Abstract
Several methyltransferases have been shown to catalyze the oxidative methylation of inorganic arsenic (iAs) in mammalian species. However, the relative contributions of these enzymes to the overall capacity of cells to methylate iAs have not been characterized. Arsenic (+3 oxidation state) methyltransferase (AS3MT) that is expressed in rat and human hepatocytes catalyzes the conversion of iAs, yielding methylated metabolites that contain arsenic in +3 or +5 oxidation states. This study used short hairpin RNA (shRNA) to knock down AS3MT expression in human hepatocellular carcinoma (HepG2) cells. In a stable clonal HepG2/A cell line, AS3MT mRNA and protein levels were reduced by 83 and 88%, respectively. In comparison, the capacity to methylate iAs decreased only by 70%. These data suggest that AS3MT is the major enzyme in this pathway, although an AS3MT-independent process may contribute to iAs methylation in human hepatic cells.
Introduction
The metabolism of inorganic arsenic (iAs)1 in humans and in other mammalian species is a complex process that yields various reactive and toxic intermediates and products. Reactions involved in this process are enzymatically catalyzed (1). Enzymes have been identified that catalyze the reduction of AsV-containing species (2–6) and the S-adenosylmethionine-dependent methylation of AsIII-containing species (7, 8). However, the significance of these enzymes for in vivo metabolism of iAs and the linkage between steps in the metabolic pathway for iAs remains unclear. Identification of a novel arsenic (+3 oxidation state) methyltransferase (AS3MT) in rat liver (9) has allowed the study of iAs methylation at the molecular level. Results of studies with recombinant rat AS3MT (rAS3MT) (10, 11) showed that this enzyme catalyzes the entire sequence of reactions that converts trivalent iAs, arsenite (iAsIII), to AsIII-or AsV-containing methylarsenic (MAs), dimethylarsenic (DMAs), and trimethylarsenic (TMAs) species that have been identified as intermediates or products in in vivo methylation of iAs in rats (12, 13). The presence of a synthetic reductant [e.g., tris-(2-carboxylethyl)phosphine] or of one of the cellular reducing systems, thioredoxin/thioredoxin reductase/NADPH, lipoic acid/thioredoxin reductase/NADPH, or glutaredoxin/glutathione/glutathione reductase/NADPH, is required for the activity of recombinant rAS3MT (11). Orthologues of rAS3MT have been identified in genomes of other mammalian species, including human (14). An orthologue of AS3MT (hAS3MT) [human arsenic (+3 oxidation state) methyltransferase] is expressed in cultured primary human hepatocytes (15) that methylate iAsIII to MAs and DMAs. MAs and DMAs, but not TMAs, have also been found as products of iAs methylation by recombinant hAS3MT (16). The methylation patterns for iAs by human hepatocytes and by recombinant hAS3MT are consistent with the results of epidemiological studies that found MAsIII, MAsV, DMAsIII, and DMAsV, but not TMAs, in the urine of individuals chronically exposed to iAs in drinking water (17–21). Notably, recent studies have linked hASMT polymorphisms to specific methylation patterns for iAs in primary cultures of human hepatocytes (15) and to specific urinary profiles of iAs metabolites found in residents of arsenosis endemic areas of Mexico (22). Studies examining the characteristics of iAs methylation by recombinant genetic variants of rAS3MT and hAS3MT are underway in several laboratories, providing new data on the associations between AS3MT genotypes and specific metabolic phenotypes (14, 23).
Despite growing evidence that links the hAS3MT-catalyzed reactions to the metabolism of iAs in humans, the relative contribution of this enzyme to the overall capacity to methylate iAs has not been determined (24). In this study, we have silenced hAS3MT expression using RNA interference (RNAi) in human hepatocellular carcinoma (HepG2) cells that constitutively express hAS3MT. Short hairpin RNA (shRNA) vectors were used to deliver small interfering RNA (siRNA) oligonucleotides complementary to hAS3MT transcript into HepG2 cells. Quantitative analysis of hAS3MT mRNA and protein levels and of the pattern of iAs metabolism in clonal HepG2/A cells that stably express hAS3MT-specific siRNA oligonucleotides show that reduced hAS3MT expression is associated with a significant reduction in the capacity to methylate iAs. This finding suggests that hAS3MT is the key enzyme in the pathway for methylation of iAs in human hepatic cells.
Experimental Procedures
Caution
iAs has been classified as a human carcinogen (25) and should be handled accordingly.
Arsenicals
iAsIII, sodium salt (>98% pure), was purchased from Sigma (St. Louis, MO). Radiolabeled carrier-free [73As]iAsIII was prepared by reduction of [73As]iAsV (Oak Ridge National Laboratory, Oak Ridge, TN) with metabisulfite–thiosulfate reagent (26). The yield of [73As]iAsIII in this reaction as determined by TLC (27) typically exceeded 95%.
Cells
HepG2 cells were obtained from the Tissue Culture Facility of the Lineberger Comprehensive Cancer Center of the University of North Carolina at Chapel Hill. Cells were maintained in minimum essential medium (MEM, Gibco BRL, Grand Island, NY) with 2 mM glutamine, 1 mM sodium-pyruvate, 0.1 mM nonessential amino acids, 1.5 g/L sodium bicarbonate, 10% fetal bovine serum, penicillin (50 units/mL), and streptomycin (50 μg/mL) (all from Gibco) at 37 °C in a humidified incubator in a 5% CO2 atmosphere.
Design and Cloning of shRNA Cassettes
The BLOCK-iT RNAi Designer algorithm (Invitrogen, Carlsbad, CA) was used to analyze hAS3MT mRNA (GenBank accession number, NM_020682) and to identify three target sequences for siRNA. Three shRNA constructs (shRNA/A, shRNA/B, and shRNA/C) that contained the corresponding target sense and antisense sequences and the loop sequence (Figure 1) were designed using the Small Interfering RNA Hairpin Oligonucleotide Sequence Designer (Clontech Laboratories, Mountain View, CA). This design ensured that shRNA constructs delivered into cells were efficiently transcribed and the transcripts folded properly to form hairpin-like structures that were recognized by cellular enzymes. Selected shRNA oligonucleotides were synthesized by Operon Biotechnologies (Huntsville, VA), purified by PAGE, and cloned into pSIREN-RetroQ vectors, following the siRNA Hairpin Oligonucleotide Sequence Designer protocol. Complete pSIREN-RetroQ-shRNA/A, /B, and /C expression vectors were sequenced to verify insert orientation and nucleotide sequence.
Figure 1.
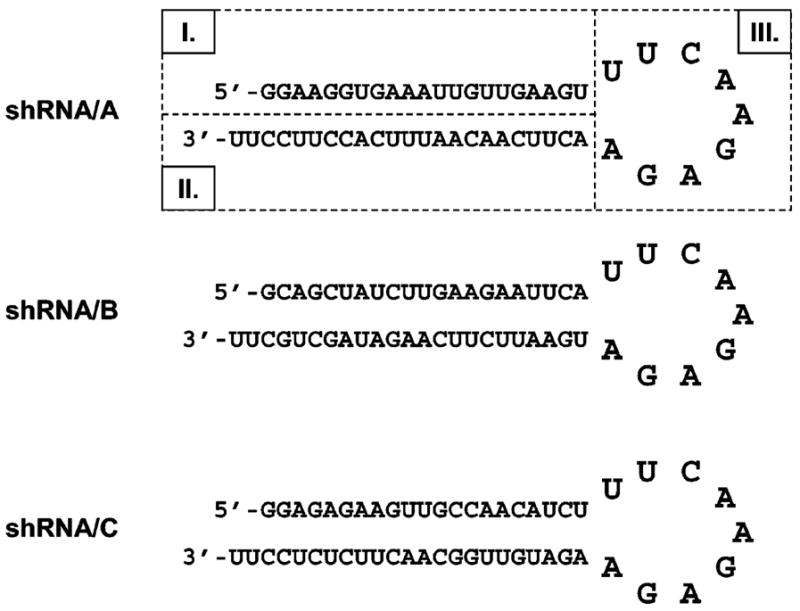
Sequences and predicted folding for three shRNA oligonucleotides designed to silence hAS3MT expression in HepG2 cells: I, target sense sequence; II, target antisense sequence (siRNA); and III, loop sequence.
Expression of shRNA Oligonucleotides in HepG2 Cells
The pSIREN-RetroQ-shRNA/A, /B, and /C expression vectors and an empty pSIREN-RetroQ vector were used for transfection of the packaging AmphoPack-293 cells (Clontech). Here, AmphoPack-293 cells were cultured in Dulbecco’s modified Eagle medium (DMEM; Gibco) supplemented with 10% FBS, penicillin (50 units/mL), and streptomycin (50 μg/mL). Cells were transfected using GeneJuice transfection reagent (Novagen, Darmstad, Germany) and selected in DMEM medium containing puromycin (1 μg/mL, Sigma). Stable AmphoPack-293 cell lines producing retroviral particles containing pSIREN-RetroQ expression vectors were established. Medium containing retroviral particles and polybrene (4 μg/mL, Sigma) was used to infect HepG2 cells. Clonal HepG2 cell lines were then selected in MEM in the presence of puromycin (2 μg/mL). Stable clonal HepG2 cell lines expressing the empty pSIREN-RetroQ vector or vectors encoding for the shRNA/A, /B, and /C were established.
Analysis of hAS3MT mRNA
hAS3MT mRNA levels in parental and clonal HepG2 cell lines were analyzed by touch-down PCR and quantified by real-time PCR as previously described (15, 27). Briefly, 1 μg of DNase-treated and purified total RNA was reverse transcribed. The following primers were used for the touch-down PCR: hAS3MT/sense, 5′-CGTCTATACGAGCCTTGAA-3′; hAS3MT/antisense, 5′-TTAGCAGCTTTTCTTTGTGC-3′. These primers amplify a 616 bp segment of hAS3MT cDNA (nucleotide no. 555–1170) in which all three sequences targeted by shRNA constructs are located. β-Actin mRNA was analyzed in parallel with hAS3MT mRNA, using the following PCR primers: sense, 5′-TCATGAAGTGTGACGTGGACATCCGC-3′; antisense, 5′-CTAGAAGCATTTGCGGTGGACGATG-3′. These primers amplify a 284 bp segment of β-actin cDNA. Quantitative PCR analysis was carried out with 100 ng of total RNA, using a computerized LightCycler Instrument (Roche Applied Science, Manheim, Germany). The set of hAS3MT-specific primers designed for touch-down PCR was also used for quantitative PCR. The PCR product was detected using LightCycler FastStart DNA MasterPlus SYBR Green I (Roche Applied Science). A hAS3MT expression (pRSET/hAS3MT) vector was used as a positive control and to generate a calibration curve for quantification of hAS3MT mRNA (10, 15, 27).
Immunoblot Analysis
The hAS3MT protein levels in parental and clonal HepG2 cells were analyzed by immunoblot using a rabbit polyclonal anti-hAS3MT antibody generously provided by Professor R. M. Weinshilboum (Mayo Clinic College of Medicine, Rochester, MN). Cell lysates were prepared in RIPA buffer as previously described (27). Aliquots of 30 μg of total protein were separated by NuPAGE in a 10% Bis-Tris gel and 3-(N-morpholino)-propanesulfonic acid (MOPS) SDS running buffer (Invitrogen) and electroblotted to polyvinylidene difluoride (PVDF) membrane. After it was blocked with 5% nonfat milk for 1 h at room temperature, the membrane was probed with a rabbit anti-hAS3MT antibody (1:2000) for 1 h at room temperature. β-Actin was analyzed in parallel with hAS3MT as a loading control, using a mouse monoclonal antibody (Abcam, Cambridge, MA). Antibody-antigen complexes were visualized by enhanced chemiluminescence detection (Amersham, Biosciences, Pittsburgh, PA) after incubation with the corresponding HRP-conjugated antibody (Santa Cruz Biotechnology, Santa Cruz, CA) for 1 h at room temperature. The chemiluminescence signal was analyzed by a FluorChem Imaging System (Alpha Innotech Corp., San Leandro, CA), using AlphaE-aseFCTM software (Alpha Innotech). The β-actin signal was used to normalize hAS3MT protein contents in parental and clonal HepG2 cell lines.
Analysis of iAs Metabolism
Metabolism of iAs was examined in parental and clonal HepG2 cells incubated with carrier-free [73As]iAsIII. Radiolabeled metabolites in cells and culture media were analyzed by TLC (28). Radioactivity associated with iAs, MAs, and DMAs was quantified with a computerized Fuji FLA-5000 imaging system (Fujifilm, Stamford, CT).
Statistical Analysis
Results of quantitative PCR analysis were statistically evaluated using GraphPad Instat software package (GraphPad Software Inc., San Diego, CA). One-way ANOVA followed by Tukey’s multiple comparison test evaluated differences between experimental groups. Differences at the p < 0.05 level were considered statistically significant.
Results and Discussion
Techniques involving the posttranscriptional silencing by RNAi have widely been used for analyses of gene functions in mammalian cells. Initially, transfection was used for the delivery of siRNA oligonucleotides complementary to target gene sequences. As compared with this approach, a relatively new methodology using shRNA offers a significant advantage in silencing longevity and delivery options. Retrovirus-delivered shRNA permits efficient delivery and immediate selection of stable knockdown cell lines. Integrated shRNA is transcribed in cells from a DNA template as a single-stranded RNA molecule. Complementary regions spaced by a small loop (Figure 1) cause the transcript to fold in a “short hairpin” conformation that resembles cellular microRNA. This molecule is a substrate for the endogenous enzyme Dicer (29). The siRNA released by shRNA digestion enters into a nuclease complex to form an RNA-induced silencing complex (RISC). RISC targets a complementary endogenous gene transcript, cleaving mRNA or suppressing translation (30, 31). Notably, an shRNA construct has recently been used by Dhankher and associates (32) to silence expression of arsenate reductase (ACR2) in Arabidopsis thaliana. ACR2 silencing resulted in a significant decrease in ACR2 protein level, loss of the capacity to reduce arsenate, and hyperaccumulation of arsenic in the shoots of the plant.
In this study, we tested three sequences in the open reading frame (ORF) of hAS3MT gene as potential targets for siRNA. Blast analysis using the BLOCK-iT RNAi Designer database (Invitrogen) confirmed that each selected sequence is unique and highly specific for hAS3MT. Selected sequences are located 885–905, 919–939, and 973–993 bp in the ORF of hAS3MT. shRNA constructs were designed containing siRNA oligonucleotides that target each selected sequence (shRNA/A, shRNA/B, and shRNA/C). A retroviral expression pSIREN-RetroQ vector delivered shRNA constructs to HepG2 cells and allowed selection of stable genetically modified HepG2 cells. Those clonal cell lines expressing shRNA/A, /B, and /C were designated as HepG2/A, HepG2/B, and HepG2/C, respectively. The clonal cell line expressing empty pSIREN-RetroQ vector was designated as HepG2/Empty.
Cell morphology, proliferation rates, hAS3MT expression, and capacity for iAs methylation were compared in parental and clonal HepG2 cells. No significant differences in cell morphology or in proliferation rates were found between parental and clonal cell lines during 12 weeks in culture (i.e., about 24 passages). Analysis of hAS3MT expression by conventional (touch-down) RT-PCR found that HepG2/A cells expressed less hAS3MT mRNA than did parental and other clonal cell lines (Figure 2a). Quantitative analysis by real-time PCR confirmed that HepG2/A cells contained only 17% of hAS3MT mRNA found in parental HepG2 cells (Figure 2b). Significantly less hAS3MT mRNA was detected in HepG2/B but not in HepG2/C or HepG2/Empty cells, indicating that the capacity to suppress hAS3MT expression varied markedly among the three shRNA constructs. Consistent with PCR data, immunoblot analysis showed that HepG2/A and HepG2/B cells contained significantly less hAS3MT protein that did HepG2, HepG2/Empty, or HepG2/C cells (Figure 3). The normalized levels of hAS3MT protein in HepG2/A and HepG2/B cells were less than 12 and 30% of that found in parental HepG2 cells, respectively. A statistically significant linear correlation (R = 0.997, p < 0.0001) between the hAS3MT mRNA level and the normalized hAS3MT protein level was found for the five HepG2 cell lines examined (data not shown). To examine how the suppression of hAS3MT expression affected capacity to methylate iAs, parental HepG2 and clonal cell lines were incubated for 72 h in the presence of carrier-free [73As]iAsIII and radiolabeled metabolites were analyzed in cell lysates and culture medium. Parental HepG2 cells methylated about 63% of iAs; MAs and DMAs that were found in both cell lysates and culture medium accounted for 10 and 53% of total 73As, respectively (Figure 4). A similar portion of iAs (58%) was methylated by HepG2/Empty and HepG2/C cells. For both cells lines, MAs and DMAs accounted for about 8 and 50% of total 73As, respectively. In contrast, HepG2/B cells methylated less than 44% and HepG2/A cells methylated less than 25% of iAs. MAs accounted for 13% of total 73As in both cell lines; DMAs represented 30 and 11% of total 73As in HepG2/B and HepG2/A, respectively. Thus, given the percentage of methylated iAs, silencing of hAS3MT expression in HepG2/A resulted in about a 60% decrease of the methylation capacity. However, on the basis of the number of methyl groups transferred to iAs (one for MAs and two for DMAs), the capacity of HepG2/A cells to methylate iAs represents only 30% of the original capacity found in HepG2 cells.
Figure 2.
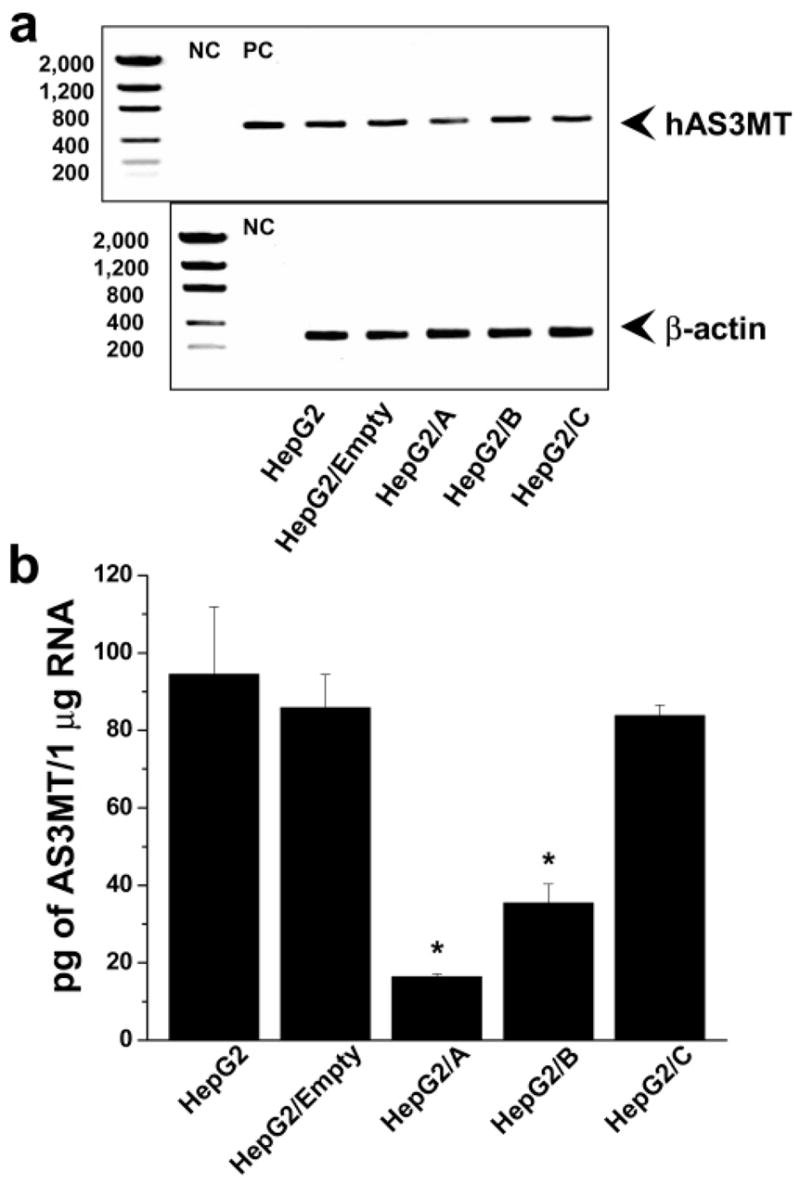
hAS3MT mRNA levels in parental and clonal HepG2 cells. (a) PCR products generated by touch-down PCR from hAS3MT and β-actin cDNA. Negative control (NC) represents a PCR reaction, in which cDNA was replaced with water. Positive control (PC) represents a PCR reaction in which cDNA was replaced with pRSET/hAS3MT expression vector. (b) hAS3MT mRNA levels quantified by real-time PCR. Mean and SD values for three independent measurements are shown. *The mean value is significantly different (p < 0.001) from that in parental HepG2 cells.
Figure 3.
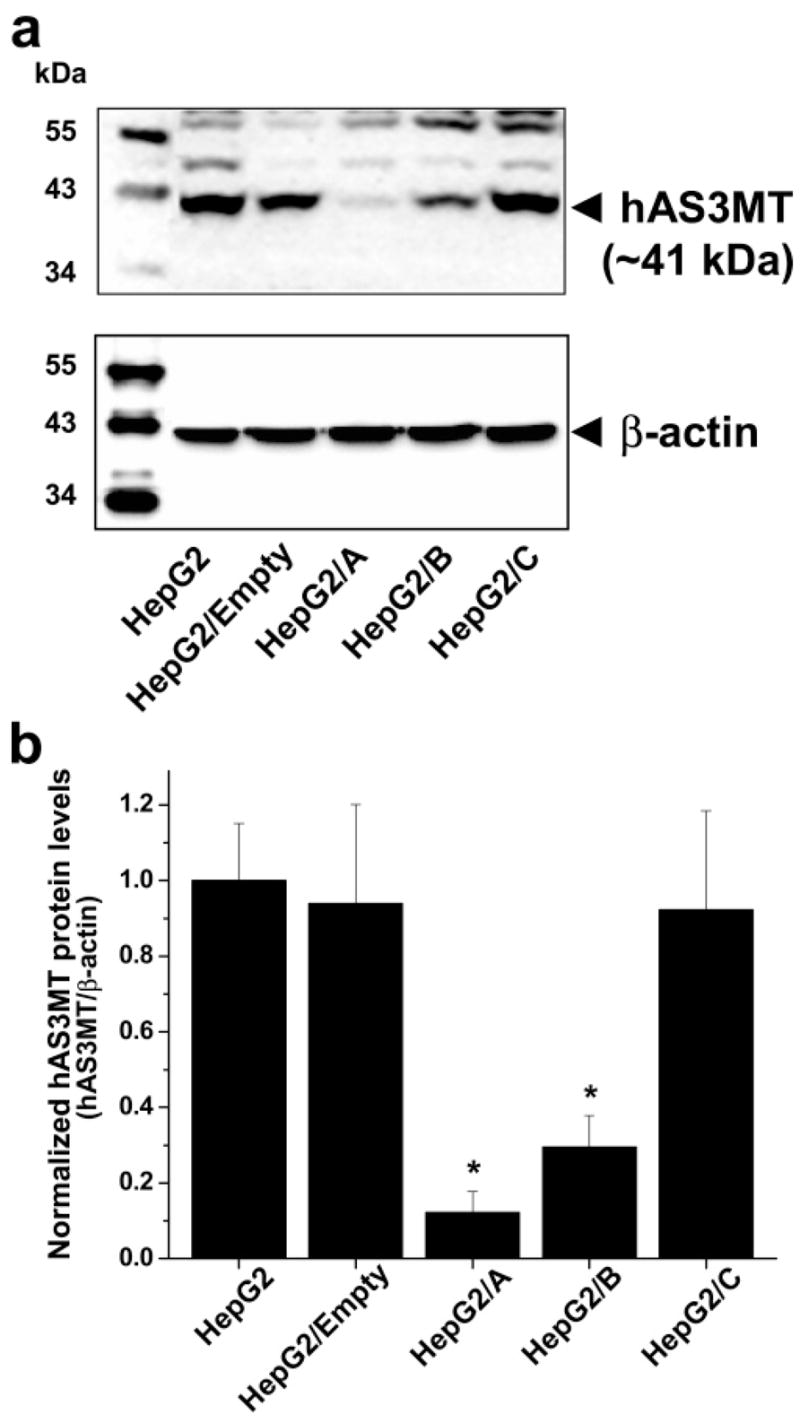
hAS3MT protein levels in parental and clonal HepG2 cells. (a) Immunoblot images of hAS3MT and β-actin. A representative image from one of three independent cultures is shown. (b) hAS3MT protein levels normalized for β-actin contents. Mean and SD values for the three independent cultures are shown. *The mean value is significantly different (p < 0.05) from that in parental HepG2 cells.
Figure 4.
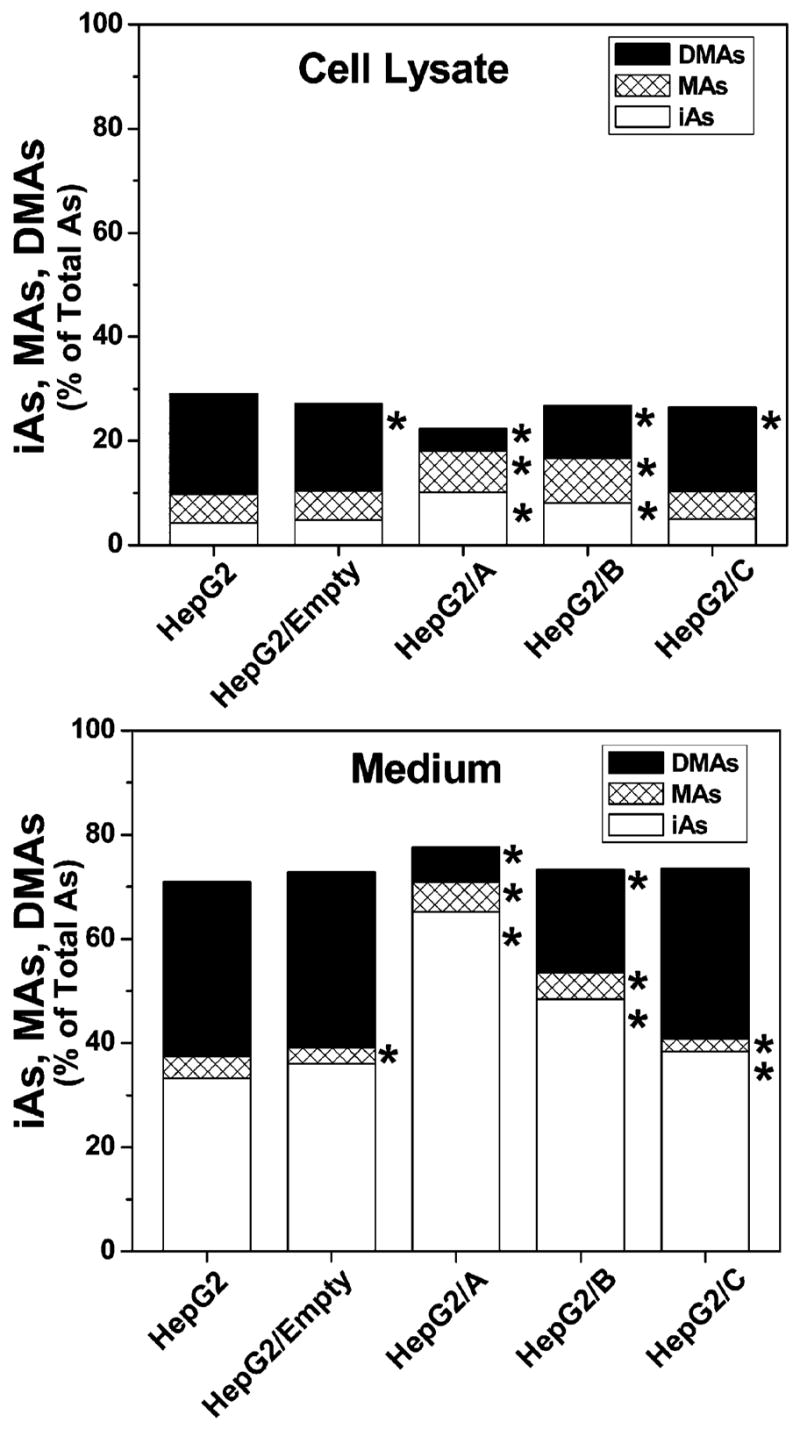
Methylation of iAsIII by parental and clonal HepG2 cells: Radiolabeled metabolites in cell lysates and media after a 72 h exposure to carrier-free [73As]iAsIII. Amounts of iAs, MAs, and DMAs are expressed as a percentage of total 73As. The mean value for three culture wells is shown for each metabolite. Although not shown, SD values did not exceed 10% of the corresponding mean value. *The mean value for a metabolite is significantly different (p < 0.05) from the corresponding value in parental HepG2 cells.
HepG2 cells have frequently been used to study metabolism of nutrients and xenobiotics in human liver (33, 34). The liver is also thought to play a key role in the metabolism of iAs. Notably, HepG2 cells are relatively efficient methylators of iAs. The rates and patterns of iAs methylation by HepG2 cells (35) resemble those described in cultured primary human hepatocytes (15). In addition, like primary human hepatocytes, HepG2 cells express hAS3MT. Thus, HepG2 cells are an appropriate cell culture model in which the enzymatic processes underlying metabolism of iAs in human liver can be examined. Because nonproliferating primary hepatocytes are not suitable for RNAi-mediated gene silencing and for selection of stable clonal cell lines, we chose HepG2 cells to examine the significance of hAS3MT expression in iAs methylation in human liver. Comparisons of hAS3MT mRNA and protein levels and the methylation patterns for iAs in parental HepG2 and clonal cells show that the capacity of these cells to methylate iAs was strongly dependent on hAS3MT expression. The quantitative association between hAS3MT silencing by RNAi and reduced capacity of HepG2 cells to methylate iAs suggest that hAS3MT may be the predominant enzyme in the pathway for iAs methylation in human liver. However, the disproportional decrease in hAS3MT mRNA (by 83%), AS3MT protein (by 88%), and methylation capacity (by 70%) in HepG2/A cells indicates that a minor AS3MT-independent mechanism may contribute to the hepatic methylation of iAs.
Acknowledgments
We thank Professor R. M. Weinshilboum, Mayo Clinic College of Medicine, for the generous gift of the rabbit anti-human AS3MT antibody. This work has been supported in part by NIH Grant ES010845 to M.S. and a Clinical Nutrition Research Center Grant DK 56350. W.X. is a postdoctoral fellow in the Curriculum in Toxicology, University of North Carolina at Chapel Hill, and is supported by Training Grant T901915 of the U.S. Environmental Protection Agencys University of North Carolina Toxicology Research Program. This manuscript has been reviewed in accordance with the policy of the National Health and Environmental Effects Research Laboratory, U.S. Environmental Protection Agency, and approved for publication. Approval does not signify that the contents necessarily reflect the views and policies of the Agency nor does mention of trade names or commercial products constitute endorsement or recommendation for use.
Footnotes
Abbreviations: shRNA, short hairpin RNA; AS3MT, arsenic (+3 oxidation state) methyltransferase; HepG2, human hepatocellular carcinoma cells; rAS3MT, rat arsenic (+3 oxidation state) methyltransferase; hAS3MT, human arsenic (+3 oxidation state) methyltransferase; iAs, inorganic arsenic; MAs, methylated arsenicals; DMAs, dimethylated arsenicals; TMAs, trimethylated arsenicals; RNAi, RNA interference; siRNA, small interfering RNA; MOPS, 3-(N-morpholino)propanesulfonic acid; PVDF, poly(vinylidene difluoride); RISC, RNA-induced silencing complex; ORF, open reading frame.
References
- 1.Thomas DJ, Waters SB, Styblo M. Elucidating the pathway for arsenic methylation. Toxicol Appl Pharmacol. 2004;198:319–326. doi: 10.1016/j.taap.2003.10.020. [DOI] [PubMed] [Google Scholar]
- 2.Gregus Z, Nemeti B. Purine nucleoside phosphorylase as a cytosolic arsenate reductase. Toxicol Sci. 2002;70:13–19. doi: 10.1093/toxsci/70.1.13. [DOI] [PubMed] [Google Scholar]
- 3.Gregus Z, Nemeti B. The glycolytic enzyme glyceral-dehyde-3-phosphate dehydrogenase works as an arsenate reductase in human red blood cells and rat liver cytosol. Toxicol Sci. 2005;85:859–869. doi: 10.1093/toxsci/kfi158. [DOI] [PubMed] [Google Scholar]
- 4.Nemeti B, Gregus Z. Reduction of arsenate to arsenite in hepatic cytosol. Toxicol Sci. 2002;70:4–12. doi: 10.1093/toxsci/70.1.4. [DOI] [PubMed] [Google Scholar]
- 5.Radabaugh TR, Sampayo-Reyes A, Zakharyan RA, Aposhian HV. Arsenate reductase II. Purine nucleoside phosphorylase in the presence of dihydrolipoic acid is a route for reduction of arsenate to arsenite in mammalian systems. Chem Res Toxicol. 2002;15:692–698. doi: 10.1021/tx0101853. [DOI] [PubMed] [Google Scholar]
- 6.Zakharyan RA, Sampayo-Reyes A, Healy SM, Tsaprailis G, Board PG, Liebler DC, Aposhian HV. Human monomethylarsonic acid (MMA(V)) reductase is a member of the glutathione-S-transferase superfamily. Chem Res Toxicol. 2001;14:1051–1057. doi: 10.1021/tx010052h. [DOI] [PubMed] [Google Scholar]
- 7.Zakharyan AR, Wu Y, Bogdan GM, Aposhian HV. Enzymatic methylation of arsenic compounds: Assay, partial purification, and properties of arsenite methyltransferase and monomethylarsonic acid methyltransferase of rabbit liver. Chem Res Toxicol. 1995;8:1029–1038. doi: 10.1021/tx00050a006. [DOI] [PubMed] [Google Scholar]
- 8.Zakharyan AR, Aposhian HV. Enzymatic reduction of arsenic compounds in mammalian systems: The rate-limiting enzyme of rabbit liver arsenic biotransformation is MMA(V) reductase. Chem Res Toxicol. 1999;12:1278–1283. doi: 10.1021/tx9901231. [DOI] [PubMed] [Google Scholar]
- 9.Lin S, Shi Q, Nix FB, Styblo M, Beck MA, Herbin-Davis KM, Hall LM, Simeonsson JB, Thomas DJ. A novel S-adenosyl-L-methionine: Arsenic(III) methyltransferase from rat liver cytosol. J Biol Chem. 2002;277:10795–10803. doi: 10.1074/jbc.M110246200. [DOI] [PubMed] [Google Scholar]
- 10.Walton FS, Waters SB, Jolley SL, LeCluyse EL, Thomas DJ, Styblo M. Selenium compounds modulate the activity of recombinant rat AsIII-methyltransferase and the methylation of arsenite by rat and human hepatocytes. Chem Res Toxicol. 2003;16:261–265. doi: 10.1021/tx025649r. [DOI] [PubMed] [Google Scholar]
- 11.Waters SB, Devesa V, Del Razo LM, Styblo M, Thomas DJ. Endogenous reductants support the catalytic function of recombinant rat cyt19, an arsenic methyltransferase. Chem Res Toxicol. 2004;17:404–409. doi: 10.1021/tx0342161. [DOI] [PubMed] [Google Scholar]
- 12.Suzuki KT, Katagiri A, Sakuma Y, Ogra Y, Ohmichi M. Distributions and chemical forms of arsenic after intravenous administration of dimethylarsinic and monomethylarsonic acids to rats. Toxicol Appl Pharmacol. 2004;198:336–344. doi: 10.1016/j.taap.2003.10.029. [DOI] [PubMed] [Google Scholar]
- 13.Cui X, Kobayashi Y, Hayakawa T, Hirano S. Arsenic speciation in bile and urine following oral and intravenous exposure to inorganic and organic arsenics in rats. Toxicol Sci. 2004;82:478–487. doi: 10.1093/toxsci/kfh265. [DOI] [PubMed] [Google Scholar]
- 14.Li J, Waters SB, Drobna Z, Devesa V, Styblo M, Thomas DJ. Arsenic (+3 oxidation state) methyltransferase and the inorganic arsenic methylation phenotype. Toxicol Appl Pharmacol. 2005;204:164–169. doi: 10.1016/j.taap.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 15.Drobna Z, Waters SB, Walton FS, LeCluyse EL, Thomas DJ, Styblo M. Interindividual variation in the metabolism of arsenic in cultured primary human hepatocytes. Toxicol Appl Pharmacol. 2004;201:166–177. doi: 10.1016/j.taap.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 16.Thomas DJ, Li J, Waters SB, Xing W, Adair BM, Drobna Z, Devesa V, Styblo M. Arsenic (+3 oxidation state) methyltransferase and methylation of arsenicals. Exp Biol Med. 2006 In press. [PMC free article] [PubMed] [Google Scholar]
- 17.Mandal BK, Ogra Y, Suzuki KT. Identification of dimethylarsinous and monomethylarsonous acids in human urine of the arsenic-affected areas in West Bengal, India. Chem Res Toxicol. 2001;14:371–378. doi: 10.1021/tx000246h. [DOI] [PubMed] [Google Scholar]
- 18.Le XC, Ma M, Lu X, Cullen WR, Aposhian V, Zheng B. Determination of monomethylarsonous acid, a key arsenic methylation intermediate, in human urine. Environ Health Perspect. 2000;108:1015–1018. doi: 10.1289/ehp.001081015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Le XC, Lu X, Ma M, Cullen WR, Aposhian HV, Zheng B. Speciation of key arsenic metabolic intermediates in human urine. Anal Chem. 2000;72:5172–5177. doi: 10.1021/ac000527u. [DOI] [PubMed] [Google Scholar]
- 20.Del Razo LM, Styblo M, Cullen WR, Thomas DJ. Determination of trivalent methylated arsenicals in biological matrices. Toxicol Appl Pharmacol. 2001;174:282–293. doi: 10.1006/taap.2001.9226. [DOI] [PubMed] [Google Scholar]
- 21.Valenzuela OL, Borja-Aburto VH, Garcia Vargas GG, Cruz-Gonzalez MB, Garcia-Montalvo EA, Calderon-Aranda ES, Del Razo LM. Urinary trivalent methylated arsenic species in a population chronically exposed to inorganic arsenic. Environ Health Perspect. 2005;113:250–254. doi: 10.1289/ehp.7519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meza MM, Yu L, Rodriguez YY, Guild M, Thompson D, Gandolfi AJ, Klimecki WT. Developmentally restricted genetic determinants of human arsenic metabolism: Association between urinary methylated arsenic and CYT19 polymorphisms in children. Environ Health Perspect. 2005;113:775–781. doi: 10.1289/ehp.7780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wood TC, Salavaggione OE, Mukherjee B, Wang L, Klumpp AF, Thomae BA, Eckloff BW, Schaid DJ, Wieben ED, Weinshilboum RM. Human arsenic methyltransferase (AS3MT) pharmacogenetics: Gene resequencing and functional genomics studies. J Biol Chem. 2006;281:7364–7373. doi: 10.1074/jbc.M512227200. [DOI] [PubMed] [Google Scholar]
- 24.Aposhian HV, Aposhian MM. Arsenic toxicology: Five questions. Chem Res Toxicol. 2006;19:1–15. doi: 10.1021/tx050106d. [DOI] [PubMed] [Google Scholar]
- 25.International Agency for Research on Cancer. IARC Monographs on the Evaluation of Carcinogenic Risk to Humanss—Overall Evaluation of Carcinogenicity: An Update of IARC Monographs 1 to 42. Suppl 7. IARC; Lyon, France: 1987. p. 100. [Google Scholar]
- 26.Reay PF, Asher CJ. Preparation and purification of 74As-labeled arsenate and arsenite for use in biological experiments. Anal Biochem. 1977;78:557–560. doi: 10.1016/0003-2697(77)90117-8. [DOI] [PubMed] [Google Scholar]
- 27.Drobna Z, Waters SB, Devesa V, Harmon AW, Thomas DJ, Styblo M. Metabolism and toxicity of As in human urothelial cells expressing rat arsenic (+3 oxidation state) methyl-transferase. Toxicol Appl Pharmacol. 2005;207:147–159. doi: 10.1016/j.taap.2004.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Styblo M, Yamauchi H, Thomas DJ. Comparative methylation of trivalent and pentavalent arsenicals. Toxicol Appl Pharmacol. 1995;135:172–178. doi: 10.1006/taap.1995.1220. [DOI] [PubMed] [Google Scholar]
- 29.Hammond SM. Dicing and slicing: The core machinery of the RNA interference pathway. FEBS Letter. 2005;579:5822–5829. doi: 10.1016/j.febslet.2005.08.079. [DOI] [PubMed] [Google Scholar]
- 30.Filipowicz W. RNAi: The nuts and bolts of the RISC machine. Cell. 2005;122:17–20. doi: 10.1016/j.cell.2005.06.023. [DOI] [PubMed] [Google Scholar]
- 31.Hutvagner G. Small RNA asymmetry in RNAi: Function in RISC assembly and gene regulation. FEBS Lett. 2005;579:5850–5857. doi: 10.1016/j.febslet.2005.08.071. [DOI] [PubMed] [Google Scholar]
- 32.Dhankher OP, Rosen BP, McKinney EC, Meagher RB. Hyperaccumulation of arsenic in the shoots of Arabidopsis silenced for arsenate reductase (ACR2) Proc Natl Acad Sci USA. 2006;103:5413–5418. doi: 10.1073/pnas.0509770102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goldring CE, Kitteringham NR, Jenkins R, Lovatt CA, Randle LE, Abdullah A, Liu X, Butler PJ, Williams DP, Metcalfe P, Berens C, Hillen W, Foster B, Simpson A, McLellan L, Park BK. Development of a transactivator in hepatoma cells that allows expression of phase I, phase II, and chemical defense genes. Am J Physiol Cell Physiol. 2006;290:104–115. doi: 10.1152/ajpcell.00133.2005. [DOI] [PubMed] [Google Scholar]
- 34.Xu J, Ma M, Purcell WM. Characterization of some cytotoxic endpoints using rat liver and HepG2 spheroids as in vitro models and their application in hepatotoxicity studies. I. Glucose metabolism and enzyme release as cytotoxic markers. Toxicol Appl Pharmacol. 2003;189:100–111. doi: 10.1016/s0041-008x(03)00089-9. [DOI] [PubMed] [Google Scholar]
- 35.Styblo M, Drobna Z, Jaspers I, Lin S, Thomas DJ. The role of biomethylation in toxicity and carcinogenicity of arsenic: A research update. Environ Health Perspect. 2002;110:767–771. doi: 10.1289/ehp.110-1241242. [DOI] [PMC free article] [PubMed] [Google Scholar]


