gamma), an upstream component of NF-kappaB signaling, after transient focal cerebral ischemia in mice. J Cereb Blood Flow Metab. 2005;25:1301–1311. doi: 10.1038/sj.jcbfm.9600123. [DOI] [PubMed] [Google Scholar]
- 49.Suska F, Gretzer C, Esposito M, Emanuelsson L, Wennerberg A, Tengvall P, et al. In vivo cytokine secretion and NF-kappaB activation around titanium and copper implants. Biomaterials. 2005;26:519–527. doi: 10.1016/j.biomaterials.2004.02.066. [DOI] [PubMed] [Google Scholar]
- 50.Ganesh L, Burstein E, Guha-Niyogi A, Louder MK, Mascola JR, Klomp LW, et al. The gene product Murr1 restricts HIV-1 replication in resting CD4+ lymphocytes. Nature. 2003;426:853–857. doi: 10.1038/nature02171. [DOI] [PubMed] [Google Scholar]
- 51.Pan Q, Bao LW, Merajver SD. Tetrathiomolybdate inhibits angiogenesis and metastasis through suppression of the NFkappaB signaling cascade. Mol Cancer Res. 2003;1:701–706. [PubMed] [Google Scholar]


