Abstract
The agr quorum-sensing system of Staphylococcus aureus modulates the expression of virulence factors in response to autoinducing peptides (AIPs). Recent studies have suggested a role for the agr system in S. aureus biofilm development, as agr mutants exhibit a high propensity to form biofilms, and cells dispersing from a biofilm have been observed displaying an active agr system. Here, we report that repression of agr is necessary to form a biofilm and that reactivation of agr in established biofilms through AIP addition or glucose depletion triggers detachment. Inhibitory AIP molecules did not induce detachment and an agr mutant was non-responsive, indicating a dependence on a functional, active agr system for dispersal. Biofilm detachment occurred in multiple S. aureus strains possessing divergent agr systems, suggesting it is a general S. aureus phenomenon. Importantly, detachment also restored sensitivity of the dispersed cells to the antibiotic rifampicin. Proteinase K inhibited biofilm formation and dispersed established biofilms, suggesting agr-mediated detachment occurred in an ica-independent manner. Consistent with a protease-mediated mechanism, increased levels of serine proteases were detected in detaching biofilm effluents, and the serine protease inhibitor PMSF reduced the degree of agr-mediated detachment. Through genetic analysis, a double mutant in the agr-regulated Aur metalloprotease and the SplABCDEF serine proteases displayed minimal extracellular protease activity, improved biofilm formation, and a strongly attenuated detachment phenotype. These findings indicate that induction of the agr system in established S. aureus biofilms detaches cells and demonstrate that the dispersal mechanism requires extracellular protease activity.
Author Summary
A biofilm is a surface-attached community of cells bound together by an extracellular matrix. In a bacterial infection, biofilm-encased cells are protected from antibiotic therapy and host immune response, and these encased cells can develop into a chronic infection. Staphylococcus aureus is a prominent bacterial pathogen known to form biofilms on many medical implants and host tissues. In this report, we demonstrate that repression of the S. aureus quorum-sensing system is required to form a biofilm, and quorum-sensing reactivation in established biofilms disperses the cells. Genetic and molecular analysis demonstrates that quorum-sensing is activated before and required for the detachment mechanism. Detachment is protease-mediated, as established biofilms are sensitive to a non-specific protease and quorum-sensing activation increases the production of extracellular proteases. Using mutations in the protease genes, we show that these secreted enzymes are required for the detachment mechanism. These findings denote that S. aureus quorum-sensing can function as a dispersal mechanism to colonize new sites, and our results suggest this mechanism could be modulated to treat recalcitrant biofilms.
Introduction
Most bacteria have an inherent ability to form surface-attached communities of cells called biofilms [1]. The opportunistic pathogen Staphylococcus aureus can form biofilms on many host tissues and implanted medical devices often causing chronic infections [2]–[5]. The challenge presented by biofilm infections is the remarkable resistance to both host immune responses and available chemotherapies [6],[7], and estimates suggest that as many as 80% of chronic bacterial infections are biofilm associated [8]. In response to certain environmental cues, bacteria living in biofilms are capable of using active mechanisms to leave biofilms and return to the planktonic (free-living) state in which sensitivity to antimicrobials is regained [9]–[11]. Therefore an improved understanding of the molecular mechanism of biofilm detachment could facilitate the discovery of innovative treatment options.
Studies on the opportunistic pathogen Pseudomonas aeruginosa indicate that cell-to-cell communication (often termed “quorum-sensing”) is required to make a robust biofilm under some growth conditions [12]. Surprisingly, the opposite is true in S. aureus, as the presence of an active quorum-sensing impedes attachment and development of a biofilm [13],[14]. The S. aureus quorum-sensing system is encoded by the accessory gene regulator (agr) locus and the communication molecule that it produces and senses is called an autoinducing peptide (AIP), which is an eight-residue peptide with the last five residues constrained in a cyclic thiolactone ring [15]. During growth, AIP is synthesized and secreted through a poorly understood mechanism that requires multiple peptidases [16],[17]. Once AIP reaches a critical concentration, it binds to a surface histidine kinase receptor, initiating a regulatory cascade that controls expression of a myriad of virulence factors, such as proteases, hemolysins, and toxins [18]. A recent study by Yarwood et. al. [19] raised the possibility that the agr quorum-sensing system is involved in biofilm detachment. This study demonstrated that bacteria dispersing from biofilms displayed high levels of agr activity, while cells in a biofilm had predominantly repressed agr systems. These findings correlate well with prior data indicating that agr deficient S. aureus strains form more robust biofilms compared to wild type strains [13],[14].
In the study presented here, we demonstrate that activation of the agr system in established biofilms is necessary for detachment. This activation could be accomplished with exogenous AIP addition or by changing nutrient availability to the biofilm. We also demonstrate that agr-mediated detachment requires the activity of extracellular proteases. Our findings suggest that agr quorum-sensing is an important regulatory switch between planktonic and biofilm lifestyles and may contribute to S. aureus dispersal and colonization of new sites.
Results
Low agr activity is important for biofilm development
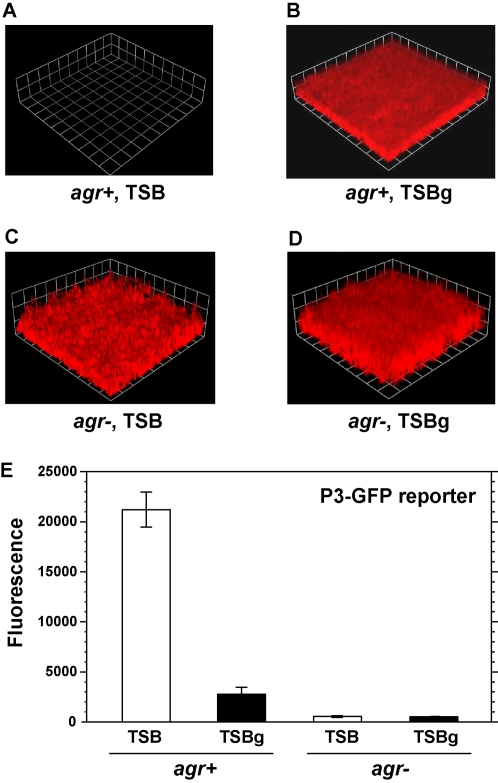
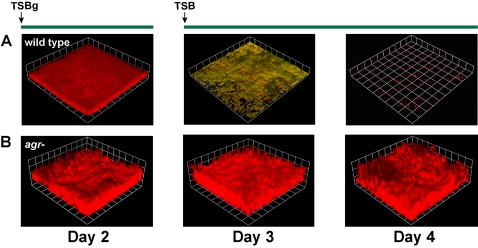
Mutations in the agr quorum-sensing system are known to improve biofilm development [13],[14]. Based on these studies, it seemed probable that there is a correlation between agr activity and biofilm formation. Regassa et al. reported that growth on rich media containing glucose represses the agr system through the nonmaintained generation of low pH [20]. Interestingly, in most published flow cell biofilm studies, one commonality is the use of growth media containing or supplemented with glucose [9], [19], [21]–[24]. In our own efforts to grow S. aureus flow cell biofilms, we found a strict dependence on glucose supplementation. For the experimental setup, a once-through, continuous culture system was employed as previously described [19],[25], and S. aureus SH1000 constitutively expressing red fluorescent protein (PsarA-RFP, plasmid pAH9) was used as the testing strain. Using 2% TSB as the growth media, SH1000 cells did not attach and develop a biofilm (Figure 1A), instead passing right through the flow cell to the effluent. However, in the presence of 0.2% glucose (TSBg), cells attached and a formed a robust biofilm (10–20 microns thick) after two days of growth, which was visually evident and monitored with confocal laser scanning microscopy (CLSM, Figure 1B). As expected, glucose strongly inhibited expression from the P3 promoter using a GFP reporter (Figure 1E), suggesting that repression of RNAIII is essential for attachment and biofilm formation. In broth culture and biofilm effluents, we observed a glucose-dependent pH decrease to the 5.5 range similar as previously reported [20],[26]. As a control, flow cell biofilms were prepared with an agr mutant strain (SH1001, Δagr::TetM) containing plasmid pAH9 (Figure 1C & D), and this strain developed a biofilm even in the absence of media supplementations (Figure 1C). As anticipated, the P3 promoter did not activate in the agr mutant (Figure 1E). Overall, these observations indicate that environmental conditions favoring low agr activity are essential for attachment and biofilm formation.
Figure 1. Low agr activity is important for S. aureus biofilm formation.
(A–D) Biofilms were grown for 2 days in either 2% TSB or 2% TSB supplemented with 0.2% glucose (referred to as “TSBg”). Biofilm integrity and RFP fluorescence were monitored with CLSM. Three dimensional image reconstructions of a z series were created with Velocity software. CLSM images are representative of three separate experiments and each side of a grid square represents 20 µM. (A) AH596 (agr+) grown in TSB. (B) AH596 grown in TSBg. (C) AH871 (agr-) grown in TSB. (D) AH871 grown in TSBg. (E) Measurement of the agr P3-GFP reporter (pDB59) activity in strains AH596 and AH871 grown in broth culture in either TSB or TSBg. Error bars show standard error of the mean (SEM).
AIP detaches S. aureus biofilms
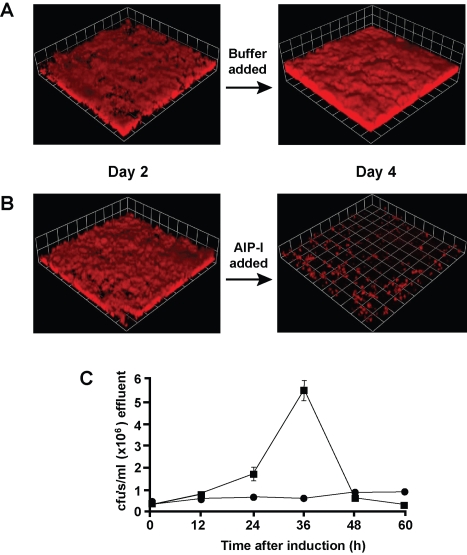
To investigate the role of the agr system in established biofilms, we developed strategies to modulate level of agr activity within a biofilm. Initially, media supplementation experiments were performed using purified AIP signal in order to place the agr system under external control. We recently developed a new method for AIP biosynthesis [27], enabling the production of sufficient signal levels for flow cell experiments. Through exogenous AIP addition, we could test wild-type strains and avoid any potential complications of constructed agr deletion mutants. For this approach, established flow cell biofilms were prepared using S. aureus SH1000 constitutively expressing RFP with plasmid pAH9. The flow cell media was supplemented with glucose to attenuate agr expression [20], allowing cell attachment and biofilm development. After two days, either 1 mL of buffer (100 mM phosphate [pH 7], 50 mM NaCl, 1 mM TCEP; Figure 2A) or 1 mL of 20 µM AIP-I in buffer (Figure 2B and Video S1) was diluted 1000-fold (50 nM final concentration) into the growth media. Using our synthesized AIP-I in dose-response curves [27], we estimate the amount of AIP-I in supernatants of TSB broth cultures (OD600 1.0–1.3) reaches approximately 400 nM (data not shown), indicating the 50 nM level used for the biofilm experiments is within a relevant concentration range. Examination with CLSM showed that the AIP-I treated biofilm sloughed off the flow cell over a period of 1–2 days (Figure 2B and Video S1), suggesting that AIP-I activated a detachment mechanism. To confirm that AIP-I caused detachment, we counted viable S. aureus cells in the effluent media (Figure 2C). The concentration of bacteria in the effluent increased markedly 24–36 hours after AIP-I addition. In contrast, the number of bacteria in the biofilm effluent without AIP-I addition remained relatively constant. Computational analysis of the detachment phenotype indicated that 91.3±4.3% of the biomass dispersed within 48 hrs of AIP-I addition.
Figure 2. Detachment of S. aureus biofilms with AIP.
Biofilms (strain AH500) were grown in flow cells for 2 days. Either (A) 1 mL of buffer (100 mM phosphate [pH 7], 50 mM NaCl, 1 mM TCEP) or (B) 1 mL of 20 µM AIP-I in buffer was diluted 1000-fold into the biofilm growth media. The biofilm integrity was monitored with CLSM for 2 more days. Each side of a grid square in the image reconstructions represents 20 µM. (C) Effect of AIP-I addition on number of detached bacteria in the effluent medium from flow cell biofilms. The plot depicts CFU/ml in effluents from biofilms, and the black squares (▪) represent AIP-I addition and the black circles (•) represent buffer addition to the biofilm. Graph shows the mean of 3 effluent collections from 1 experiment, error bars show SEM.
AIP-mediated biofilm detachment is a general phenomenon
Among S. aureus strains, there are four types of agr quorum-sensing systems. Each of these agr systems, referred to as agr-I through agr-IV, recognizes a unique AIP structure (AIP-I through AIP-IV). Through an intriguing mechanism of chemical communication, these varying quorum-sensing systems can be subdivided into three cross-inhibitory groups: agr-I/IV, agr-II, and agr-III. The activating signals of each group cross-inhibits the alternative signal receptors with surprising potency, a phenomenon termed “bacterial interference” [15]. Since AIP-I and AIP-IV differ by only one amino acid and function interchangeably [28], they are grouped together in the classification scheme, although this assignment has been controversial [29],[30].
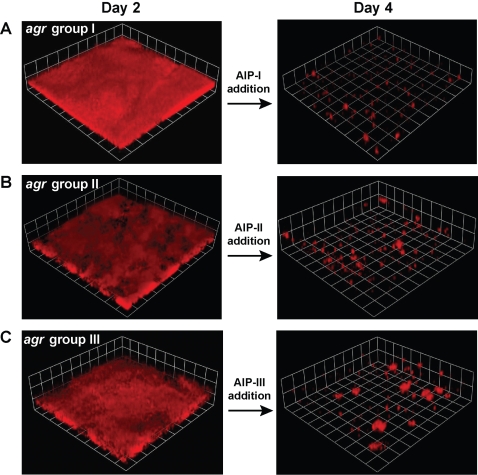
To determine the generality of the detachment mechanism, we examined the effect of AIP addition using S. aureus strains representing different agr groups. The strains tested were (i) FRI1169, agr-I, toxic shock syndrome isolate [31]; (ii) SA502a (ATCC27217), nasal isolate and prototype agr-II strain [15],[32]; and (iii) ATCC25923, clinical agr-III isolate [9]. When the correct AIP signal was added to 2-day old biofilms of each strain (FRI1169, AIP-1; SA502a, AIP-II; ATCC25923, AIP-III), signal addition resulted in robust detachment of each biofilm over a period of 48 hours (Figure 3). These findings indicate biofilm detachment is a general S. aureus phenomenon that occurs in laboratory strains and clinical isolates, and functions across diverse agr systems.
Figure 3. Effect of AIP addition to biofilms from S. aureus strains representing different agr classes.
Biofilms were grown in flow cells for 2 days and indicated AIP was added (50 nM final concentration) to the growth media. Biofilm integrity was monitored with CLSM. Each side of a grid square in the image reconstructions represents 20 µM, and red color is from propidium iodide stain present in growth medium. (A) Biofilm of strain FRI1169 (agr Type I) treated with AIP-I. (B) Biofilm of strain SA502A (agr Type II) treated with AIP-II. (C) Biofilm of strain ATCC25923 (agr Type III) treated with AIP-III.
The timing and requirement of the agr system in detachment
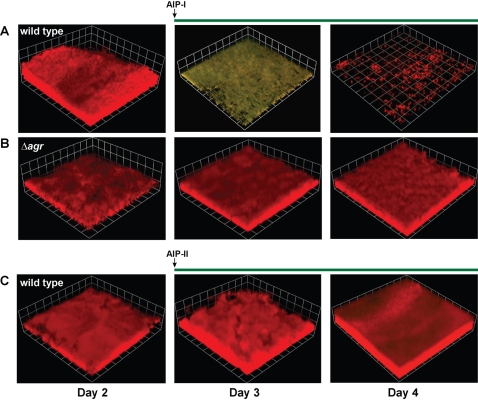
If AIP was promoting biofilm detachment via the agr system, we predicted that agr expression would be induced prior to detachment and an agr deficient mutant would not detach in response to AIP. To determine whether the agr system is activated prior to biofilm detachment, a dual fluorescent-labeled SH1000 strain was constructed with a constitutive RFP (PsarA-RFP, pAH9) and an agr responsive GFP reporter (PagrP3-GFP, pDB59). After two days of biofilm growth, we added AIP-I to the biofilm flow medium and this resulted in strong induction of the GFP reporter (Figure 4A), indicating activation of the agr system. As shown, the GFP reporter was clearly activated before dispersal of the biofilm cells. By the fourth day, all cells with detectable GFP expression detached from the biofilm. These observations provide convincing evidence that AIP activates the agr system prior to biofilm dispersal.
Figure 4. Expression of agr P3 promoter in biofilms after AIP addition.
Dual-labeled biofilms (PsarA-RFP, PagrP3-GFP) were grown for 2 days, and AIP-I (50 nM final) was added to the growth media. Biofilm integrity and RFP/GFP fluorescence were monitored with CLSM at day 3 and 4. Greenish yellow color indicates expression of the agr P3-GFP reporter (pDB59). (A) Addition of AIP-I to an agr type I wild type strain (AH596) or (B) agr deficient strain (AH861). (C) Addition of interfering AIP-II to an agr type-I strain biofilm (AH596). CLSM image reconstructions are representative of three separate experiments and each side of a grid square represents 20 µM.
To further investigate the role of the agr system, we utilized a mutant strain with a complete deletion of the agr locus (SH1001). Unlike the wild type strain (Figure 4A), the agr mutant biofilm harboring the same dual reporters did not respond to AIP-I treatment, as evidenced by a lack of GFP induction, and the mutant biofilm did not disperse (Figure 4B). Similarly, addition of an inhibitory AIP (50 nM AIP-II) to the dual-labeled SH1000 biofilm failed to induce GFP expression, and again, the biofilm did not disperse (Figure 4C). Taken together, these data demonstrate that an active agr quorum-sensing system is necessary for AIP-mediated biofilm dispersal.
Changing environmental conditions can induce detachment
We have demonstrated that low agr activity is important for biofilm formation and that activation of the agr system in established biofilms induces detachment. Considering changes to the physiochemical environment may occur in vivo, we investigated whether an alteration in nutrient availability could reproduce the detachment phenotype. Again, two day flow cell biofilms were prepared with the dual-labeled strain (AH596) in TSBg (Figure 5A). The glucose was removed and significant activation of the P3 promoter was apparent by monitoring GFP levels using CLSM (Figure 5A), supporting our previous result (Figure 1A). Once the agr system was activated, robust detachment from the flow cell was observed and monitored with CLSM (Figure 5A). An agr deletion mutant did not respond to glucose depletion (Figure 5B), indicating the detachment phenotype was dependent upon a functional agr system. These findings demonstrated that glucose depletion can disperse an S. aureus biofilm and again the detachment occurred through an agr-dependent mechanism. These experimental observations mirrored those with AIP addition and further support the apparent inverse correlation between agr activity and biofilm formation.
Figure 5. Effect of changing growth conditions on agr-mediated biofilm detachment.
Dual-labeled biofilms (PsarA-RFP, PagrP3-GFP) of (A) agr positive strain AH596 and (B) agr mutant strain AH871 were grown for 2 days in TSBg. Glucose was removed from the growth media and the biofilm was grown an additional 2 days. Biofilm integrity and RFP/GFP fluorescence were monitored with CLSM. CLSM image reconstructions are representative of three separate experiments and each side of a grid square represents 20 µM.
Detached S. aureus cells regain antibiotic sensitivity
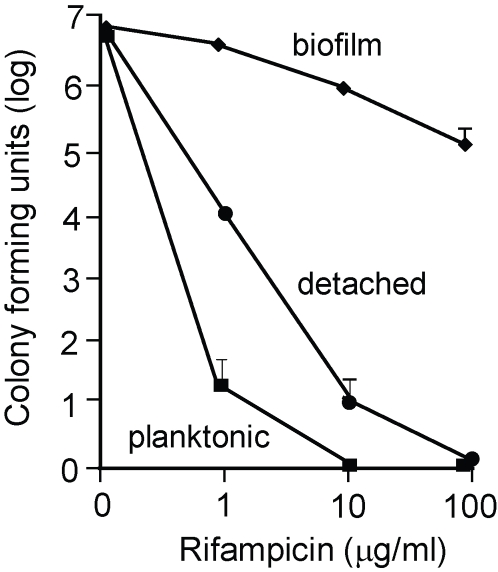
Biofilm growth of S. aureus increases resistance to antimicrobials when compared to the planktonic growth mode [9],[19]. This biofilm mediated resistance hinders treatment of many chronic S. aureus biofilm related infections, including endocarditis, osteomyelitis, and indwelling medical device infections [3],[33]. Therefore, we asked whether AIP-dispersed bacteria regained sensitivity to a clinically relevant antibiotic, rifampicin. To test this, we collected detached cells from an AIP-treated biofilm effluent and compared resistance to intact biofilms exposed to different levels of rifampicin. Similar to previous antibiotic susceptibility results [19], even at the highest concentration tested (100 µg/ml), the level of rifampicin killing was <2 log units of the biofilm biomass (Figure 6). In contrast, the viability of detached cells displayed a different antibiotic response. At 10 µg/ml rifampicin, a 6 log decrease of viable cells was detected, and at 100 µg/ml, complete killing of the detached cells was observed (Figure 6). The AIP-detached cells were more resistant than broth culture to comparable levels of rifampicin, suggesting parts of the detached biofilm may remain in emboli that are known to possess elevated antibiotic resistance [9]. These observations demonstrated that S. aureus cells detached from a biofilm regain susceptibility to a clinical antibiotic.
Figure 6. Susceptibility of biofilm and detached bacteria to rifampicin killing.
S. aureus SH1000 biofilm bacteria (black diamonds) were grown in flow cells containing removable coupons, allowing multiple replicate biofilms to be exposed to rifampicin and surviving CFU's to be determined. Detached bacteria (black circles) were collected from flow cell effluents of biofilms exposed to AIP-I. As a control, planktonic bacteria (black squares) were treated with the same level of rifampicin. Graph show the mean of three experiments; error bars show SEM.
The role of PIA in biofilm detachment
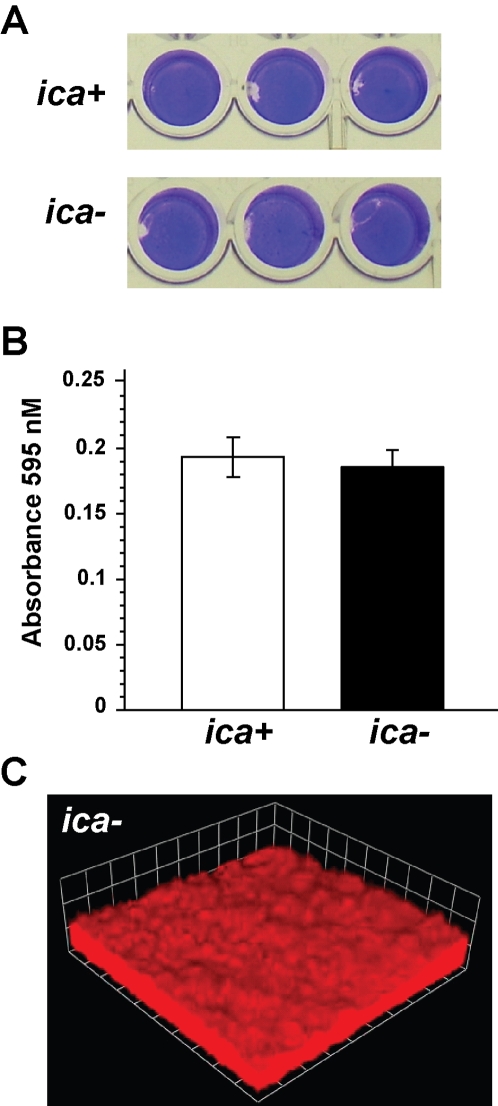
S. aureus possesses the icaRADBC locus that is required to synthesize and generate an exopolysaccharide, which is referred to as the polysaccharide intracellular adhesin or PIA (also called PNAG). S. aureus is known to form biofilms through both ica-dependent and ica-independent mechanisms [34],[35]. To gain insight on the biofilm detachment mechanism, we sought to distinguish whether our S. aureus biofilms were dependent on PIA. In strain SH1000, we constructed an Δica::Tet deletion mutant (strain AH595) using generalized transduction and confirmed the mutation with PCR and sequencing. In microtiter biofilm assays, we were unable to identify a biofilm phenotype (Figure 7A and 7B). Similarly in flow cell biofilms, we did not observe a defect in the ability of strain AH595 to form a biofilm (Figure 7C). No difference was observed compared to flow cell biofilms of SH1000 grown in parallel (data not shown). While SH1000 is a derivative of 8325-4, and there are reports that the ica locus is required for 8325-4 derived strains to make a biofilm [36], the ica locus was not required for biofilm formation under our experimental conditions. Similar to our observations, an ica mutant of the clinical S. aureus isolate UAMS-1 displays no defect in microtiter and flow cell biofilm assays [22]. In contrast, when proteinase K was added to SH1000, biofilms were unable to develop in the microtiter plate format (data not shown), indicating the biofilms are forming through an ica-independent mechanism. These findings suggest that PIA is unlikely to have a role in biofilm detachment in the SH1000 strain background.
Figure 7. Role of ica locus in biofilm development.
(A) Microtiter biofilms of ica positive strain SH1000 and ica deletion mutant AH595. (B) Quantitation of microtiter biofilms. (C) Representative CLSM image of flow cells biofilms of strain AH595 grow in TSBg for 2 days. Each side of a grid square represents 20 µM, and red color is from propidium iodide stain present in growth medium.
Investigating the biofilm detachment mechanism
Knowing the agr system is essential for biofilm detachment, what agr regulated products are responsible for the dispersal phenotype? In S. aureus strains that produce ica-independent biofilms, proteinase K eliminates adherence and biofilm formation [35], [37]–[39], perhaps through cleavage of surface structures. S. aureus is coated with cell wall attached proteins that mediate adherence to a variety of substrates [40], and some of these adhesins, such as biofilm associated protein (BAP) and SasG are important for biofilm formation [41],[42]. It is also known that some surface adhesins, such as protein A and fibronectin-binding protein, are cleaved by the native S. aureus secreted proteases [43],[44]. Considering the agr system regulates the secreted proteases [45],[46], we hypothesized that increased expression of extracellular proteases could be responsible for biofilm detachment.
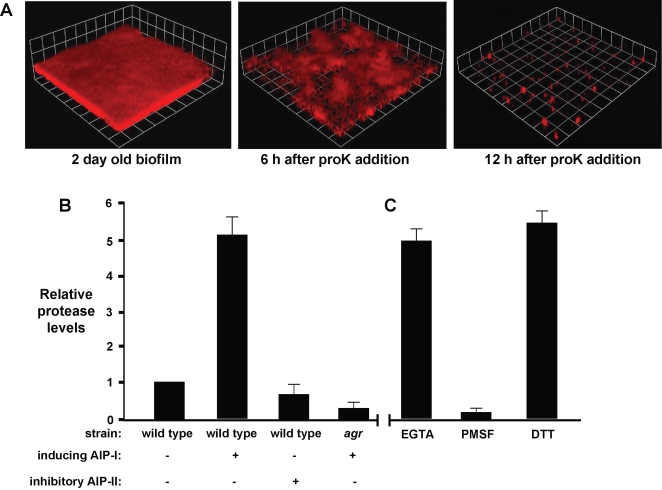
If S. aureus proteases have a role in detachment, proteinase K should be able to disperse an established biofilm. To test this proposal, proteinase K (2 µg/mL) was added to a SH1000 biofilm and resulted in rapid detachment over 12 hrs (Figure 8A). With this preliminary observation, we measured the levels of protease activity in effluents from biofilms with and without AIP-I addition using Azocoll (azo dye impregnated collagen) reagent. As shown in Figure 8B, we detected a baseline level of protease activity in biofilm effluents without AIP-I addition and referenced other measurements to this baseline. With the addition of activating AIP-I, the protease activity increased approximately five-fold compared to a biofilm with no AIP-I treatment. As anticipated, addition of inhibitory AIP-II reduced the level of proteolytic activity in the effluent. Similarly, an agr mutant biofilm supplemented with activating AIP-I displayed very low levels of extracellular proteases (Figure 8B).
Figure 8. Effect of Proteinase K on biofilms and measurement of extracellular protease activity in AIP-detached biofilms.
(A) Proteinase K (proK, 2 µg/ml) was added to a 2 day old biofilm (strain AH500) and the biofilm integrity was monitored with CLSM at 6 and 12 hr. (B) Levels of protease activity detected in biofilm effluent collected from wild type (SH1000) or Δagr (SH1001) biofilms supplemented with indicated AIP's. Protease activity was referenced to wild-type without AIP addition. (C) The effect of inhibitors or activators on the proteolytic activity in an AIP-I detached biofilm effluent. Activity assay was supplemented with either the metalloprotease inhibitor EGTA (1 mM), serine protease inhibitor PMSF (10 µM), or the reducing agent DTT (1 mM). Error bars show SEM.
There are 10 known extracellular proteases produced by most S. aureus strains and expression of all these enzymes is controlled by the agr system [18],[45],[46]. These 10 proteases include the metalloprotease aureolysin (aur), two cysteine proteases (scpA and sspB), and seven serine proteases (sspA (V8) and splABCDEF) [47]. To elucidate what class(es) of proteases are prevalent in AIP-treated biofilms, the effluent from a detaching biofilm was assayed for protease activity in the presence of protease inhibitors or activating agents. The addition of EGTA, an inhibitor of the metalloprotease aureolysin [16], had a negligible effect on overall protease activity (Figure 8C). The addition of PMSF, a potent serine protease inhibitor, however reduced overall protease activity to almost undetectable levels. Lastly, the addition of DTT, a reducing agent used to activate thiol proteases [48], did not significantly change protease activity in the effluents. These results suggest that serine proteases are the dominant, detectable secreted protease in AIP-treated biofilms.
Protease activity is required for biofilm detachment
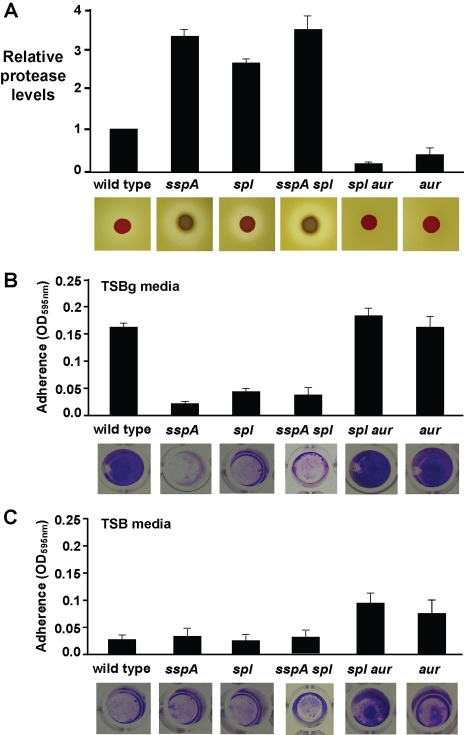
With our observation that serine proteases are abundant in detaching biofilms, we examined the effect of a serine protease inhibitor on AIP-mediated detachment. The addition of 10 µM PMSF in combination with AIP-I to an S. aureus biofilm significantly reduced the level of detachment compared with AIP-I alone (Figure 9A vs. B). However, 48.8% (±5.2) of the biomass still detached indicating that serine proteases are necessary but not sufficient for complete detachment. To further examine the mechanism, knock-out mutations were constructed in the genes encoding the V8 (SspA) and SplABCDEF serine proteases. Surprisingly, sspA::Tet and Δspl::Erm single mutants, and an sspA::Tet Δspl::Erm double mutant, all increased extracellular protease levels (Figure 10A) and eliminated biofilm formation under microtiter plate conditions (Figure 10B & 10C).
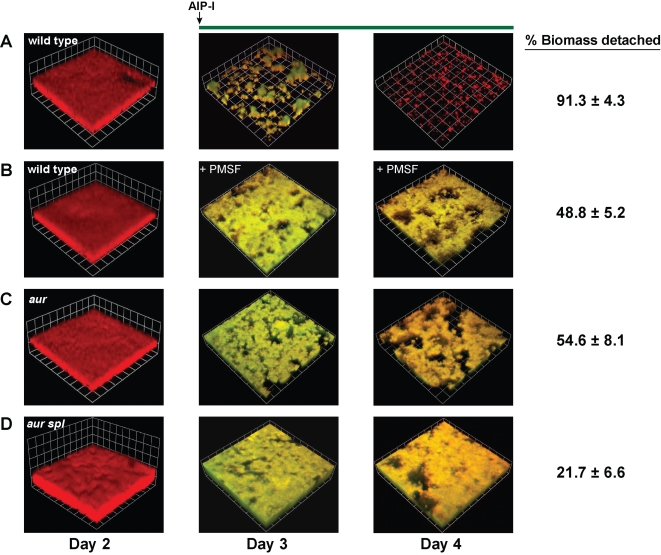
Figure 9. Effect of a serine protease inhibitor and protease deficient mutants on AIP-I mediated biofilm detachment.
Columns show CLSM reconstructions of biofilms at day 2, day 3 and day 4. Biofilms were grown for 2 days and the growth media was supplemented with AIP-I or AIP-I+PMSF as indicated. Greenish yellow color indicates expression of the agr P3-GFP reporter, and the red color is from propidium iodide present in the growth medium. (A) Wild type biofilm (AH462) supplemented with 50 nM AIP-I. (B) Wild type biofilm (AH462) supplemented with 50 nM AIP-I and 10 µM PMSF. (C) Aureolysin (Δaur) mutant biofilm (AH789) supplemented with 50 nM AIP-I. (D) Aureolysin Spl (Δaur Δspl) double mutant biofilm (AH788) supplemented with 50 nM AIP-I. CSLM reconstructions are representative of three separate experiments and each side of a grid square represents 20 µM. Percent biomass detached was calculated by COMSTAT analysis comparing biomass at day 2 to biomass at day 4.
Figure 10. Extracellular protease activity and biofilm formation of protease mutants.
(A) Relative protease levels detected in wild type and protease mutants grown in broth culture. Images show bacterial colonies and zones of clearing caused by protease activity on milk agar plates. (B–C) Biofilm formation of wild type and protease mutants in wells of microtiter plates. Graphs show quantitation of biofilm biomass attached to microtiter plate grown in either (B) TSBg or (C) TSB. Images below each graph are of crystal violet stained biofilms in wells of microtiter plates.
To block other extracellular proteases, a mutation was constructed in the gene encoding aureolysin (Aur). Aur is a metalloprotease that is required to initiate a zymogen activation cascade [49],[50], starting with the V8 protease [51], which in turn activates the SspB cysteine protease [52]. The activation mechanism of the ScpA cysteine protease remains unresolved [49]. In contrast to the serine protease mutations, introduction of the Δaur deletion into S. aureus reduced extracellular protease levels (Figure 10A) and did not affect biofilm formation (Figure 10B). Interestingly, under conditions of high agr activity, the Δaur deletion displayed improved biofilm formation versus wild-type (Figure 10C). In biofilm detachment tests, the Δaur mutant reduced AIP-mediated detachment, but 54.6% (±8.1) of the biomass still detached (Figure 9C). Considering the Spl proteases are not zymogens [53], we examined the combined effects of the Aur cascade and the Spl proteases by constructing an Δaur Δspl::Erm double mutant. The Δaur Δspl strain possessed very low levels of extracellular protease activity (Figure 10A) and had a minor enhancement in biofilm formation (Figure 10B). Similar to the Δaur mutant, the Δaur Δspl double mutant also displayed improved biofilm formation versus wild-type under conditions of high agr activity (Figure 10C). After AIP-I addition, only 21.7% (±6.6) of the Δaur Δspl mutant biomass detached in comparison to 91.3 (±4.3) of the wild-type strain (Figure 9D). These experiments indicate that the extracellular proteases have anti-biofilm properties and they demonstrate that agr-mediated biofilm detachment requires the activity of these proteases.
Discussion
The majority of studies on biofilm detachment have focused on factors capable of initiating the process, such as nutrient availability [54],[55], nitric oxide exposure [56], oxygen tension [57], iron salts [58], chelators [59], and signaling molecules [60]–[63]. Alternatively, detachment studies have addressed effector gene products that contribute to the dissolution of the biofilm, including surfactants [10],[13],[64],[65], hydrolases [66],[67], proteases [37]–[39], and DNase [68]. Here were do both, by demonstrating that the increasing AIP levels or lowering available glucose can function as a S. aureus biofilm detachment signal by activating the agr quorum-sensing system, resulting in increased levels of extracellular proteases needed for the detachment mechanism. Importantly, agr-mediated detachment also restores antibiotic sensitivity to the released bacteria, suggesting the mechanism could be a target for treating biofilm infections.
These results are in accord with previous studies showing that agr mutants have a propensity to form biofilms [13],[14] and that cells actively expressing agr leave biofilms at a high frequency [19]. Our findings also explain why S. aureus biofilm formation requires glucose supplementation to growth media. Unless the agr system is repressed or inactivated, or the enzymes mediating detachment are inhibited, S. aureus will remain in a planktonic state. The presence of glucose is known to represses RNAIII through a nonmaintained pH decrease to ∼5.5 [20], resulting from the secretion of acidic metabolites. The RNAIII repression is not due to glucose itself, but results from the mild acid conditions [26] and can be mimicked with other carbon sources, such as galactose [20], that also lower the media pH. In microtiter biofilm experiments, we found these alternative pH-lowering carbon sources could substitute for glucose in facilitating biofilm formation (data not shown). The molecular mechanism through which low pH inhibits RNAIII expression remains to be determined. In the host, many niches colonized by S. aureus are maintained in lower pH ranges, such as the skin and vaginal tract [26], colonization sites that repress agr function could promote biofilm formation.
Based on our findings, we propose that the S. aureus agr quorum-sensing system controls the switch between planktonic and biofilm lifestyles. When the agr system is repressed, cells have a propensity to attach to surfaces and form biofilms as detachment factors are produced at low levels. In our detachment model, dispersal of cells from an established biofilm requires reactivation of the agr system and occurs through a protease-mediated, ica-independent mechanism. Yarwood et al. demonstrated through time-course, flow cell studies that reactivation of agr does occur in a biofilm [19], presumably through autonomous AIP production that reaches local concentrations high enough to activate agr. Under these fixed conditions, the agr system may function primarily as a mechanism to detach clumps (also called emboli) that seed new colonization sites.
In the experiments presented herein, we have employed growth conditions that tip the balance of the agr system, allowing an investigation into full agr reactivation within an established biofilm. This delicate balance can be offset with an increase in local AIP concentration or through changing environmental conditions, both situations that induce agr and result in massive dispersion of the cells. Biofilms are dynamic and dispersal is always operating [11], but accelerated detachment has been observed in response to changing environmental conditions, such as oxygen levels [57],[69], nutrient depletion [54], changing nutrient composition [55], or increased concentration of quorum-sensing signals [61]. An S. aureus biofilm growing in vivo is likely to encounter a changing physiochemical environment, which could serve as a cue to induce accelerated detachment through an agr-mediated mechanism.
S. aureus has been reported to form biofilms through an ica-dependent mechanism suggesting that PIA could have a role in detachment [34],[36]. We observed no defect in microtiter or flow cell biofilm formation using an ica mutant of SH1000 (Figure 7). Our findings support the growing evidence that PIA is not a major matrix component of S. aureus biofilms, as exogenous addition of dispersin B, an N-acetyl-glucosaminidase capable of degrading PIA, has little effect on established biofilms of SH1000 and other S. aureus strains [70]. In contrast, dispersin B does detach S. epidermidis biofilms indicating a more significant role for PIA in the S. epidermidis matrix structure [70]. Our experiments with proteinase K and the S. aureus proteases indicate that some proteinaceous material is important for SH1000 biofilm integrity, and this result supports a number of recent studies demonstrating that proteases can inhibit biofilm formation or detach established biofilms from many S. aureus strains [35], [37]–[39]. It is not clear whether agr-mediated detachment will function in S. aureus strains that produce an ica-dependent biofilm.
In this study, we document a role for the Aur and Spl proteases in biofilm detachment. Global expression analysis has shown that activation of the agr quorum-sensing system results in up-regulation of extracellular proteases (Aur, SplABCDEF, ScpA, SspAB) and down-regulation of many surface proteins [45],[46]. However, the target of these agr controlled proteases is not clear. One potential target is the surface adhesins, and possible candidates include the surface proteins Atl, Bap, and SasG, all of which have reported roles in biofilm formation [41], [42], [71]–[73]. Atl is additionally known to require proteolytic processing for activation, and this processing is PMSF inhibited [74]. Other possibilities include microbial surface components recognizing adhesive matrix molecules (MSCRAMMs), which are important for adherence to the extracellular matrices of mammalian cells [40]. Also, the S. aureus secreted proteases are known to activate lipase (Sal-1 and Sal-2) precursors [75] and process other secreted enzymes, such as staphylococcal nuclease [76],[77].
In addition to proteases, there may be other agr regulated factors that contribute to biofilm detachment. Surfactant-like molecules, such as δ-toxin, are induced by the agr system and may exert dispersal effects on biofilms [13],[78]. There is growing evidence that extracellular DNA (eDNA) is an important S. aureus biofilm matrix component [24],[70], and expression of staphylococcal nuclease is reported to be under control of the agr system [18]. Thus, while agr induced proteases are required for the detachment phenotype, the agr controlled expression of an array of factors (proteases, nuclease, surfactants) may also contribute to the biofilm detachment mechanism.
There is increasing interest in understanding how bacteria detach from biofilms and initiate colonization of new surfaces. The regulation of quorum-sensing systems may be one mechanism by which many bacteria control biofilm formation and dispersal. Quorum-sensing has been implicated in dispersal of biofilms formed by Yersinia pseudotuberculosis [79], Rhodobacter sphaeroides [80], Pseudomonas aureofaciens [81], Xanthomonas capmestris [62], and Serratia marceascens [61]. However, homoserine lactone signals play a divergent role in Pseudomonas aeuruginosa [12], Pseudomonas fluorescens [82], and Burkholderia cepacia [83], where the active versions of these quorum-sensing systems are necessary for biofilm formation and robustness under some growth conditions. In both cases, it appears quorum-sensing plays a significant role in biofilm development and determining the environmental stimuli that modulate quorum-sensing activity will provide insight on bacterial colonization, detachment, and dispersal to new sites.
Materials and Methods
Strains and growth conditions
The bacterial strains and plasmids used in this study are described in Table 1. S. aureus or Escherichia coli were grown in tryptic soy broth (TSB) or on tryptic soy agar (TSA) with the appropriate antibiotics for plasmid selection or maintenance (erythromycin 10 µg/ml; chloramphenicol 10 ug/ml; tetracycline 5 ug/ml) and incubated at 37°C. Plasmid DNA was prepared from E. coli and transformed by electroporation into S. aureus RN4220 as described [84]. Plasmids were moved from RN4220 into other S. aureus strains by transduction with bacteriophage α80 [85] or by purifying the plasmid DNA and transformed by electroporation into appropriate strains. To move sspA and splABCDEF mutations into appropriate genetic backgrounds, phage transduction with α80 was used as described [85]. To construct the Δaur mutation, the pKOR1-aur plasmid was used as described [16]. Fluorescence measurements with S. aureus strains containing pDB59 were performed as previously described [27].
Table 1. Strain and plasmid list.
| Strain or plasmid | Genotype | Resistance | Source or reference |
| Escherichia coli | |||
| DH5α-E | Cloning strain | None | Invitrogen |
| AH394 | ER2566/ΔgshA::cat | Cam | [27] |
| AH426 | AH394/pDnaB8–AIPI | Amp | [27] |
| AH495 | AH394/pDnaB8–AIPII | Amp | [27] |
| AH496 | AH394/pDnaB8–AIPIII | Amp | [27] |
| Staphylococcus aureus | |||
| RN4220 | Restriction mutant of 8325-4 | None | [85] |
| SH1000 | rsbU positive derivative of 8325-4, agr Type I | None | [89] |
| SH1001 | SH1000/Δagr::tet | Tet | [89] |
| FRI1169 | agr Type I | None | [31] |
| SA502A | agr Type II | None | [15] |
| ATCC25923 | agr Type III | None | ATCC |
| KB600 | Δspl::erm | Erm | [90] |
| SP6391 | sspA::erm | Erm | [50] |
| DU1126 | sspA::tet | Tet | [91] |
| MN8 | Δica::tet | Tet | [92] |
| AH462 | SH1000/pDB59 | Cam | [16] |
| AH500 | SH1000/pAH9 | Erm | This work |
| AH595 | SH1000/Δica::tet | Tet | This work |
| AH596 | SH1000/pDB59+pAH9 | Cam, Erm | This work |
| AH703 | SH1000/Δaur | None | This work |
| AH741 | SH1000/sspA::erm | Erm | This work |
| AH751 | SH1000/Δspl::erm | Erm | This work |
| AH750 | SH1000/Δaur Δspl::erm | Erm | This work |
| AH788 | AH750/pDB59 | Cam, Erm | This work |
| AH789 | AH703/pDB59 | Cam | This work |
| AH860 | SH1000/Δspl::erm sspA::tet | Erm, Tet | This work |
| AH861 | SH1001/pDB59+pAH9 | Cam, Erm | This work |
| Plasmids | |||
| pDB59 | P3-GFP reporter | Amp, Cam | [19] |
| pAH9 | sarA promoter P1-RFP | Amp, Erm | This work |
| pDNAB8-AIPI | AIP-I intein plasmid | Amp | [27] |
| pDNAB8-AIPII | AIP-II intein plasmid | Amp | [27] |
| pDNAB8-AIPIII | AIP-III intein plasmid | Amp | [27] |
| pKOR1-aur | aur knockout vector | Amp, Cam | [16] |
Construction of an RFP reporter plasmid
The sarA P1 promoter region was PCR amplified from SH1000 genomic DNA with oligonucleotides (for 5′-GTTGTTAAGCTTCTGATATTTTTGACTAAACCAAATGC-3′, rev 5′-GTTGGATCCGATGCATCTTGCTCGATACATTTG-3′), digested with HindIII and BamHI, and cloned into the erythromycin shuttle plasmid pCE107 [19]. The mCherry (RFP) gene was PCR amplified from pRSET-mCherry [86] with oligonucleotides incorporating a 5′ ribosome binding site and KpnI site and a 3′ EcoRI site (for 5′-GTTGGTACCTAGGGAGGTTTTAAACATGGTGAGCAAGGGCGAGGAGG-3′, rev 5′-GTTGAATTCTTACTTGTACAGCTCGTCCATGCC-3′). The mCherry fragment was cut with KpnI and EcoRI and cloned downstream of the sarA promoter to generate a constitutive RFP expressing plasmid called pAH9.
Monitoring protease activity
Milk agar plates for detection of protease activity consisted of 3 g/L Tryptic Soy broth, 20 g/L non-fat dry milk, and 15 g/L agar. To determine relative protease activities of strains, assays were performed as described previously using the Azocoll (Calbiochem) reagent [48]. For measuring protease levels in biofilm effluents, 100 mL of effluent was collected on ice (∼12 hours) after AIP addition to the biofilm medium. Cells were removed from the effluents through centrifugation and filtering, and ammonium sulfate was added to 60% over one hour at 4°C to concentrate proteins. The precipitated proteins were pelleted by centrifugation at 19,000 rpm for 30 min, and the pellet was washed and resuspended in 1 ml with 10 mM Tris pH 7.5. For the protease assay, the reaction mixture was supplemented with either 1 mM EGTA, 200 µM PMSF, or 1 mM DTT to gauge relative levels of protease classes.
Biofilm experiments
Microtiter plate biofilms were performed as described [87] except that the plates were incubated at 37°C with shaking at 200 rpm for 12 hours. For flow cell experiments, AIPs were generated using the DnaB intein method, and the AIP concentrations were determined as previously described [27]. AIPs stocks (∼20 µM) were stored in 100 mM phosphate [pH 7], 50 mM NaCl, 1 mM tris(2-carboxyethyl) phosphine (TCEP) and were diluted into the biofilm flow medium to a final concentration of 50 nM. When required, 5 µg/ml of erythromycin and/or chloramphenicol were added to the flow cell media to maintain plasmids. The growth medium for flow cell biofilms consisted of 2% TSB plus 0.2% glucose unless otherwise indicated. Flow cell biofilm experiments and confocal microscopy were performed as previously described [19]. Flow cells were inoculated with overnight cultures diluted 1:100 in sterile water and laminar flow (170 µl/min) was initiated after one hour incubation. Confocal microscopy was performed using a Radiance 2100 system (Biorad) with a Nikon Eclipse E600 microscope. Confocal images were processed using Velocity software (Improvision, Lexington, Mass.). Biofilm biomass was quantified with the COMSTAT program [88] and percent biomass detached was calculated by subtracting biomass present at day 4 from day 2. To quantitate the number of bacteria detaching from a biofilm, 1 ml of flow cell effluent was collected on ice at indicated time points. The collected effluent was vortexed and sonicated in a water bath for 10 minutes to break up clumps, and serial dilutions were plated on TSA plates to determine colony forming units (CFUs). For the Proteinase K detachment experiments, the enzyme (Sigma-Aldrich) was suspended in water and added to the media reservoir at a final concentration of 2 µg/ml.
Antibiotic sensitivity
S. aureus biofilms were grown for two days in a flow chamber lined with removable polycarbonate coupons (Flow Cell FC271, Biosurface Technologies, Bozeman MT). Biofilm effluents were collected on ice ∼24 hours after AIP-I addition. In parallel, coupons with biofilm growth were removed from flow cells not exposed to AIP-I. Both detached bacteria and the biofilms were exposed to the indicated levels of rifampicin for six hours. Subsequently, cells were vortexed, and the coupons were sonicated in a water bath to break up the biofilm or cell clumps. Serial dilutions were plated on TSA to determine surviving CFU's.
Supporting Information
CLSM time course of AIP mediated detachment.
(0.95 MB AVI)
Acknowledgments
We thank K. Bayles, M. Smeltzer, S. Foster, D. Bartels, and G. O'Toole for providing plasmids and strains. We thank M. Thoendel for performing the AIP concentration measurements.
Footnotes
The authors have declared that no competing interests exist.
B. R. Boles was supported by NIH Training Grant No. T32 AI07511. This work was supported by a Cystic Fibrosis Foundation pilot grant and a Roy J. Carver Charitable Trust medical research initiative grant.
References
- 1.Davey ME, O'Toole GA. Microbial biofilms: from ecology to molecular genetics. Microbiol Mol Biol Rev. 2000;64:847–867. doi: 10.1128/mmbr.64.4.847-867.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Furukawa S, Kuchma SL, O'Toole GA. Keeping their options open: acute versus persistent infections. J Bacteriol. 2006;188:1211–1217. doi: 10.1128/JB.188.4.1211-1217.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parsek MR, Singh PK. Bacterial biofilms: an emerging link to disease pathogenesis. Annu Rev Microbiol. 2003;57:677–701. doi: 10.1146/annurev.micro.57.030502.090720. [DOI] [PubMed] [Google Scholar]
- 4.Harris LG, Richards RG. Staphylococci and implant surfaces: a review. Injury. 2006;37(Suppl 2):S3–14. doi: 10.1016/j.injury.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 5.Costerton JW. Biofilm theory can guide the treatment of device-related orthopaedic infections. Clin Orthop Relat Res. 2005:7–11. doi: 10.1097/00003086-200508000-00003. [DOI] [PubMed] [Google Scholar]
- 6.Patel R. Biofilms and antimicrobial resistance. Clin Orthop Relat Res. 2005:41–47. doi: 10.1097/01.blo.0000175714.68624.74. [DOI] [PubMed] [Google Scholar]
- 7.Leid JG, Shirtliff ME, Costerton JW, Stoodley AP. Human leukocytes adhere to, penetrate, and respond to Staphylococcus aureus biofilms. Infect Immun. 2002;70:6339–6345. doi: 10.1128/IAI.70.11.6339-6345.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davies D. Understanding biofilm resistance to antibacterial agents. Nat Rev Drug Discov. 2003;2:114–122. doi: 10.1038/nrd1008. [DOI] [PubMed] [Google Scholar]
- 9.Fux CA, Wilson S, Stoodley P. Detachment characteristics and oxacillin resistance of Staphyloccocus aureus biofilm emboli in an in vitro catheter infection model. J Bacteriol. 2004;186:4486–4491. doi: 10.1128/JB.186.14.4486-4491.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boles BR, Thoendel M, Singh PK. Rhamnolipids mediate detachment of Pseudomonas aeruginosa from biofilms. Mol Microbiol. 2005;57:1210–1223. doi: 10.1111/j.1365-2958.2005.04743.x. [DOI] [PubMed] [Google Scholar]
- 11.Hall-Stoodley L, Stoodley P. Biofilm formation and dispersal and the transmission of human pathogens. Trends Microbiol. 2005;13:7–10. doi: 10.1016/j.tim.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 12.Davies DG, Parsek MR, Pearson JP, Iglewski BH, Costerton JW, et al. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science. 1998;280:295–298. doi: 10.1126/science.280.5361.295. [DOI] [PubMed] [Google Scholar]
- 13.Vuong C, Saenz HL, Gotz F, Otto M. Impact of the agr quorum-sensing system on adherence to polystyrene in Staphylococcus aureus. J Infect Dis. 2000;182:1688–1693. doi: 10.1086/317606. [DOI] [PubMed] [Google Scholar]
- 14.Beenken KE, Blevins JS, Smeltzer MS. Mutation of sarA in Staphylococcus aureus limits biofilm formation. Infect Immun. 2003;71:4206–4211. doi: 10.1128/IAI.71.7.4206-4211.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ji G, Beavis R, Novick RP. Bacterial interference caused by autoinducing peptide variants. Science. 1997;276:2027–2030. doi: 10.1126/science.276.5321.2027. [DOI] [PubMed] [Google Scholar]
- 16.Kavanaugh JS, Thoendel M, Horswill AR. A role for type I signal peptidase in Staphylococcus aureus quorum-sensing. Mol Microbiol. 2007;65:780–798. doi: 10.1111/j.1365-2958.2007.05830.x. [DOI] [PubMed] [Google Scholar]
- 17.Qiu R, Pei W, Zhang L, Lin J, Ji G. Identification of the putative staphylococcal AgrB catalytic residues involving the proteolytic cleavage of AgrD to generate autoinducing peptide. J Biol Chem. 2005;280:16695–16704. doi: 10.1074/jbc.M411372200. [DOI] [PubMed] [Google Scholar]
- 18.Novick RP. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol Microbiol. 2003;48:1429–1449. doi: 10.1046/j.1365-2958.2003.03526.x. [DOI] [PubMed] [Google Scholar]
- 19.Yarwood JM, Bartels DJ, Volper EM, Greenberg EP. Quorum sensing in Staphylococcus aureus biofilms. J Bacteriol. 2004;186:1838–1850. doi: 10.1128/JB.186.6.1838-1850.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Regassa LB, Novick RP, Betley MJ. Glucose and nonmaintained pH decrease expression of the accessory gene regulator (agr) in Staphylococcus aureus. Infect Immun. 1992;60:3381–3388. doi: 10.1128/iai.60.8.3381-3388.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rupp CJ, Fux CA, Stoodley P. Viscoelasticity of Staphylococcus aureus biofilms in response to fluid shear allows resistance to detachment and facilitates rolling migration. Appl Environ Microbiol. 2005;71:2175–2178. doi: 10.1128/AEM.71.4.2175-2178.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beenken KE, Dunman PM, McAleese F, Macapagal D, Murphy E, et al. Global gene expression in Staphylococcus aureus biofilms. J Bacteriol. 2004;186:4665–4684. doi: 10.1128/JB.186.14.4665-4684.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caiazza NC, O'Toole GA. Alpha-toxin is required for biofilm formation by Staphylococcus aureus. J Bacteriol. 2003;185:3214–3217. doi: 10.1128/JB.185.10.3214-3217.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rice KC, Mann EE, Endres JL, Weiss EC, Cassat JE, et al. The cidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus. Proc Natl Acad Sci U S A. 2007;104:8113–8118. doi: 10.1073/pnas.0610226104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davies DG, Chakrabarty AM, Geesey GG. Exopolysaccharide production in biofilms: substratum activation of alginate gene expression by Pseudomonas aeruginosa. Appl Environ Microbiol. 1993;59:1181–1186. doi: 10.1128/aem.59.4.1181-1186.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weinrick B, Dunman PM, McAleese F, Murphy E, Projan SJ, et al. Effect of mild acid on gene expression in Staphylococcus aureus. J Bacteriol. 2004;186:8407–8423. doi: 10.1128/JB.186.24.8407-8423.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malone CL, Boles BR, Horswill AR. Biosynthesis of Staphylococcus aureus autoinducing peptides using the Synechocystis DnaB mini-intein. Appl Environ Microbiol In Press. 2007 doi: 10.1128/AEM.00912-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jarraud S, Lyon GJ, Figueiredo AM, Gerard L, Vandenesch F, et al. Exfoliatin-producing strains define a fourth agr specificity group in Staphylococcus aureus. J Bacteriol. 2000;182:6517–6522. doi: 10.1128/jb.182.22.6517-6522.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goerke C, Kummel M, Dietz K, Wolz C. Evaluation of intraspecies interference due to agr polymorphism in Staphylococcus aureus during infection and colonization. J Infect Dis. 2003;188:250–256. doi: 10.1086/376450. [DOI] [PubMed] [Google Scholar]
- 30.McDowell P, Affas Z, Reynolds C, Holden MT, Wood SJ, et al. Structure, activity and evolution of the group I thiolactone peptide quorum-sensing system of Staphylococcus aureus. Mol Microbiol. 2001;41:503–512. doi: 10.1046/j.1365-2958.2001.02539.x. [DOI] [PubMed] [Google Scholar]
- 31.Sloane R, de Azavedo JC, Arbuthnott JP, Hartigan PJ, Kreiswirth B, et al. A toxic shock syndrome toxin mutant of Staphylococcus aureus isolated by allelic replacement lacks virulence in a rabbit uterine model. FEMS Microbiol Lett. 1991;62:239–244. doi: 10.1016/0378-1097(91)90164-6. [DOI] [PubMed] [Google Scholar]
- 32.Shinefield HR, Ribble JC, Boris M, Eichenwald HF. Bacterial interference: its effect on nursery-acquired infection with Staphylococcus aureus. I. Preliminary observations on artificial colonzation of newborns. Am J Dis Child. 1963;105:646–654. [PubMed] [Google Scholar]
- 33.Costerton W, Veeh R, Shirtliff M, Pasmore M, Post C, et al. The application of biofilm science to the study and control of chronic bacterial infections. J Clin Invest. 2003;112:1466–1477. doi: 10.1172/JCI20365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O'Gara JP. ica and beyond: biofilm mechanisms and regulation in Staphylococcus epidermidis and Staphylococcus aureus. FEMS Microbiol Lett. 2007;270:179–188. doi: 10.1111/j.1574-6968.2007.00688.x. [DOI] [PubMed] [Google Scholar]
- 35.Toledo-Arana A, Merino N, Vergara-Irigaray M, Debarbouille M, Penades JR, et al. Staphylococcus aureus develops an alternative, ica-independent biofilm in the absence of the arlRS two-component system. J Bacteriol. 2005;187:5318–5329. doi: 10.1128/JB.187.15.5318-5329.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cramton SE, Gerke C, Schnell NF, Nichols WW, Gotz F. The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect Immun. 1999;67:5427–5433. doi: 10.1128/iai.67.10.5427-5433.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chaignon P, Sadovskaya I, Ragunah C, Ramasubbu N, Kaplan JB, et al. Susceptibility of staphylococcal biofilms to enzymatic treatments depends on their chemical composition. Appl Microbiol Biotechnol. 2007;75:125–132. doi: 10.1007/s00253-006-0790-y. [DOI] [PubMed] [Google Scholar]
- 38.O'Neill E, Pozzi C, Houston P, Smyth D, Humphreys H, et al. Association between methicillin susceptibility and biofilm regulation in Staphylococcus aureus isolates from device-related infections. J Clin Microbiol. 2007;45:1379–1388. doi: 10.1128/JCM.02280-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rohde H, Burandt EC, Siemssen N, Frommelt L, Burdelski C, et al. Polysaccharide intercellular adhesin or protein factors in biofilm accumulation of Staphylococcus epidermidis and Staphylococcus aureus isolated from prosthetic hip and knee joint infections. Biomaterials. 2007;28:1711–1720. doi: 10.1016/j.biomaterials.2006.11.046. [DOI] [PubMed] [Google Scholar]
- 40.Clarke SR, Foster SJ. Surface adhesins of Staphylococcus aureus. Adv Microb Physiol. 2006;51:187–224. doi: 10.1016/S0065-2911(06)51004-5. [DOI] [PubMed] [Google Scholar]
- 41.Corrigan RM, Rigby D, Handley P, Foster TJ. The role of Staphylococcus aureus surface protein SasG in adherence and biofilm formation. Microbiology. 2007;153:2435–2446. doi: 10.1099/mic.0.2007/006676-0. [DOI] [PubMed] [Google Scholar]
- 42.Trotonda MP, Manna AC, Cheung AL, Lasa I, Penades JR. SarA positively controls bap-dependent biofilm formation in Staphylococcus aureus. J Bacteriol. 2005;187:5790–5798. doi: 10.1128/JB.187.16.5790-5798.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Karlsson A, Saravia-Otten P, Tegmark K, Morfeldt E, Arvidson S. Decreased amounts of cell wall-associated protein A and fibronectin-binding proteins in Staphylococcus aureus sarA mutants due to up-regulation of extracellular proteases. Infect Immun. 2001;69:4742–4748. doi: 10.1128/IAI.69.8.4742-4748.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McGavin MJ, Zahradka C, Rice K, Scott JE. Modification of the Staphylococcus aureus fibronectin binding phenotype by V8 protease. Infect Immun. 1997;65:2621–2628. doi: 10.1128/iai.65.7.2621-2628.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dunman PM, Murphy E, Haney S, Palacios D, Tucker-Kellogg G, et al. Transcription profiling-based identification of Staphylococcus aureus genes regulated by the agr and/or sarA loci. J Bacteriol. 2001;183:7341–7353. doi: 10.1128/JB.183.24.7341-7353.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ziebandt AK, Becher D, Ohlsen K, Hacker J, Hecker M, et al. The influence of agr and sigmaB in growth phase dependent regulation of virulence factors in Staphylococcus aureus. Proteomics. 2004;4:3034–3047. doi: 10.1002/pmic.200400937. [DOI] [PubMed] [Google Scholar]
- 47.Dubin G. Extracellular proteases of Staphylococcus spp. Biol Chem. 2002;383:1075–1086. doi: 10.1515/BC.2002.116. [DOI] [PubMed] [Google Scholar]
- 48.Fournier B, Klier A, Rapoport G. The two-component system ArlS-ArlR is a regulator of virulence gene expression in Staphylococcus aureus. Mol Microbiol. 2001;41:247–261. doi: 10.1046/j.1365-2958.2001.02515.x. [DOI] [PubMed] [Google Scholar]
- 49.Shaw L, Golonka E, Potempa J, Foster SJ. The role and regulation of the extracellular proteases of Staphylococcus aureus. Microbiology. 2004;150:217–228. doi: 10.1099/mic.0.26634-0. [DOI] [PubMed] [Google Scholar]
- 50.Rice K, Peralta R, Bast D, de Azavedo J, McGavin MJ. Description of staphylococcus serine protease (ssp) operon in Staphylococcus aureus and nonpolar inactivation of sspA-encoded serine protease. Infect Immun. 2001;69:159–169. doi: 10.1128/IAI.69.1.159-169.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Drapeau GR. Role of metalloprotease in activation of the precursor of staphylococcal protease. J Bacteriol. 1978;136:607–613. doi: 10.1128/jb.136.2.607-613.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Massimi I, Park E, Rice K, Muller-Esterl W, Sauder D, et al. Identification of a novel maturation mechanism and restricted substrate specificity for the SspB cysteine protease of Staphylococcus aureus. J Biol Chem. 2002;277:41770–41777. doi: 10.1074/jbc.M207162200. [DOI] [PubMed] [Google Scholar]
- 53.Popowicz GM, Dubin G, Stec-Niemczyk J, Czarny A, Dubin A, et al. Functional and structural characterization of Spl proteases from Staphylococcus aureus. J Mol Biol. 2006;358:270–279. doi: 10.1016/j.jmb.2006.01.098. [DOI] [PubMed] [Google Scholar]
- 54.Hunt SM, Werner EM, Huang B, Hamilton MA, Stewart PS. Hypothesis for the role of nutrient starvation in biofilm detachment. Appl Environ Microbiol. 2004;70:7418–7425. doi: 10.1128/AEM.70.12.7418-7425.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sauer K, Cullen MC, Rickard AH, Zeef LA, Davies DG, et al. Characterization of nutrient-induced dispersion in Pseudomonas aeruginosa PAO1 biofilm. J Bacteriol. 2004;186:7312–7326. doi: 10.1128/JB.186.21.7312-7326.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barraud N, Hassett DJ, Hwang SH, Rice SA, Kjelleberg S, et al. Involvement of nitric oxide in biofilm dispersal of Pseudomonas aeruginosa. J Bacteriol. 2006;188:7344–7353. doi: 10.1128/JB.00779-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thormann KM, Saville RM, Shukla S, Spormann AM. Induction of rapid detachment in Shewanella oneidensis MR-1 biofilms. J Bacteriol. 2005;187:1014–1021. doi: 10.1128/JB.187.3.1014-1021.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Musk DJ, Banko DA, Hergenrother PJ. Iron salts perturb biofilm formation and disrupt existing biofilms of Pseudomonas aeruginosa. Chem Biol. 2005;12:789–796. doi: 10.1016/j.chembiol.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 59.Banin E, Brady KM, Greenberg EP. Chelator-induced dispersal and killing of Pseudomonas aeruginosa cells in a biofilm. Appl Environ Microbiol. 2006;72:2064–2069. doi: 10.1128/AEM.72.3.2064-2069.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Morgan R, Kohn S, Hwang SH, Hassett DJ, Sauer K. BdlA, a chemotaxis regulator essential for biofilm dispersion in Pseudomonas aeruginosa. J Bacteriol. 2006;188:7335–7343. doi: 10.1128/JB.00599-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rice SA, Koh KS, Queck SY, Labbate M, Lam KW, et al. Biofilm formation and sloughing in Serratia marcescens are controlled by quorum sensing and nutrient cues. J Bacteriol. 2005;187:3477–3485. doi: 10.1128/JB.187.10.3477-3485.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dow JM, Crossman L, Findlay K, He YQ, Feng JX, et al. Biofilm dispersal in Xanthomonas campestris is controlled by cell-cell signaling and is required for full virulence to plants. Proc Natl Acad Sci U S A. 2003;100:10995–11000. doi: 10.1073/pnas.1833360100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thormann KM, Duttler S, Saville RM, Hyodo M, Shukla S, et al. Control of formation and cellular detachment from Shewanella oneidensis MR-1 biofilms by cyclic di-GMP. J Bacteriol. 2006;188:2681–2691. doi: 10.1128/JB.188.7.2681-2691.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Irie Y, O'Toole GA, Yuk MH. Pseudomonas aeruginosa rhamnolipids disperse Bordetella bronchiseptica biofilms. FEMS Microbiol Lett. 2005;250:237–243. doi: 10.1016/j.femsle.2005.07.012. [DOI] [PubMed] [Google Scholar]
- 65.Davey ME, Caiazza NC, O'Toole GA. Rhamnolipid surfactant production affects biofilm architecture in Pseudomonas aeruginosa PAO1. J Bacteriol. 2003;185:1027–1036. doi: 10.1128/JB.185.3.1027-1036.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kaplan JB, Velliyagounder K, Ragunath C, Rohde H, Mack D, et al. Genes involved in the synthesis and degradation of matrix polysaccharide in Actinobacillus actinomycetemcomitans and Actinobacillus pleuropneumoniae biofilms. J Bacteriol. 2004;186:8213–8220. doi: 10.1128/JB.186.24.8213-8220.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kaplan JB, Ragunath C, Ramasubbu N, Fine DH. Detachment of Actinobacillus actinomycetemcomitans biofilm cells by an endogenous beta-hexosaminidase activity. J Bacteriol. 2003;185:4693–4698. doi: 10.1128/JB.185.16.4693-4698.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. Extracellular DNA required for bacterial biofilm formation. Science. 2002;295:1487. doi: 10.1126/science.295.5559.1487. [DOI] [PubMed] [Google Scholar]
- 69.Applegate DH, Bryers JD. Effects of carbon and oxygen limitations and calcium concentrations on biofilm removal processes. Biotechnol Bioeng. 1991;37:15–25. doi: 10.1002/bit.260370105. [DOI] [PubMed] [Google Scholar]
- 70.Izano EA, Amarante MA, Kher WB, Kaplan JB. Differential roles of poly-N-acetylglucosamine surface polysaccharide and extracellular DNA in Staphylococcus aureus and Staphylococcus epidermidis biofilms. Appl Environ Microbiol. 2008;74:470–476. doi: 10.1128/AEM.02073-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cucarella C, Solano C, Valle J, Amorena B, Lasa I, et al. Bap, a Staphylococcus aureus surface protein involved in biofilm formation. J Bacteriol. 2001;183:2888–2896. doi: 10.1128/JB.183.9.2888-2896.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Biswas R, Voggu L, Simon UK, Hentschel P, Thumm G, et al. Activity of the major staphylococcal autolysin Atl. FEMS Microbiol Lett. 2006;259:260–268. doi: 10.1111/j.1574-6968.2006.00281.x. [DOI] [PubMed] [Google Scholar]
- 73.Heilmann C, Hussain M, Peters G, Gotz F. Evidence for autolysin-mediated primary attachment of Staphylococcus epidermidis to a polystyrene surface. Mol Microbiol. 1997;24:1013–1024. doi: 10.1046/j.1365-2958.1997.4101774.x. [DOI] [PubMed] [Google Scholar]
- 74.Oshida T, Sugai M, Komatsuzawa H, Hong YM, Suginaka H, et al. A Staphylococcus aureus autolysin that has an N-acetylmuramoyl-L-alanine amidase domain and an endo-beta-N-acetylglucosaminidase domain: cloning, sequence analysis, and characterization. Proc Natl Acad Sci U S A. 1995;92:285–289. doi: 10.1073/pnas.92.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gotz F, Verheij HM, Rosenstein R. Staphylococcal lipases: molecular characterisation, secretion, and processing. Chem Phys Lipids. 1998;93:15–25. doi: 10.1016/s0009-3084(98)00025-5. [DOI] [PubMed] [Google Scholar]
- 76.Suciu D, Inouye M. The 19-residue pro-peptide of staphylococcal nuclease has a profound secretion-enhancing ability in Escherichia coli. Mol Microbiol. 1996;21:181–195. doi: 10.1046/j.1365-2958.1996.6211341.x. [DOI] [PubMed] [Google Scholar]
- 77.Davis A, Moore IB, Parker DS, Taniuchi H. Nuclease B. A possible precursor of nuclease A, an extracellular nuclease of Staphylococcus aureus. J Biol Chem. 1977;252:6544–6553. [PubMed] [Google Scholar]
- 78.Kong KF, Vuong C, Otto M. Staphylococcus quorum sensing in biofilm formation and infection. Int J Med Microbiol. 2006;296:133–139. doi: 10.1016/j.ijmm.2006.01.042. [DOI] [PubMed] [Google Scholar]
- 79.Atkinson S, Throup JP, Stewart GS, Williams P. A hierarchical quorum-sensing system in Yersinia pseudotuberculosis is involved in the regulation of motility and clumping. Mol Microbiol. 1999;33:1267–1277. doi: 10.1046/j.1365-2958.1999.01578.x. [DOI] [PubMed] [Google Scholar]
- 80.Puskas A, Greenberg EP, Kaplan S, Schaefer AL. A quorum-sensing system in the free-living photosynthetic bacterium Rhodobacter sphaeroides. J Bacteriol. 1997;179:7530–7537. doi: 10.1128/jb.179.23.7530-7537.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang Z, Pierson LS., 3rd A second quorum-sensing system regulates cell surface properties but not phenazine antibiotic production in Pseudomonas aureofaciens. Appl Environ Microbiol. 2001;67:4305–4315. doi: 10.1128/AEM.67.9.4305-4315.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Allison DG, Ruiz B, SanJose C, Jaspe A, Gilbert P. Extracellular products as mediators of the formation and detachment of Pseudomonas fluorescens biofilms. FEMS Microbiol Lett. 1998;167:179–184. doi: 10.1111/j.1574-6968.1998.tb13225.x. [DOI] [PubMed] [Google Scholar]
- 83.Huber B, Riedel K, Hentzer M, Heydorn A, Gotschlich A, et al. The cep quorum-sensing system of Burkholderia cepacia H111 controls biofilm formation and swarming motility. Microbiology. 2001;147:2517–2528. doi: 10.1099/00221287-147-9-2517. [DOI] [PubMed] [Google Scholar]
- 84.Schenk S, Laddaga RA. Improved method for electroporation of Staphylococcus aureus. FEMS Microbiol Lett. 1992;73:133–138. doi: 10.1016/0378-1097(92)90596-g. [DOI] [PubMed] [Google Scholar]
- 85.Novick RP. Genetic systems in staphylococci. Methods Enzymol. 1991;204:587–636. doi: 10.1016/0076-6879(91)04029-n. [DOI] [PubMed] [Google Scholar]
- 86.Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, et al. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 87.Shanks RM, Donegan NP, Graber ML, Buckingham SE, Zegans ME, et al. Heparin stimulates Staphylococcus aureus biofilm formation. Infect Immun. 2005;73:4596–4606. doi: 10.1128/IAI.73.8.4596-4606.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Heydorn A, Nielsen AT, Hentzer M, Sternberg C, Givskov M, et al. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology. 2000;146 (Pt 10):2395–2407. doi: 10.1099/00221287-146-10-2395. [DOI] [PubMed] [Google Scholar]
- 89.Horsburgh MJ, Aish JL, White IJ, Shaw L, Lithgow JK, et al. sigmaB modulates virulence determinant expression and stress resistance: characterization of a functional rsbU strain derived from Staphylococcus aureus 8325-4. J Bacteriol. 2002;184:5457–5467. doi: 10.1128/JB.184.19.5457-5467.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Reed SB, Wesson CA, Liou LE, Trumble WR, Schlievert PM, et al. Molecular characterization of a novel Staphylococcus aureus serine protease operon. Infect Immun. 2001;69:1521–1527. doi: 10.1128/IAI.69.3.1521-1527.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Blevins JS, Beenken KE, Elasri MO, Hurlburt BK, Smeltzer MS. Strain-dependent differences in the regulatory roles of sarA and agr in Staphylococcus aureus. Infect Immun. 2002;70:470–480. doi: 10.1128/IAI.70.2.470-480.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Maira-Litran T, Kropec A, Abeygunawardana C, Joyce J, Mark G, 3rd, et al. Immunochemical properties of the staphylococcal poly-N-acetylglucosamine surface polysaccharide. Infect Immun. 2002;70:4433–4440. doi: 10.1128/IAI.70.8.4433-4440.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
CLSM time course of AIP mediated detachment.
(0.95 MB AVI)