Abstract
The presenilin proteins PS-1 and PS-2 are crucially involved in Alzheimer disease (AD), but their molecular functions are not known. They are integral membrane proteins, but whether they can be expressed at the surface of cells has been in dispute. Here we show by immunofluorescence experiments, using anti-peptide antibodies specific for either PS-1 or PS-2, that live cultured DAMI cells and differentiated human NT2N neuronal cells are specifically immunolabeled for their endogenous as well as transfected presenilins, although the cells cannot be immunolabeled for their intracellular tubulin, unless they are first fixed and permeabilized. These and other results establish that portions of the presenilins are indeed expressed at the surfaces of these cells. These findings support our previous proposal that the presenilins on the surface of a cell engage in intercellular interactions with the β-amyloid precursor protein on the surface of a neighboring cell, as a critical step in the molecular and cellular mechanisms that lead to AD.
Keywords: β-amyloid precursor protein, ligand-receptor interactions, immunofluorescence
Early onset, or familial, Alzheimer disease (FAD) is thought to be a mutationally induced acceleration of the more common, nonfamilial, form of the disease (AD). Mutations in only three genes account for all known cases of FAD. These genes encode the β-amyloid precursor protein (β-APP) (for a review, see ref. 1) and the two presenilin proteins, PS-1 (2) and PS-2 (3), the latter two closely homologous to one another, but structurally unrelated to β-APP. All three are integral proteins of membranes. The involvement of the gene for β-APP in FAD confirms the evidence (4) that the oligopeptides called β-amyloid (Aβ), the main constituents of the neuritic plaques that are characteristic of AD, are important to the etiology of the disease. Aβ is derived by an as yet unknown proteolytic process from β-APP. While the finding that the gene for β-APP is implicated in FAD was therefore not altogether a surprise, the involvement of the genes for the presenilins was entirely unanticipated. These proteins had previously gone undiscovered, and their physiological functions are unknown. This genetic information, however, strongly suggests that the three proteins, β-APP, PS-1, and PS-2, in their nonmutated forms are all somehow directly involved in the generation of the Aβ that is thought to bring about nonfamilial AD. If this is so, then the question which clearly holds the key to understanding the molecular basis of AD is: by what mechanisms are the three proteins involved?
Some time ago, we published (5) an explicit hypothesis about the functional relationship between β-APP and the presenilins, and how these proteins might together operate to produce the Aβ relevant to AD. We proposed that all three proteins are expressed at, and in part protrude from, the plasma membranes of their respective cells, and further that the exterior-facing domain of β-APP on one cell (say, a neuron) binds specifically to the exposed domain of PS-1, or alternatively, its homologue PS-2, on a neighboring cell. This molecular binding, in which β-APP may be considered the membrane-bound ligand for its specific PS receptor, creates a close adhesion between the two cells (6), which triggers an incorporation of contacting membrane elements into the interior of the β-APP-expressing neuronal cell, in the form of double-membrane-bounded vesicles. (A precedent for this mechanism has been studied in great detail in ref. 7.) Once inside the neuron, these vesicles, containing intact β-APP and PS-1 or PS-2, fuse with secondary lysosomes (7, 8) in which, it is proposed, the β-APP is internalized and broken down by specific proteases to Aβ; the latter is eventually extruded from the neuron to form the neuritic plaques in the AD brain. In other words, we proposed that the production of the Aβ that is relevant to AD only occurs as a consequence of specific cell–cell interactions in the brain that are mediated by the intercellular binding of β-APP either to PS-1 or to PS-2. This would explain why mutant forms of these and only these proteins are involved in FAD.
For this proposal to be even tenable, all three proteins must be expressible at the surfaces of the appropriate cells. To investigate this, we carried out immunofluorescence experiments (9) with cultured cells transfected with the cDNAs of each of the three proteins. The immunolabeling results were entirely consistent with their cell surface expression. However, a number of other laboratories have reported that in their transfected cell culture systems the presenilins were confined to the cell interior, primarily in the endoplasmic reticulum (ER) and in the Golgi apparatus (10–16); only one other report of their cell surface expression (17) has appeared. We therefore have re-investigated this critical matter in greater detail and with additional controls. We report in this paper unequivocal evidence by immunofluorescence experiments of the cell surface expression of PS-1 and PS-2 in two types of human cells in culture, one of which is a differentiated neuronal cell line. This is true for both the endogenously expressed PS-1 and PS-2, as well as for the two proteins expressed in transfected cells.
MATERIALS AND METHODS
Cell Culture and Transfections.
DAMI cells (American Type Culture Collection, CRL 9792) were cultured and transiently transfected with pcDNA3 constructs of full-length PS-1 and PS-2 exactly as described (9). NTera 2/C1-D1 (undifferentiated NT2), a human embryonal carcinoma cell line, was obtained from the American Type Culture Collection (CRL 1973) and differentiated into 99% neuronal cells in the presence of retinoic acid and mitotic inhibitors on matrigel-coated surfaces exactly as described (18).
Antibodies.
Polyclonal anti-peptide antibodies, specific for PS-1 and PS-2 as described (9), were raised in rabbits to the following peptide sequences: PS1, DSHLGPHRST [residues 345–354 (ref. 2, misnumbered in ref. 9)] a sequence in the exoplasmic loop between transmembrane domains 6 and 7 in the seven-transmembrane spanning topography (2); PS-2, ESPTPRSCQEGR [residues 24–35 (3)], a sequence in the N-terminal region.
Immunofluorescent Labeling.
Live, or fixed and permeabilized, transfected and untransfected DAMI cells were first reacted in suspension with one of the following primary antibodies: rabbit polyclonal anti-peptide antisera to PS-1 (1:200 dilution), or the antisera to PS-2 (1:100 dilution), or a mouse monoclonal antibody (Sigma) to α-tubulin (1:500 dilution) in PBS containing 3% BSA for 20 min at room temperature. After washing with PBS three times by centrifugation, the cells were resuspended in 3% BSA in PBS and the appropriate one of the following secondary antibodies were added: either dichlorotriazinylamino fluorescein-conjugated affinity-purified donkey anti-rabbit IgG, or fluorescein isothiocyanate-conjugated affinity-purified donkey anti-mouse IgG (1.5 mg/ml, 1:100; Jackson ImmunoResearch) at room temperature for 15 min. Cells were washed with PBS as described above, and were examined by immunofluorescent microscopy in the presence of 90% glycerol-phenylene diamine (1 mg/ml in 100 mM Tris⋅HCl, pH 8.5).
NT2N cells were grown on matrigel-coated glass coverslips, and immunolabeling reactions were carried out on attached cells. Live, or fixed and permeabilized, untransfected NT2N cells were either indirectly single labeled as for the DAMI cells with the primary antibodies to either PS-1, PS-2, or α-tubulin, followed by the appropriate fluorescein-tagged secondary antibodies; or were indirectly double-labeled with the primary antibodies to one of the presenilin proteins together with the primary antibody to α-tubulin, followed after washing with a mixture of the two fluorescent-labeled secondary antibodies, the dichlorotriazinylamino fluorescein-donkey anti-rabbit IgG and a tetramethylrhodamine B isothiocyanate-conjugated affinity-purified goat anti-mouse IgG (Jackson ImmunoResearch). Antibody dilutions for both primary and secondary antibodies, incubation times, and washing conditions for the attached cells were identical to those used above for DAMI cells labeled in suspension.
Fixation and Permeabilization.
Cells were fixed for 5 min in 3% paraformaldehyde in PBS, followed by permeabilization for 5 min in the presence of 0.05% Nonidet P-40 and 0.05% Triton X-100 in 3% paraformaldehyde.
Fluorescence Microscopy.
Immunofluorescent microscopy was performed using oil immersion with a ×60 objective lens. The slides were viewed using fluorescein isothiocyanate and tetramethylrhodamine B isothiocyanate filters and a Zeiss Photoscope III instrument, or with Nomarski optics.
Trypsin Treatment of Live DAMI Cells.
DAMI cells were detached by sharply striking the culture flasks a few times and cells were incubated in PBS containing 1:40 dilution of trypsin EDTA solution (1:250; Irvine Scientific) at 37° for 15 min. Trypsin was then inactivated by the addition of medium containing horse serum and immunolabeling was carried out as described.
In Vitro Translations of PS-1 and PS-2 mRNA.
pcDNA3 plasmid harboring full-length cDNA for PS-1 or PS-2 was linearized with BamHI, and capped RNA for PS-1 and PS-2 was generated in vitro using T7 RNA polymerase according to the supplier’s instructions (Stratagene), and translated in vitro using rabbit reticulocyte lysate (Promega) according to the manufacturer’s protocols.
Western Blot Hybridization.
Extracts of transfected and untransfected DAMI cells were made in lysis buffer (50 mM Tris⋅HCl, pH 8.0/150 mM NaCl/1 mM phenylmethylsulfonyl fluoride/1 μg/ml antipain/0.1 μg/ml leuptin/0.1 μg/ml pepstatin) by sonication on ice (three bursts of 20 sec each). Proteins (100 μg/lane) from whole cell extracts or from the in vitro translation mixtures were separated on 10% SDS/polyacrylamide gels in the presence of 8 M urea and electrophoretically transferred onto nitrocellulose. Filters were blocked with 5% BSA in PBS and incubated with an appropriate dilution of primary antibodies to either PS-1 or PS-2 for 16 h at room temperature. Filters were then washed once with PBS, once in 0.05% Nonidet P-40 in PBS, and once again with PBS, followed by an incubation for 1.5 h with horseradish peroxidase-conjugated goat anti-rabbit IgG (1:2000). Washing steps were repeated as described, and the filter-bound horseradish peroxidase activity was detected with 4-chloro-1-naphthol.
Oligopeptide Inhibition of Antibody Labeling.
The specific inhibition by excess oligopeptide of either immunofluorescent labeling or of the immunoblotting was carried out for the PS-1 using the soluble oligopeptide–multiple antigen peptide (MAP) complex and for PS-2 using the soluble oligopeptide–keyhole limpet hemocyanin (KLH) conjugate that served as the immunogens for raising the anti-PS-1 and anti-PS-2 antibodies, respectively (Research Genetics). Each conjugate was added to a concentration of 2 mM in the oligopeptide.
RESULTS
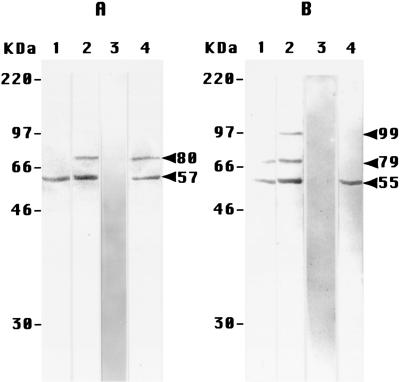
The specificity of the anti-peptide antibodies used in this study is critical to the work, and was examined in immunoblotting experiments (Fig. 1). The antibodies to PS-1 immunoblotted a single band of 57 kDa apparent molecular weight (MA) in the in vitro translation product of PS-1 mRNA (Fig. 1A, lane 1). This MA is close to that expected (53 kDa) for intact PS-1 (2). The same 57-kDa band was immunoblotted in extracts of both PS-1-transfected DAMI cells (Fig. 1A, lane 2) and untransfected DAMI cells (Fig. 1A, lane 4), but was somewhat more intensely labeled in the former. In addition, the antibody blotted a band of MA 80 kDa in both extracts, but did not significantly label any other (and in particular no smaller) components. The immunoblotting of both 57- and 80-kDa bands in the transfected extract (Fig. 1A, lane 3) and the untransfected extract (not shown) was completely inhibited by an excess of the soluble oligopeptide specific for the anti-PS-1 antibodies, suggesting that the larger MA band in the extracts was a modified or aggregated form of PS-1. These results together, therefore, demonstrate that these antibodies were indeed specifically directed to PS-1.
Figure 1.
Immunoblotting experiments on extracts of DAMI cells with the anti-peptide antibodies to PS-1 (A) and PS-2 (B). The apparent MA values on the left of each panel were obtained from Rainbow (Amersham) MA standards. (A) Lanes: 1, the in vitro translation product of PS-1 mRNA, showing only the single band of 57 kDa (arrow, at right of A); 2, PS-1 transfected cells, showing the same 57-kDa band and one additional band at 80 kDa; 3, PS-1 transfected cells (lane 2), immunoblotted in the presence of an excess of the specific soluble PS-1–MAP oligopeptide complex; 4, untransfected cells. (B) Lanes: 1, the in vitro translation product of PS-2 mRNA, showing a major band at 55 kDa, and another band at 79 kDa; 2, PS-2 transfected cells, showing the same two bands in lane 1, plus an additional minor band at 99 kDa; 3, PS-2 transfected cells (lane 2), immunoblotted in the presence of an excess of the soluble specific PS-2 oligopeptide–KLH conjugate; 4, untransfected cells, showing only the one band at 55 kDa.
Parallel results were obtained with the extracts of the DAMI cells labeled with antibodies to PS-2 (Fig. 1B). These antibodies immunoblotted predominantly a single band of MA 55 kDa in the product of the in vitro translation of PS-2 mRNA (Fig. 1B, lane 1), with a slightly smaller MA than for PS-1, as expected (3). With extracts of the transfected cells, the same major band was detected (Fig. 1B, lane 2), along with two minor bands of larger MA, 79 and 99 kDa. The former of these was also observed in the in vitro translation product (Fig. 1B, lane 1). Only the single 55-kDa band was observed in the untransfected extracts (Fig. 1B, lane 4); no lower MA bands were detected in either of the extracts. The immunoblotting of all of these bands (shown for the transfected extracts in Fig. 1B, lane 3) was inhibited by an excess of the soluble oligopeptide to which the anti-PS-2 antibodies were raised. The one or two minor bands of larger MA observed in Fig. 1B therefore probably represent modified or aggregated forms of PS-2. These results establish that the antibodies raised to the PS-2 oligopeptide were indeed specific for PS-2.
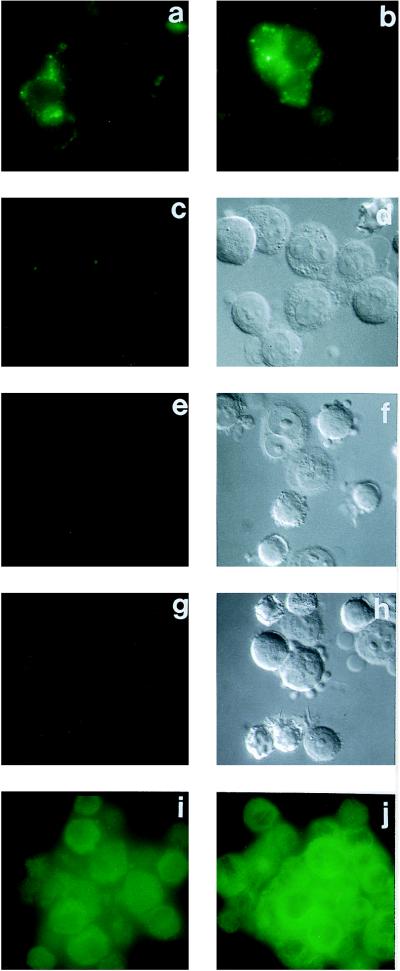
The first set of immunofluorescence experiments reported here involve cultured DAMI cells and labeling with the anti-peptide antibodies specific for an epitope on PS-2 (Fig. 1B). Untransfected live DAMI cells (not shown), and live DAMI cells transfected with the pcDNA3 vector alone (Fig. 2a), uniformly showed a similar distinct but low intensity of labeling. Live DAMI cells that had been transfected with PS-2 showed a large fraction (60–80%) with a significant increase of PS-2 immunolabeling (Fig. 2b) as compared with the untransfected controls (Fig. 2a). This increased surface labeling of the transfected cells was completely inhibited in the presence of an excess of the soluble specific oligopeptide–KLH conjugate to which the anti-PS-2 antibodies were raised (Fig. 2 c and d). To demonstrate unequivocally that the PS-2 immunolabeling of the live transfected cells in Fig. 2b was at the cell surface, the following additional experiments were performed. Treatment of the live transfected cells with trypsin prior to the addition of the antibodies largely eliminated the subsequent immunolabeling (Fig. 2 e and f), signifying that the epitope of PS-2 that was being labeled was on the cell surface because it was susceptible to removal by proteolysis of the intact cell. Furthermore, live transfected cells, such as were immunolabeled for PS-2 (Fig. 2b), uniformly showed no immunolabeling for tubulin (Fig. 2 g and h), although such cells after fixation and permeabilization were, as expected, all intensely immunolabeled for tubulin (Fig. 2j). These experiments establish that the interior of the live transfected DAMI cells was inaccessible to the antibodies to tubulin, and therefore that the immunolabeling observed with the antibodies to PS-2 (Fig. 2b) had to be confined to the cell surface. If the fixed and permeabilized PS-2 transfected DAMI cells were immunolabeled for PS-2 (Fig. 2i), they showed a diffuse intracellular staining that was more intense than, and easily discriminated from, the PS-2 surface labeling of the live cells (Fig. 2b).
Figure 2.
Immunofluorescence microscopic labeling with live, or with fixed and permeabilized, DAMI cells, using as the primary antibody reagent either the anti-PS-2 antibodies of Fig. 1B, or anti-tubulin antibodies in single indirect labeling experiments, with fluorescein-tagged secondary antibodies. (a) Live cells, transfected with the pcDNA3 vector alone, labeled with anti-PS-2. (b) Live cells, PS-2 transfected, labeled with anti-PS-2. (c) Live cells, PS-2 transfected, labeled with anti-PS-2 in the presence of an excess of soluble specific PS-2–KLH oligopeptide conjugate. (d) Nomarski image of c. (e) Live cells, PS-2 transfected, treated with trypsin before anti-PS-2 labeling. (f) Nomarski image of e. (g) Live cells, PS-2 transfected, labeled with anti-tubulin. (h) Nomarski image of g. (i) Fixed and permeabilized cells, PS-2 transfected, labeled with anti-PS-2. (j) Fixed and permeabilized cells, PS-2 transfected, labeled with anti-tubulin.
Closely similar results to those obtained with the antibodies to PS-2 (Fig. 2) were observed if the same kinds of experiments were carried out with the antibodies to PS-1 (not shown, but see ref. 9).
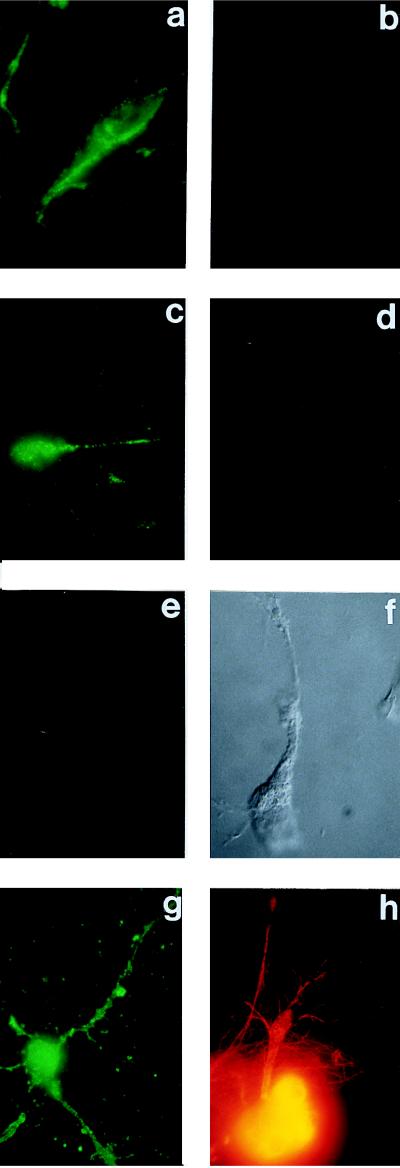
The second set of representative immunofluorescence experiments reported here were carried out with cultured differentiated human NT2N neuronal cells that were labeled with anti-peptide antibodies directed to an epitope on PS-1 (Fig. 1A). In this set only untransfected NT2N cells are shown, their immunolabeling for PS-1 (Fig. 3 a and c) being more intense than that for PS-2 on the live untransfected DAMI cells (Fig. 2a). This immunolabeling was specific for PS-1 because it was completely prevented (Fig. 3 e and f) by an excess of the soluble specific oligopeptide–MAP complex to which the anti-PS-1 antibodies were raised. Furthermore, in double immunofluorescence experiments of the live neurons with the anti-PS-1 antibodies and anti-tubulin antibodies (two examples shown in Fig. 3 a and b and in Fig. 3 c and d), no tubulin immunolabeling was detected on the cells labeled for PS-1, but as expected, if the live cells were first fixed and permeabilized, the immunolabeling for tubulin was intense (Fig. 3h). (Double immunolabeling was performed with the neuronal cells but was not necessary with the DAMI cells because of the comparative fragility of the neurons. It was important, therefore, to show that the same live neuron that was immunolabeled for PS-1 was not labeled for tubulin.) These experiments are unequivocal in demonstrating a cell surface expression of endogenous PS-1 on these live untransfected neurons. If the cells were first fixed and permeabilized, they showed intense intracellular immunolabeling for PS-1 (Fig. 3g). Trypsin experiments with the live neurons, such as were carried out with the DAMI cells (Fig. 2 e and f), were not feasible because trypsin treatment detached the neurons from the substratum.
Figure 3.
Immunofluorescence microscopic labeling with either live, or fixed and permeabilized, untransfected NT2N neuronal cells, using as the primary antibody reagents either the anti-PS-1 antibodies of Fig. 1A, or anti-tubulin antibodies, or both in double-labeling experiments on the same cells. In these latter experiments, the anti-PS-2 labeling used a fluoresceinated secondary antibody, and the anti-tubulin labeling a rhodamine-tagged secondary antibody. (a and b) Live cells, double labeling with anti-PS-1 and anti-tubulin, respectively. (c and d) Another example of a and b, respectively. (e) Live cell, single labeling with anti-PS-1 in the presence of an excess of the soluble specific PS-1–MAP oligopeptide complex. (f) Nomarski image of e. (g) Fixed and permeabilized cell, single labeling with anti-PS-1. (h) Fixed and permeabilized cell, single labeling with anti-tubulin.
DISCUSSION
The results described herein firmly establish that the presenilins are in part expressed at the surfaces of two types of cultured human cells. Two anti-peptide antibodies, one raised to an epitope of PS-1 and the other to one of PS-2, were used in this study. Their specificities were established by immunoblotting experiments of DAMI cell extracts (Fig. 1), which showed that antibody labeling mainly of the appropriate monomer MA bands of the two presenilins occurred, and that this labeling was inhibited by an excess of the soluble oligopeptide to which the antibody was raised. Using these antibodies, live untransfected DAMI cells and live untransfected neurons in culture, as well as live transfected DAMI cells, all impermeable to antibodies to tubulin, were immunofluorescently labeled specifically with either of the antibodies to PS-1 or PS-2. Therefore, the conclusion is that cell surface expression of the presenilins can and does occur. This confirms the results of other experiments described in a previous report (9). Of particular significance to the brain and to AD, we have shown that untransfected neuronal cells have part of their normal endogenous presenilins accessible at the cell surface (Fig. 3).
A number of other studies (10–16), however, have inferred that the presenilins are exclusively intracellular proteins, confined to the membranes of the ER and Golgi apparatus. That the largest amount of a cell surface integral membrane protein would be found in the interior of a steady-state cell is, of course, to be expected. We have observed this with the presenilins as well (Figs. 2i and 3g). This distribution between the cell interior and surface is the consequence of the rates of intracellular transport of such integral membrane proteins from the ER through the Golgi apparatus to the plasma membrane (see ref. 19). To detect the relatively small amount of a membrane protein expressed at a cell surface, it must therefore be experimentally discriminated from the much larger intracellular pool, as we have done in this and the previous (9) paper by immunofluorescence experiments on live cultured cells. For cells within intact tissues, such as the brain (20), cell surface expression of a membrane protein is even more difficult to establish than for cultured cells, and may require high resolution immunoelectron microscopic analysis. In any event, it is not clear from the previous studies cited (10–16) how, or whether, cell surface expression of the presenilins was explicitly ruled out.
On the other hand, it is also possible that the presenilins may in certain cells or particular circumstances be expressed inside a cell but not at the cell surface. This might depend on the presence of auxiliary proteins in the cell type examined (see ref. 21) and possibly be influenced by the particular cDNA constructs used to transfect the presenilins into the cells. The DAMI cells used in our studies were chosen because of their unusual property of not expressing β-APP (9, 22), and the neuronal cells were utilized because of their relevance to the brain. Other investigators have used a variety of other cultured cells and cDNA constructs in their studies.
Another factor in the apparent absence of surface expression of the presenilins in the other studies cited (10–16) is the extensive proteolytic cleavage of the presenilins that was often detected in these studies, but not in ours (Fig. 1). We believe that this proteolytic cleavage is an abnormal event and is not physiologically relevant to AD (see below). In any event, in cells in which this artifactual cleavage did occur, it might have precluded the normal integration of the presenilin proteins into the ER membrane and their subsequent intracellular transport to the cell surface.
For these several reasons, we do not believe that the cell surface expression of the presenilins that we have securely demonstrated in this paper is brought into question by others’ studies purporting to indicate the absolute exclusion of presenilins from cell surfaces.
In a previous paper (5), and as stated in the Introduction, we proposed that β-APP and either PS-1 or PS-2 bind to one another specifically and intercellularly via their extracellular domains that are exposed on their respective cell surfaces; and further, that this intercellular interaction is obligatory to the subsequent formation from β-APP of the Aβ that is the chief component of the neuritic plaques in AD brains. If the presenilins were indeed excluded from cell surfaces, this proposal would be ruled out. Our demonstration of their cell surface expression, therefore, makes our proposal at least tenable.
Further support for our proposal is derived from our previous demonstration (9) that β-APP-transfected DAMI cells formed specific cell aggregates with cells transfected with either PS-1 or PS-2 under conditions where the β-APP transfected cells alone, despite their surface expression of endogenous presenilins, did not produce cell–cell aggregates (figure 1 A and B, and figure 1 K and L of ref. 9). [These aggregation conditions were chosen to take advantage of the greater surface expression of the PS proteins in PS-transfected DAMI cells as compared with untransfected cells (Fig. 2b compared with Fig. 2a in this paper). If the β-APP transfected cells alone were incubated for longer times, they eventually did indeed form aggregates, presumably because of a low level of β-APP–PS-1 or β-APP–PS-2 intercellular binding.] These results showed that β-APP binds specifically to either PS-1 or PS-2, and can do so between two cells expressing these proteins on their respective cell surfaces, as our proposal (5) requires. Using detergent-solubilized proteins and immunoprecipitation techniques, others (23) have recently confirmed our finding that β-APP binds to presenilins, but by the nature of their experiments, they were unable to address the possibility that such binding could occur intercellularly and thereby generate a cell–cell adhesion.
The results in this and the previous (9) paper also bear on the topography of the presenilin molecules in the membrane bilayer. The anti-peptide antibodies used in these studies (9) were raised to the sequence of residues 345–354 (2) of PS-1 and 24–35 (3) of PS-2. The former is within the large intramolecular loop, and the latter within the N-terminal domain, of the respective presenilins. Our immunofluorescence results demonstrate that these two epitopes are exposed at the cell surface. They are, therefore, entirely consistent with the seven-transmembrane spanning topography originally proposed (2, 3) for the presenilins, but are wholly inconsistent with the six- or eight-transmembrane spanning topography proposed by others (12). More extensive and as yet unpublished studies with a battery of eight anti-peptide antibodies raised to different sequences in the PS-1 and PS-2 molecules have indeed confirmed their seven-transmembrane spanning topography.
It should be noted that the intramolecular proteolytic cleavage of the presenilins observed by others in cell and tissue extracts (10–16), referred to earlier, arises in a domain of the molecule that in the seven-transmembrane-spanning topography is exposed at the cell surface. Inside the cell, this domain would therefore normally be expected to face the lumen of the ER and of the Golgi apparatus where it would be inaccessible to cytoplasmic proteases. Indeed, the smaller fragments expected from this proteolysis were not observed in the immunoblots of our untransfected or transfected cell extracts (Fig. 1). An explanation of the cytoplasmic cleavage of the presenilins observed by others could involve their abnormal integration into the ER membrane. Normally, that integration occurs cotranslationally—i.e., the more NH2-terminal parts of the polypeptide chain become inserted, probably in a stepwise fashion (24, 25)—while simultaneously the more C-terminal parts are being translated within the ribosome, until translation is completed and the C terminus is released from the ribosome. If this stepwise integration of the presenilins into the ER membrane is retarded or is incomplete for whatever reasons, regions of the molecules that are normally rapidly cotranslationally translocated into the lumen of the ER may remain exposed in the cytoplasm and be susceptible to proteolytic cleavage (see ref. 26).
Acknowledgments
We thank Mr. Chau Do for his excellent technical assistance. This work was supported by the National Institutes of Health Grant 2RO1-NS27850 to N.N.D.
ABBREVIATIONS
- AD
Alzheimer disease
- FAD
familial AD
- β-APP
β-amyloid precursor protein
- Aβ
β-amyloid
- ER
endoplasmic reticulum
- MAP
multiple antigen peptide
- KLH
keyhole limpet hemocyanin
References
- 1.Selkoe D J. Annu Rev Cell Biol. 1994;10:373–403. doi: 10.1146/annurev.cb.10.110194.002105. [DOI] [PubMed] [Google Scholar]
- 2.Sherrington R, Rogaev E I, Liang Y, Rogaeva E A, Levesque G, et al. Nature (London) 1995;375:754–760. [Google Scholar]
- 3.Levy-Lahad E, Wijsman E M, Nemens E, Anderson L, Goddard K A, Weber J L, Bird T D, Schellenberg G D. Science. 1995;269:970–973. doi: 10.1126/science.7638621. [DOI] [PubMed] [Google Scholar]
- 4.Glenner G, Wong C. Biochem Biophys Res Commun. 1984;120:885–890. doi: 10.1016/s0006-291x(84)80190-4. [DOI] [PubMed] [Google Scholar]
- 5.Dewji N N, Singer S J. Science. 1996;271:159–160. doi: 10.1126/science.271.5246.159. [DOI] [PubMed] [Google Scholar]
- 6.Singer S J. Science. 1992;255:1671–1677. doi: 10.1126/science.1313187. [DOI] [PubMed] [Google Scholar]
- 7.Bailey C H, Chen M, Keller F, Kandel E. Science. 1992;256:645–647. doi: 10.1126/science.1585177. [DOI] [PubMed] [Google Scholar]
- 8.Cagan R L, Kramer H, Hart A C, Zipursky S L. Cell. 1992;69:393–399. doi: 10.1016/0092-8674(92)90442-f. [DOI] [PubMed] [Google Scholar]
- 9.Dewji N N, Singer S J. Proc Natl Acad Sci USA. 1996;93:12575–12580. doi: 10.1073/pnas.93.22.12575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kovacs D M, Fausett H J, Page K J, Kim T-W, Moir R D, Merriam D E, Hollister R D, Hallmark O G, Mancini R, Felsenstein K M, Hyman B T, Tanzi R E, Wasco W. Nat Med. 1996;2:224–227. doi: 10.1038/nm0296-224. [DOI] [PubMed] [Google Scholar]
- 11.Cook D G, Sung J C, Golde T E, Felsenstein K M, Wojczyk B S, Tanzi R E, Trojanowski J W, Lee V M, Doms R W. Proc Natl Acad Sci USA. 1996;93:9223–9228. doi: 10.1073/pnas.93.17.9223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thinakaran G, Borchelt D R, Lee M K, Slunt H H, Spitzer L, Kim G, Ratovitsky T, Davenport F, Norstedt C, Seeger M, Hardy J, Levey A I, Gandy S E, Jenkins N A, Copeland N G, Price D L, Sisodia S S. Neuron. 1996;17:181–190. doi: 10.1016/s0896-6273(00)80291-3. [DOI] [PubMed] [Google Scholar]
- 13.De Strooper B, Beullens M, Contreras B, Levesque L, Craessaerts K, Cordell B, Moechars D, Bollen M, Fraser P, St. George-Hyslop P S, Van Leuvan F. J Biol Chem. 1997;272:3590–3598. doi: 10.1074/jbc.272.6.3590. [DOI] [PubMed] [Google Scholar]
- 14.Kim T-W, Pettingill W H, Hallmark O G, Moir R D, Wasco W, Tanzi R E. J Biol Chem. 1997;272:11006–11010. doi: 10.1074/jbc.272.17.11006. [DOI] [PubMed] [Google Scholar]
- 15.Podlisny M B, Citron M, Amarante P, Sherrington R, Xia W, Zhang J M, Diehl T, Levesque G, Fraser P, Haass C, Koo E H M, Seubert P, St. George-Hyslop P, Teplow D B, Selkoe D J. Neurobiol Dis. 1997;3:325–337. doi: 10.1006/nbdi.1997.0129. [DOI] [PubMed] [Google Scholar]
- 16.Tomita T, Marayama K, Saido T, Hideaki K, Shinozaki K, Tokuhiro S, Capell A, Walter J, Grunberg J, Haass C, Iwatsubo T, Obata K. Proc Natl Acad Sci USA. 1997;94:2025–2030. doi: 10.1073/pnas.94.5.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takashima A, Sato M, Mercken M, Tanaka S, Kondo S, Honda T, Sato K, Maruyama M, Noguchi K, Nakazato Y, Takahashi H. Biochem Biophys Res Commun. 1996;227:423–426. doi: 10.1006/bbrc.1996.1523. [DOI] [PubMed] [Google Scholar]
- 18.Pleasure S, Page C, Lee V M-Y. J Neurosci. 1992;12:1802–1815. doi: 10.1523/JNEUROSCI.12-05-01802.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bergmann J E, Singer S J. J Cell Biol. 1983;97:1777–1787. doi: 10.1083/jcb.97.6.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moussaoui S, Czech C, Pradier L, Blanchard V, Bonici B, Gohin M, Imperato A, Revah F. FEBS Lett. 1996;383:219–222. doi: 10.1016/0014-5793(96)00250-5. [DOI] [PubMed] [Google Scholar]
- 21.Helekar S A, Patrick J. Proc Natl Acad Sci USA. 1997;94:5432–5437. doi: 10.1073/pnas.94.10.5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Querfurth H W, Selkoe D J. Biochemistry. 1994;33:4550–4561. doi: 10.1021/bi00181a016. [DOI] [PubMed] [Google Scholar]
- 23.Weidemann A, Paliga K, Durrwang U, Czech C, Evin G, Masters C L, Beyreuther K. Nat Med. 1997;3:328–332. doi: 10.1038/nm0397-328. [DOI] [PubMed] [Google Scholar]
- 24.Blobel G. Proc Natl Acad Sci USA. 1980;77:1496–1500. doi: 10.1073/pnas.77.3.1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Singer S J. Annu Rev Cell Biol. 1990;6:247–296. doi: 10.1146/annurev.cb.06.110190.001335. [DOI] [PubMed] [Google Scholar]
- 26.Heymann J A W, Subramaniam S. Proc Natl Acad Sci USA. 1997;94:4966–4971. doi: 10.1073/pnas.94.10.4966. [DOI] [PMC free article] [PubMed] [Google Scholar]