Abstract
β-catenin is remarkably multifunctional, acting in adhesion, cytoskeletal regulation, and Wnt signaling. In this issue, Xing et al. (2008) present the full-length structure of β-catenin, providing a clearer picture of how these terminal regions modulate β-catenin activities.
Natural selection prefers to tinker rather than invent, and thus cells often use one protein for multiple purposes. β-catenin is a prototypical example. At the cell surface, β-catenin binds cadherins, mediating cell adhesion and helping organize cortical actin. In the nucleus, a cadherin-independent pool of β-catenin transduces Wnt signals by interacting with T cell factor (TCF)-family transcription factors to activate target genes. Defining the molecular nature of β-catenin/cadherin and β-catenin/TCF complexes is critical because of their basic biological roles and their clinical implications. β-catenin/cadherin has tumor-suppressive activity while β-catenin/TCF can promote onco-genesis. Thus, structural information may facilitate development of strategies to specifically inhibit β-catenin oncogenic activities, while sparing its adhesive ones.
β-catenin presents a fascinating structural problem: how can one protein engage diverse adhesive and signaling ligands using a single binding interface (Figure 1A)? The Xu and Weis labs previously presented several illuminating structures of the central Armadillo (Arm) repeats bound to various ligands (Gooding et al., 2004). However, these structures did not include the N- or C-terminal regions flanking the Arm repeats, known to bind the adhesive/cytoskeletal regulatory partner, α-catenin, as well as several transcriptional regulatory partners. Interest in the β-catenin structure is further increased by data suggesting that different β-catenin isoforms/conformations might act in signaling and adhesion (Brembeck et al., 2004; Gottardi and Gumbiner, 2004), and that terminal regions of β-catenin might fold back on the central Arm repeats, regulating access to certain ligands (Castano et al., 2002; Cox et al., 1999; Gottardi and Gumbiner, 2004).
Figure 1. Partners and Structure of β-Catenin.
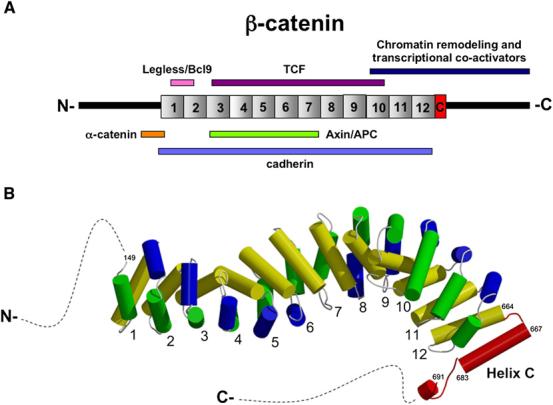
(A) Line diagram of β-catenin arm repeat and terminal regions with key binding partners.
(B) Structure of full-length β-catenin revealing HelixC (red); unstructured N- and C-terminal regions shown as dotted lines. See Xing et al. (2008) for details.
In this issue, the Xu lab presents the structure of a full-length β-catenin (Xing et al., 2008). In comparison with previous structures of β-catenin's central Arm repeats, the full-length structure reveals new helical domains flanking Arm repeats 1 and 12, showing that the central structured region of β-catenin extends beyond the original 12 repeats. The most N- and C-terminal regions remain unstructured (Figure 1B). The revelation of both structured and unstructured regions within the termini has important implications for how these regions modulate β-catenin function.
In the N-terminal region, the first Arm repeat differs from the others in that helix 1 and 2 (of the three helices comprising each repeat) are fused into a single elongated helix with a kink (Graham et al., 2000; Huber and Weis, 2001). By comparing the current and past structures, a significant hinge motion around Arg151 is observed in a region that binds the junctional partner α-catenin, as well as the transcriptional coactivator complex Legless (Bcl9)/Pygopus. This hinge region is just C-terminal to the minimally defined α-catenin binding domain, and N-terminal to Asp162 and Asp164, which are required for Legless/Bcl9 recruitment. These data suggest that α-catenin and/or Legless may affect, or be affected by the dynamic properties of this region. This dynamism fits nicely into a new adhesion model, which suggests that α-catenin exists in a dynamic equilibrium between a pool bound to cadherin/β-catenin and free α-catenin that regulates actin assembly (Drees et al., 2005; Yamada et al., 2005). This new structural information will also be critical for understanding how these regions may work as a “switch” between adhesive and signaling forms of β-catenin. It will be exciting to learn how phosphorylation at nearby residue Tyr142 in β-catenin by src/hepatocyte growth factor receptor (HGFR) kinases might affect this kinked helix, since modification at this site reduces α-catenin binding (Brembeck et al., 2004). Moreover, since phosphorylation at Tyr142 enhances β-catenin/TCF's transcription function (Brembeck et al., 2004), it will be important to learn whether this modification affects recruitment of the Legless/Pygopus coactivator complex to promote transcription, or alters α-catenin's ability to attenuate β-catenin transcription.
At the C terminus, a new structural element (HelixC) “caps” the end of the Arm repeats, packing against repeat 12 to shield hydrophobic residues that would otherwise be exposed. This is particularly satisfying for several reasons. First, it resolves a discrepancy between the original bioinformatics definition of Arm repeats (Peifer et al., 1994), which predicted 13 repeats in β-catenin, and the crystal structure, which only included 12 (Huber et al., 1997). The predicted 13th Arm repeat is now encompassed by HelixC. The structure also explains functional studies demonstrating that sequences just distal to Arm Repeat 12, comprising HelixC, are essential for transcriptional activity. For example, two truncation mutations in Drosophila Armadillo, armXM19 and armH8.6, differ in their Wnt signaling abilities. ArmXM19, truncated almost precisely after the Arm repeats, is null for signaling but fully functional for adhesion. In contrast, armH8.6, which exhibits residual signaling ability, retains the Arm repeats and HelixC, but deletes the rest of the C terminus. Further evidence that HelixC is critical for signaling but not adhesive function comes from analysis of β-catenin evolution. C. elegans evolved distinct β-catenins that separate adhesion and signaling functions: Hmp-2 interacts with cadherin, while wrm-1 and bar-1 bind TCFs and regulate transcription (Korswagen et al., 2000). Strikingly, only these latter two signaling forms of β-catenin retain HelixC (Schneider et al., 2003). This suggests that HelixC is dispensible for cell-cell adhesion, implying therapies targeting it may specifically inhibit Wnt signaling.
The C-terminal region of β-catenin recruits both effectors and inhibitors of transcriptional regulation (Städeli et al., 2006). Affinity-precipitation suggests that many of these components specifically require the terminal Arm repeat region of β-catenin (including HelixC), as few interactors are observed to bind unstructured residues distal to HelixC (Sierra et al., 2006). Xing et al. nicely show that HelixC is required for efficient binding to the signaling inhibitor, Chibby, underscoring that therapeutic approaches targeting HelixC may be useful to inhibit β-catenin's role in transcription. A functionally related inhibitor, ICAT (Inhibitor of β-catenin and TCF), binds the C-terminal Arm repeats and competes with both TCF and p300 binding. By superimposing the β-catenin/ICAT cocrystal with the HelixC structure, Xing et al. infer that ICAT, like Chibby, will make important interactions with HelixC that ultimately antagonize transcriptional activation.
While this study reveals HelixC as the key structural element required for β-catenin-mediated transcription, unstructured sequences distal to HelixC also contribute to signaling (Cox et al., 1999), although how remains poorly defined. Unstructured domains can contribute to functional diversity, and can evolve more quickly due to structural freedom (Romero et al., 2006). Consistent with this, the disordered C-terminal region is conserved among β-catenins within vertebrates or within insects, but diverges between these groups (Peifer and Wieschaus, 1993), suggesting that it mediates lineage-specific interactors. In this regard, a second zebrafish β-catenin was discovered (zβ-cat2) that is 92% identical to the original zβ-cat1 (Bellipanni et al., 2006). Interestingly, loss of zβ-cat2 gives a headless phenotype despite the fact that zβ-cat1 is broadly expressed. Why zβ-cat1 cannot compensate is not yet clear, but the greatest region of dissimilarity covers a 20 amino acid stretch within the unstructured C terminus. Perhaps this diverged region promotes functional specialization.
Some have suggested that the N and C termini can directly engage the Arm repeats of β-catenin and regulate binding to certain ligands (e.g., unphosphorylated E-cadherin; Castano et al., 2002; Cox et al., 1999). However, the termini remain unstructured in the full-length structure, ruling out tight intramolecular interactions. To explore the possibility of transient weak interactions between the termini and Arm-repeats, Xing et al. employed NMR spectroscopy. The β-catenin Arm-repeats did not significantly affect the NMR spectrum of N15-labeled β-catenin C terminus. However, as these experiments are done “in trans,” it remains formally possible that a C terminus tethered “in cis” could compete more effectively. Considering the acidic nature of the terminal regions compared with a positively charged Arm repeat groove, Xing et al. speculate that the terminal regions may effectively “shield” the Arm repeats from nonspecific interactions. Although formal proof for such a model is lacking, β-catenin constructs containing the C terminus exhibit reduced binding affinity to Axin or Adenomatous Polyposis Coli tumor suppression protein (APC; Choi et al., 2006; binding to E-cadherin was not affected). Thus the C terminus may inhibit Arm repeat binding to weak β-catenin binding partners, such as unphosphorylated Axin and APC.
The full-length β-catenin structure reveals new structural boundaries for the central ligand-binding region, and clarifies the unstructured nature of the N and C termini. Given the importance of these regions in β-catenin stabilization, adhesive, and transcriptional activities, coupled with the challenge of learning to inhibit some but not all β-catenin activities, it will be important to build on this study and use new techniques to bring the dynamic nature of these terminal regions of β-catenin into view.
REFERENCES
- Bellipanni G, Varga M, Maegawa S, Imai Y, Kelly C, Myers AP, Chu F, Talbot WS, Weinberg ES. Development. 2006;133:1299–1309. doi: 10.1242/dev.02295. [DOI] [PubMed] [Google Scholar]
- Brembeck FH, Schwarz-Romond T, Bakkers J, Wilhelm S, Hammerschmidt M, Birchmeier W. Genes Dev. 2004;18:2225–2230. doi: 10.1101/gad.317604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castano J, Raurell I, Piedra JA, Miravet S, Dunach M, Garcia de Herreros A. J. Biol. Chem. 2002;277:31541–31550. doi: 10.1074/jbc.M204376200. [DOI] [PubMed] [Google Scholar]
- Choi HJ, Huber AH, Weis WI. J. Biol. Chem. 2006;281:1027–1038. doi: 10.1074/jbc.M511338200. [DOI] [PubMed] [Google Scholar]
- Cox RT, Pai LM, Kirkpatrick C, Stein J, Peifer M. Genetics. 1999;153:319–332. doi: 10.1093/genetics/153.1.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drees F, Pokutta S, Yamada S, Nelson WJ, Weis WI. Cell. 2005;123:903–915. doi: 10.1016/j.cell.2005.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gooding JM, Yap KL, Ikura M. Bioessays. 2004;26:497–511. doi: 10.1002/bies.20033. [DOI] [PubMed] [Google Scholar]
- Gottardi CJ, Gumbiner BM. J. Cell Biol. 2004;167:339–349. doi: 10.1083/jcb.200402153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham TA, Weaver C, Mao F, Kimelman D, Xu W. Cell. 2000;103:885–896. doi: 10.1016/s0092-8674(00)00192-6. [DOI] [PubMed] [Google Scholar]
- Huber AH, Weis WI. Cell. 2001;105:391–402. doi: 10.1016/s0092-8674(01)00330-0. [DOI] [PubMed] [Google Scholar]
- Huber AH, Nelson WJ, Weis WI. Cell. 1997;90:871–882. doi: 10.1016/s0092-8674(00)80352-9. [DOI] [PubMed] [Google Scholar]
- Korswagen HC, Herman MA, Clevers HC. Nature. 2000;406:527–532. doi: 10.1038/35020099. [DOI] [PubMed] [Google Scholar]
- Peifer M, Wieschaus E. J. Mol. Evol. 1993;36:224–233. doi: 10.1007/BF00160477. [DOI] [PubMed] [Google Scholar]
- Peifer M, Berg S, Reynolds AB. Cell. 1994;76:789–791. doi: 10.1016/0092-8674(94)90353-0. [DOI] [PubMed] [Google Scholar]
- Romero PR, Zaidi S, Fang YY, Uversky VN, Radivojac P, Oldfield CJ, Cortese MS, Sick-meier M, LeGall T, Obradovic Z. Proc. Natl. Acad. Sci. USA. 2006;103:8390–8395. doi: 10.1073/pnas.0507916103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider SQ, Finnerty JR, Martindale MQ. J. Exp. Zoolog. B Mol. Dev. Evol. 2003;295:25–44. doi: 10.1002/jez.b.6. [DOI] [PubMed] [Google Scholar]
- Sierra J, Yoshida T, Joazeiro CA, Jones KA. Genes Dev. 2006;20:586–600. doi: 10.1101/gad.1385806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Städeli R, Hoffmans R, Basler K. Curr. Biol. 2006;16:R378–R385. doi: 10.1016/j.cub.2006.04.019. [DOI] [PubMed] [Google Scholar]
- Xing Y, Takemaru K-I, Liu J, Berndt JD, Zheng JJ, Moon RT, Xu W. Structure. 2008;16 doi: 10.1016/j.str.2007.12.021. this issue, ■■■–■■■. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada S, Pokutta S, Drees F, Weis WI, Nelson WJ. Cell. 2005;123:889–901. doi: 10.1016/j.cell.2005.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]


