Abstract
Cyclin dependent kinase 5 (cdk5) has been implicated in Alzheimer's disease (AD) pathogenesis. Here we demonstrate that overexpression of p25, an activator of cdk5, led to increased levels of BACE1 mRNA and protein in vitro and in vivo. A p25/cdk5 responsive region containing multiple sites for Signal Transducer and Activator of Transcription (STAT1/3) was identified in the BACE1 promoter. STAT3 interacts with the BACE1 promoter, and p25 overexpressing mice had elevated levels of pSTAT3 and BACE1, whereas cdk5 deficient mice had reduced levels. Furthermore, mice with a targeted mutation in the STAT3 cdk5 responsive site had lower levels of BACE1. Increased BACE levels in p25 overexpressing mice correlated with enhanced amyloidogenic processing that could be reversed by a cdk5 inhibitor. These data demonstrate a novel pathway by which p25/cdk5 increases the amyloidogenic processing of APP through STAT3-mediated transcriptional control of BACE1 that could have implications for AD pathogenesis.
Introduction
Cdk5 is a proline directed serine/threonine kinase that is activated by association with its regulators p35 or p39, or their corresponding cleaved C-terminal fragments p25 or p29 respectively (Lee et al., 2000; Tsai et al., 1994). Several proteins with diverse functions have been identified as cdk5 substrates, including tau (Baumann et al., 1993; Kobayashi et al., 1993), β-catenin (Kesavapany et al., 2001), Nudel (Sasaki et al., 2000), FAK (Xie et al., 2003), Synapsin 1 (Matsubara et al., 1996), ErbB (Fu et al., 2001), Retinoblastoma protein (Hamdane et al., 2005), MEF2 (Gong et al., 2003) and STAT3 (Fu et al., 2004). Most of these substrates are active in the central nervous system (CNS) and have been implicated in the regulation of physiological activities, such as microtubule and actin dynamics, transportation, cell adhesion, axon guidance, secretion and neuronal migration (reviewed in Dhavan and Tsai, 2001). Cdk5 activation has also been demonstrated in non-neuronal tissues with a role in myogenesis, haematopoietic cell differentiation, spermatogenesis and insulin secretion (Cruz and Tsai, 2004).
AD is the most common form of dementia in the elderly. The disease is characterized neuropathologically by the accumulation and deposition of amyloid β–peptide (Aβ) into neuritic plaques, and by the formation of intracellular neurofibrillary tangles (NFT) containing hyperphosphorylated tau. Kinases such as cdk5 and Glycogen Synthase Kinase-3 (GSK-3) significantly enhance tau hyperphosphorylation (Maccioni et al., 2001; Planel et al., 2002) and amyloid-β precursor protein (APP) processing in vivo (Phiel et al., 2003; Cruz et al., 2006; Rockenstein et al., 2007), and they may be important therapeutic targets for AD treatment or prevention.
Plaque-associated Aβ is generated following cleavage of APP by β-site APP Cleaving Enzyme 1 (BACE1) (Vassar et al., 1999) and β-secretase (De Strooper et al., 1998; Edbauer et al., 2003). BACE1 appears to be the only β-secretase responsible for Aβ generation as BACE1 deficient mice do not generate Aβ (Luo et al., 2001). BACE1 undergoes glycosylation, endosomal targeting and activation by pro-peptide cleavage as it matures (Capell et al., 2000; Huse et al., 2000) and several pathways impact the level of BACE1 in the brain including phosphorylation at Ser498 by Casein Kinase 1 (Walter et al., 2001), lysosomal targeting (Koh et al., 2005) and ubiquitin-mediated degradation (Qing et al., 2004). BACE1 is responsive to various physiological and pathological situations including ischemia (Wen et al., 2004), hypoxia (Sun et al., 2006), cytokines (Hong et al., 2003), oxidative stress (Tamagno et al., 2005) and cholesterol content (Ghribi, 2006). The level and activity of BACE1 protein is increased in AD patient brains (Fukumoto et al., 2002; Stockley et al., 2006), possibly due to elevation of BACE around plaques (Zhao et al., 2007).
The promoter of the BACE1 gene has been characterized (Christensen et al., 2004; Sambamurti et al., 2004) and specific regulatory domains have been located by deletion analysis (Ge et al., 2004). The promoter has characteristics common to both constitutive and inducible expression, and contains both negative and positive domains, separated from the transcription seat by a long, neutral domain (Ge et al., 2004). In addition, putative transcription factor sites such as those for SP1 (Christensen et al., 2004) and STAT6 (Sambamurti et al., 2004) have been identified. An active SP1 site is over 1kb upstream of the +1 transcription start (TSS), indicating the possibility of other active, distal sites of gene regulation (Ge et al., 2004). Notably, it has been determined that the BACE1 promoter is differentially regulated according to cell type (Lahiri et al., 2006), and that its regulation differs from other members of the BACE family, such as BACE2 (Maloney et al., 2006).
In the current study, we investigated the role of p25/cdk5 in the regulation of BACE1, and the generation of Aβ. We demonstrate that p25 over-expression in mice leads to increased cdk5 activity that correlates with increased BACE1 and Aβ levels. Conversely, BACE1 and Aβ levels were reduced following administration of a cdk5 inhibitor. The identification of a functional, p25/cdk5 responsive element in the promoter of the BACE1 gene indicates that BACE1 can be regulated by cdk5 through transcriptional control, with STAT3 being a likely mediator. We therefore propose a novel, signaling pathway by which BACE1 is regulated in response to cdk5 activity in vivo. These findings identify one mechanism by which aberrant p25/cdk5 activity could enhance Aβ accumulation and suggest that inhibitors of cdk5 are candidates for therapeutic development.
Results
Elevated p25 correlates with BACE1 accumulation and enhanced APP processing in vitro
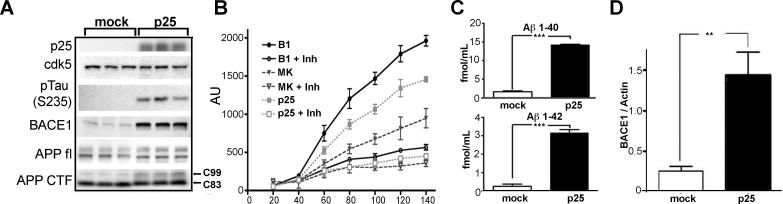
To examine the effect of p25 on BACE and APP metabolism, SH-SY5Y cells stably transfected with a tetracycline-inducible p25 construct (p25), or empty vector (mock) (Hamdane et al., 2003) (figure 1A), were assessed. Both mock and p25 transfected cells had high levels of cdk5, but lacked p35 protein. The level of BACE1 protein was greatly enhanced in induced p25 cells. BACE1 cleaves APP to generate a secreted APPβ fragment and a membrane bound carboxy-terminal fragment (C99, β-CTF). β-cleavage of APP was significantly enhanced as shown by accumulation of the β-CTF in the induced p25 cells. Compared with the mock cells, the levels of full-length APP (APP fl) and α-CTF (C83) were unchanged although a slight shift in mobility was seen for both in the p25 induced cells.
Figure 1. p25 over-expression correlates with increased APP processing, BACE1 mRNA, protein and activity, in human neuroblastoma SH-SY5Y cells.
Panel A: Representative immunoblots from mock and p25 stably transfected SH-SY5Y cells after induction. The panel shows p25, cdk5, pTau(S235) (a cdk5 substrate), BACE1 (∼70kDa), full length APP (APP fl∼110 kDa) and APP-CTFs including α-CTF (C83), and β-CTF (C99). Brain lysate from a BACE1 knock-out mouse was run alongside to confirm signal specificity (not shown). Data are shown in triplicate. Panel B: FRET assay for BACE1 activity in lysates from mock (MK) and p25 stably transfected (p25) SH-SY5Y cells. Controls for assay validation included a BACE1 transfected HEK-293 cell line (B1). All samples were assayed with, or without EMD 565788, a specific BACE1 inhibitor (Inh). The signal from p25 cells was significantly higher than similarly treated mock cells at all time points after 60 minutes (p<0.01). Panel C: ELISA results for Aβ40 and 42 from 18h-conditioned medium from mock transfected and induced p25 cells. Panel D: The level of BACE1 mRNA from mock and p25 SH-SY5Y cells was quantified by qPCR using a 3′ probe. Data are shown following normalization to actin mRNA levels. Similar results were obtained using the 5′ probe (data not shown). All results were analyzed with Student's t-test, *p<0.05 ** p<0.01 *** p<0.001.
To assess whether BACE1 activity was enhanced, a FRET based activity assay (figure 1B) was performed. As a positive control, BACE1 over-expressing HEK293 cells (B1) were included. Each sample was treated with a specific BACE1 inhibitor (EMD 565788) (Stachel et al., 2004). All samples showed similar residual activity, presumably due to the activity of non-specific, contaminating acidic proteases. Our data showed that BACE1 activity was significantly enhanced in both the positive control and the p25 transfected cells. To assess whether increased BACE1 activity resulted in increased Aβ production, endogenous Aβ40 and 42 levels were measured in conditioned media from the cultured cells (figure 1C). The levels of both Aβ40 and 42 were significantly increased (p<0.001) in media from induced p25 cells. These data demonstrate that endogenous BACE1 in p25 transfected cells was active and had enhanced activity towards its endogenous physiological substrate, APP.
To examine why BACE1 accumulates in response to elevated p25/cdk5 activity, we assessed the level of BACE1 mRNA and protein in p25 transfected cells. Quantitative real-time PCR (qPCR) showed that the level of BACE1 mRNA from p25 transfected SH-SY5Y cells was ∼6 fold higher than mock cells after normalization to actin mRNA (figure 1D). Different probes recognizing 5′ or 3′ BACE1 mRNA showed a similar fold of induction (data not shown).
Identification of cdk5 responsive sites in the BACE1 promoter
To examine whether the increase in BACE1 mRNA was due to an effect on BACE1 synthesis through up-regulation of its promoter activity, two sets of promoter constructs were examined. The first was a stable transfected, rat pheochromocytoma (PC12) cell line expressing 3.2 kb of the BACE1 promoter regulating the expression of the reporter gene, firefly luciferase (figure 2A). Levels of reporter protein were assessed in cells transfected with p25, cdk5 or empty vector. Renilla luciferase, or GFP control plasmids were cotransfected to normalize for transfection efficiency. As expected, transfection of cdk5 alone without activator had no significant effect on the regulation of the BACE promoter activity. Transfection with p25 led to 1.7-fold increase in reporter gene transcription, compared with vector alone. To identify which region of the promoter was responding to p25/cdk5, two deletion constructs, BACE1P6 (−1056/+364, +1 being the transcription start site) and BACE1P8 (−327/+364), containing portions of the BACE1 promoter regulating expression of the reporter CAT were transiently co-transfected with p25-GFP, or empty vector into N2a cells (figure 2B). After normalization, the level of CAT generated from the BACE1P6 construct was ∼2 fold higher in p25-transfected cells than in mock transfected cells. Levels of CAT from the BACE1P8 construct were not significantly different from mock transfected cells and the difference between these two constructs suggested that regions present in BACE1P6, but not BACE1P8 were responsive to p25 leading to increased activity of the BACE1 promoter (figure 2B). Mapping of the responsive region on the promoter revealed numerous potential transcriptional regulation sites for STAT1/3 and MEF2 (figure 2C).
Figure 2. p25 over-expression enhanced BACE1 promoter transcription activity.
Panel A: Schematic of the 3.2kb BACE1 promoter/luciferase fusion clone, indicating the position of +1 transcription start site. PC12 cells, stably transfected with the BACE1-pGL4.14 construct, were transiently transfected with a p25-GFP expression construct, a cdk5 expression construct, or mock vector. P25-GFP over-expression increased activity of the BACE1 promoter as determined by firefly luciferase activity. All cells were co-transfected with pRL-SV40 renilla luciferase control vector. Cdk5 over-expression without activator did not significantly affect activity of the BACE1 promoter. Over-expression of p25-GFP and cdk5 did not result in significant toxicity as determined by LDH assay (data not shown). Panel B: Schematic of the BACE1P6 and BACE1P8 CAT fusion clones, indicating the position of +1 transcription start site. Normalized CAT levels in N2a cells transiently transfected with p25/GFP or mock vector, and co-transfected with promoter constructs P6/CAT or P8/CAT. Levels of CAT were normalized to co-transfected GFP levels, which were similar among all groups. Data show n=3 wells per cell group. Transfections and assays were repeated in triplicate with essentially similar results. Panel C: An illustration of the BACE1 promoter region and 5′–UTR from −1056 to +364. Positive and neutral non–induced functional elements are indicated. The positions of the BACE1P6 and BACE1P8 inserts are indicated, as are the locations of putative STAT1/3/6 and MEF2 transcription factor binding sites. Note the absence of all putative sites in the BACE1P8 sequence. ** p<0.01.
Confirmation of the role of STAT3 in the regulation of BACE1 transcription
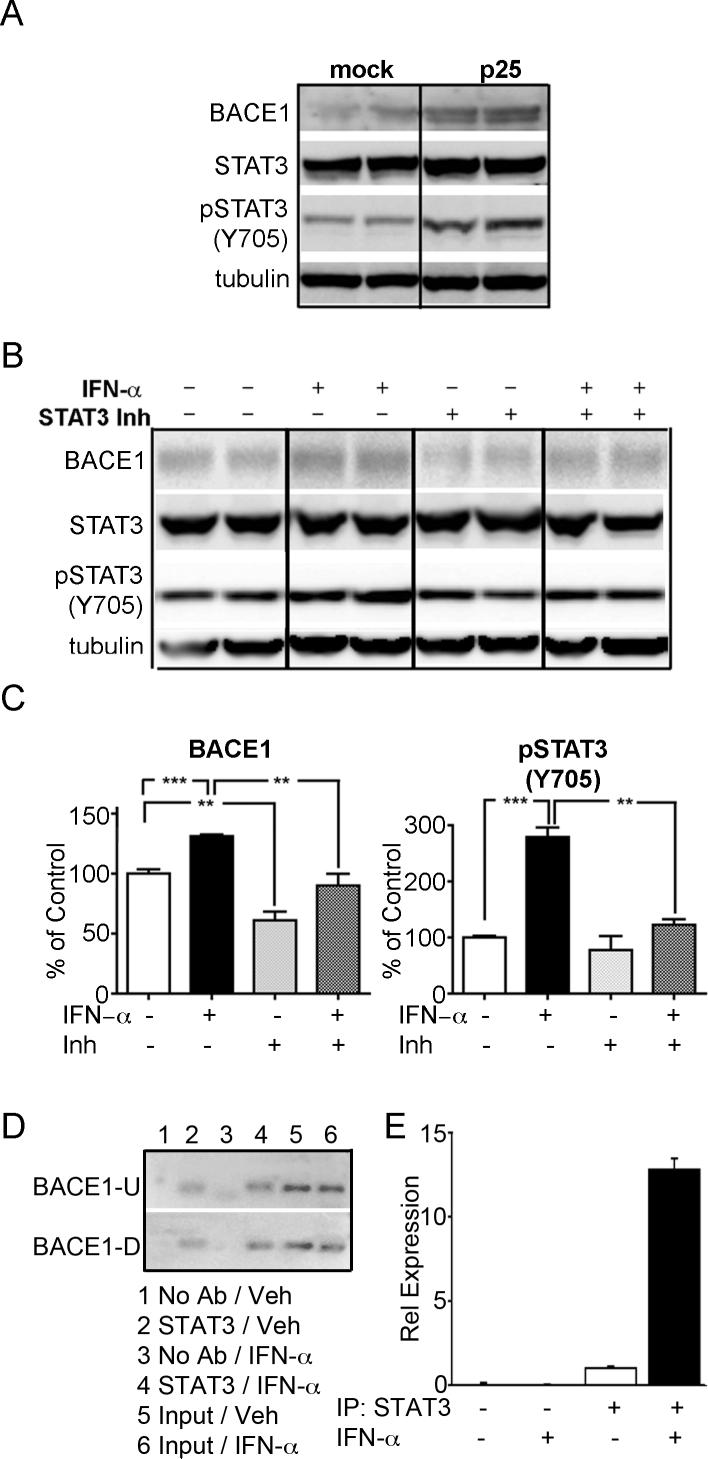
Both STAT3 and MEF2 are cdk5 responsive transcription factors known to regulate transcription of numerous target genes ( Gong et al., 2003; Fu et al., 2004). However, phosphorylation of MEF2 by cdk5 suppresses its transcription activity (Gong et al., 2003) and we did not examine its role in BACE1 transcription further. Cytoplasmic STAT3 is phosphorylated at the serine 727 site by several kinases including cdk5 (Fu et al., 2004). Phosphorylation at this site induces dimer formation, leading to phosphorylation at multiple tyrosine sites, including tyrosine 705. This then leads to translocation of STAT3 to the nucleus (Brierley and Fish, 2005). As experiments in figure 3 focused on the nuclear association of STAT3 with the BACE promoter, pSTAT3(Y705) rather than pSTAT3(S727) was monitored in these experiments. To confirm that enhanced p25/cdk5 led to increased levels of pSTAT3(Y705), induced p25 cells were examined. As expected, p25 cells showed higher levels of BACE1 compared to mock transfected cells, and this correlated with increased pSTAT3(Y705) (figure 3A). We then examined whether BACE1 was upregulated by interferon–α (INF-α)- a cytokine that upregulates phosphorylation of STAT3 at tyr705 (reviewed in Brierley and Fish, 2005). SH-SY5Y cells treated with INF-α demonstrated enhanced levels of BACE1, and this correlated with increased pSTAT3(Y705) (figure 3B). To demonstrate that the cytokine was mediating its effect on BACE1 through pSTAT3, an inhibitor of STAT3 dimerization (EMD 573095) was added to the culture medium. The inhibitor reduced levels of pSTAT3(Y705), as well as BACE1 in both control cells, and cells induced with INF-α (figure 3b). Graphical representation of the data is shown in figure 3C.
Figure 3. Altered STAT3 phosphorylation and BACE1 expression in mice.
Panel A: Representative immunoblots of BACE1, STAT3, pSTAT3(Y705) and tubulin from mock and p25 stably transfected SH-SY5Ycells. Panel B: Immunoblots of mock SH-SY5Y cells following treatment with interferon-α (IFN-α) and/or a cell permeable STAT3 inhibitor (Inh). Panel C: Statistical analyses of the level of BACE1 and pSTAT3(Y705) are represented as bar graphs which show an increase in relevant protein levels following IFN-α treatment, and a decrease following exposure to the STAT3 inhibitor. Panel D: Images of agarose gels following ChIP using two different sets of primers (BACE1-U, BACE1-D). PCR was performed following immunoprecipitation with, or without (No Ab), the STAT3 antibody in the presence, or absence of IFN-α. Total sheared DNA from the same cells (Input) was used as a positive control. Panel E: ChIP samples were also examined by qPCR using the BACE1-U primers, with the relative expression levels graphically represented following normalization. Vehicle treated cells immunoprecipitated with STAT3 antibody expression was set to1. ** p<0.01 *** p<0.001.
To examine whether STAT3 binds BACE1 promoter DNA, chromatin immunoprecipitation (ChIP) analysis was performed (figure 3D). DNA from SH-SY5Y cells that had been treated with vehicle or with INF-α was cross-linked and mechanically sheared. Chromatin was then immunoprecipitated with a STAT3 antibody that recognizes total STAT3, and was subjected to PCR using primers from two regions of the BACE1 promoter – a region upstream that contains multiple STAT1/3 sites (BACE-U primer set) and a region downstream that contains a single STAT1/3 site (BACE-D primer set). Different primers recognizing the promoter of the interleukin-8 gene (IL-8), a known STAT3 target gene (Gharavi et al., 2007), were used as a positive control (data not shown). The negative control was chromatin incubated without STAT3 antibody. Figure 3D shows that a BACE1 PCR product was only generated from DNA incubated with the STAT3 antibody. Both primer sets amplified a band of the expected size (188−197 bp) in chromatin incubated with antibody, and in the total DNA (“input”) positive control. To further confirm and quantify these results, qPCR was performed on the same samples using the BACE-U primer set (figure 3E), BACE-D primer set and the IL-8 primer set (data not shown). Samples that were incubated without STAT3 antibody failed to generate amplimers. Vehicle treated cells showed a lower level of enrichment following IP with the STAT3 antibody compared to ∼12 fold increase in cells induced with INF-α. Both primer sets for BACE1 showed a similar level of amplification when compared by qPCR (data not shown) indicating that both the cluster and single STAT1/3 sites are targeted by STAT3. These data support the deletion analysis, indicating that STAT3 and the BACE1 promoter show a physical interaction under physiological conditions, and that induction of pSTAT3 by INF-α significantly enhanced the association of STAT3 with BACE1 promoter DNA.
Elevated p25/cdk5 activity leads to hyperphosphorylation of STAT3 and enhanced BACE1 in vivo
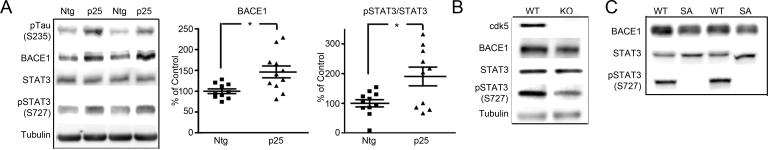
To confirm the effect of p25/cdk5 on BACE1 and APP processing in vivo, we examined mice that overexpress human p25 (Ahlijanian et al., 2000). STAT3 phosphorylation was examined in homozygous p25 mice at 4 days of age, compared with strain-matched, non-transgenic (Ntg) mice of the same age in four different sets of mice. This age was chosen because overall phosphorylation levels are high in neonatal animals (Mawal-Dewan et al., 1994). P25 mice showed enhanced levels of pTau(S235), a site used to monitor cdk5 activity, compared to Ntg mice. We then examined p25 and Ntg mice for pSTAT3. pSTAT3(S727) was significantly elevated when normalized to levels of total STAT3 protein in all sets of p25 mice. The level of BACE1 was significantly elevated in p25 mice compared to Ntg controls in two sets of mice, and a trend to increase was seen in the other two sets. A representative set is shown in figure 4A, with Ntg and p25 mice (n=10−11).
Figure 4. Enhanced cdk5 activity correlates with increased Aβ levels and APP processing in young p25 over-expressing mice.
Panel A: Representative immunoblot analysis of pTau(S235), BACE1, STAT3, pSTAT3(S727) and tubulin (loading control) in brain lysate from Ntg and p25 mice (n=10−11 for each group, two mice shown). BACE1 and pSTAT3/STAT3 levels were significantly higher in p25 mice compared with Ntg mice, as shown in the scatterplots. Panel B: Immunoblot analysis of cdk5 deficient mice compared with wild type mice. Reduced levels of BACE1 and pSTAT3(S727) were observed in cdk5 null mice (KO), compared with wild type control mice (WT); tubulin is shown as loading control. Several embryonic (day 16.5) brains were pooled to generate lysate. Panel C: Immunoblot analysis of BACE1 levels in mice with targeted ablation of the cdk5 responsive site at STAT3(727) (SA). STAT3 SA mice showed no detectable pSTAT3(S727). BACE1 was significantly reduced in homozygous SA mice at 4 days of age compared to strain and age matched controls (n=5 for SA mice, n=6 for WT, two mice for each shown). * p<0.05.
Loss of cdk5 activity reduces pSTAT3 and BACE1 levels
We next examined whether loss of cdk5 activity reduced levels of BACE1 in vivo, through the examination of cdk5 deficient mice (Ohshima et al., 1996). As cdk5 deficient mice are not viable, embryos were taken at E16.5 for analysis. Brain lysate from cdk5 deficient embryos showed ∼30% reduction in BACE1 levels, and ∼70% reduction in pSTAT3(S727) levels (figure 4B) compared to wild type controls.
Targeted mutation of pSTAT3(S727) leads to reduced BACE1
To further confirm that phosphorylation of STAT3 impacts BACE1 synthesis, we examined a line of mice in which the serine site of STAT3 at position Ser727 was substituted for alanine (SA allele) (Wen and Darnell, 1997; Shen et al., 2004) (figure 4C). Homozygous SA/SA mice are viable and phenotypically normal. Brain tissue from mice at 4 days of age was extracted and the level of BACE1 was compared with age, bodyweight and strain matched WT controls. The mean level of BACE1 in STAT3 SA mice (n=5) was reduced to 71% of WT levels (n=6), (figure 4C). To test whether BACE levels were significantly different between targeted and WT mice, ANOVA analysis was performed. The results showed that the level of BACE1 was significantly different between WT and targeted mice (p<0.05) (see supplementary data table 1 for statistical analysis).
An inhibitor of cdk5 reduced levels of pSTAT(S727) and BACE1 in vivo
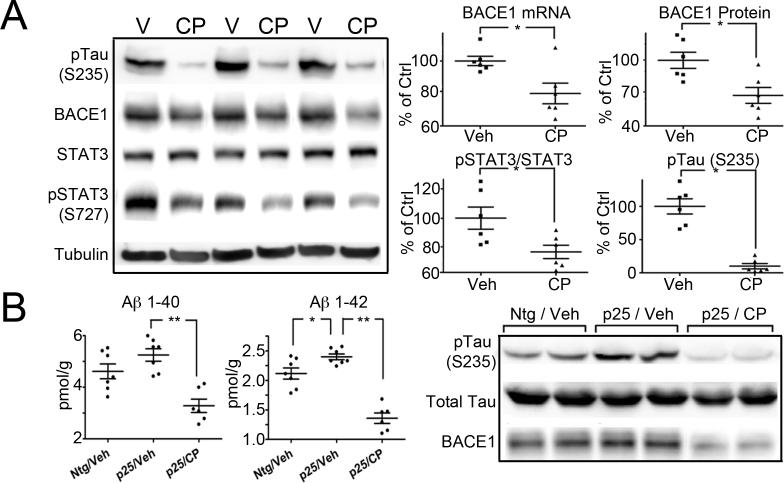
The brain permeable cdk5 inhibitor CP-681301 (CP, Pfizer, unpublished) was administered to Ntg or p25 mice by subcutaneous injection. Inhibition of cdk5 was monitored through phosphorylation of tau at S235. CP-681301 treatment of young Ntg mice (n=5−6) led to significant reduction of BACE1 mRNA and protein that correlated with reduced pSTAT3(S727) (figure 5A). Young p25 mice administered the inhibitor under similar conditions also showed a significant decrease in BACE1, as well as Aβ40/42 (two sets tested); a representative set is shown in figure 5B (see also supplementary table 2) (n=6−7). Aβ is turned over very rapidly in vivo. To confirm that the reduction of Aβ seen with inhibitor did not reflect overall reduced metabolism due to drug toxicity, we examined the levels of p35, a protein with a turnover time similar to Aβ (less than one hour) as well as tubulin (a long-lived protein). Levels of p35 were not significantly different between vehicle and CP treated mice (data not shown) suggesting that the reduction of Aβ was due to effects on APP processing, and not overall toxicity. In most sets of inhibitor-treated animals, reduced APP levels were observed, however the decrease did not reach statistical significance consistently (data not shown).
Figure 5. Inhibition of cdk5 activity reduces Aβ production and BACE1 levels in vivo.
Panel A: Representative immunoblots of pTau(S235), BACE1, STAT3, pSTAT3(S727) and tubulin in brain lysate from vehicle (V) and CP-681301 (CP) treated Ntg mice. BACE1 mRNA and protein levels, together with pSTAT3(S727)/STAT3 and pTauS235 are shown as scatterplots. N=6 in each group. There was a significant difference (p<0.05) between vehicle and CP-681301 treated mice for the proteins of interest. Panel B: Three groups of young (5 day old) mice were analyzed for Aβ and BACE1 levels: Ntg mice treated with vehicle (Veh; n=7), p25 mice treated with vehicle (p25/veh; n=7) and p25 mice treated with CP-681301 (p25/CP; n=6). Aβ40 and 42 levels were elevated in p25 mice relative to Ntg mice. Administration of CP-681301 to p25 mice significantly reduced both peptides (p<0.01). BACE1 was elevated in p25 mice relative to Ntg mice but was significantly reduced by CP-681301. The relative activity of cdk5 was shown by phosphorylation of Tau at the S235 site. Total tau levels indicated that equivalent levels of protein were loaded. All results were analyzed with Student's t-test, * p<0.05 ** p<0.01.
BACE1 synthesis and APP processing is also modulated by cdk5 in adult mice
ELISA analysis showed that the levels of both Aβ40 (p<0.01) and Aβ42 (p<0.05) were significantly increased in adult p25 mice compared to littermate Ntg (n=7−9) (figure 6A and supplementary table 3). Similarly, both protein and mRNA levels of BACE1 were significantly higher than in the Ntg control mice (p<0.001) (figure 6A). Four sets of adult Ntg mice (n=5 per group) were administered the cdk5 inhibitor CP-681301 for two days. In all four sets, CP-681301 treatment reduced Aβ and BACE1 levels significantly (figure 6B, see supplementary table 4 for ELISA data).
Figure 6. Cdk5 activity correlates with increased APP processing and BACE1 synthesis in adult p25 over-expressing mice.
Panel A: ELISA analysis of soluble murine Aβ40 and Aβ42 from Ntg and p25 mice at 18−22 months of age. Levels of both Aβ40 (p<0.01) and Aβ42 (p<0.05) were significantly increased in p25 mice compared to Ntg controls. P25 mice also had significantly elevated BACE1 mRNA and protein levels compared to Ntg. Representative immunoblots of p35/25, BACE1, APP-full length, APP α-CTF and β-actin (loading control) from Ntg and p25 mice are shown. Panel B: Effects of the cdk5 inhibitor CP-681301 on Aβ and BACE1 protein levels in adult Ntg mice. Levels of Aβ40 (p<0.01), Aβ42 (p<0.001) and BACE1 (P<0.05) were significantly reduced in CP-681301 treated (CP) mice, compared to vehicle treated (V) controls. A representative immunoblot of BACE1 and β-actin (loading control) is shown in the right hand panel. * P<0.05 ** p<0.01 *** p<0.001.
As the level of murine β-CTF is very low in Ntg mice, we examined the effect of enhanced cdk5 activity on human APP in double-transgenic mice that express both p25 and mutant, human APP (figure 7). The level of full-length APP was comparable between p25/APP and APP littermates, but the level of β-CTF (C99) was significantly elevated relative to α-CTF (C83) in p25/APP mice (figure 7A). Consistent with this being a predominantly β-secretase effect, the level of sAPPβ was significantly elevated in p25/APP mice. BACE1 was also significantly increased in p25/APP mice, compared with APP mice. The level of sAPPβ was significantly increased but the level of sAPPα was not significantly changed (figure 7B). Amyloid load was assessed in the p25/APP mice and their APP littermates to determine if p25-mediated elevation of Aβ accelerated the formation of amyloid plaques. Although plaques were detected in all mice, amyloid load was highly variable and no overt difference was observed (data not shown). ELISA data revealed a trend to increase in both soluble and formic acid extracted (total) Aβ in the double-transgenic p25/APP mice (n=6) relative to APP littermates (n=6), but the increase was not statistically significant (see supplementary data table 5 for ELISA values).
Figure 7. Enhanced cdk5 activity correlates with altered APP processing in p25/APP.
Panel A: APP processing was examined in double-transgenic p25/APP mice and single transgenic APP littermates. Levels of full length APP (APP fl) were not altered but there was an increase in the level of β-CTF (C99) relative to α-CTF (C83) in the p25/APP mice compared to controls (immunoblot shows bands recognized by antibody C1/6.1 from an intact membrane). Panel B: The level of α-secretase cleaved sAPPα fragments were unchanged in p25/APP mice compared to APP controls, but the fragments generated by BACE1 cleavage (sAPPβ) were significantly increased in p25/APP mice compared to controls. BACE1 was also increased in the double-transgenic mice. β-actin was used as a loading control (n=7 p25/APP, n=6 APP, 2 mice of each group shown). Panel C: Scatterplots of sAPPβ and BACE1 levels (values are normalized to APP group average = 1) were significantly higher (p<0.05) in the p25/APP mice than the single transgenic group. * p<0.05.
Discussion
We have shown that BACE1 is transcriptionally upregulated in response to enhanced cdk5 activity mediated by p25. The cdk5 target, STAT3, binds BACE1 promoter DNA and the level of pSTAT3 correlates with the level of BACE1 both in vivo and in vitro suggesting that STAT signaling is one pathway by which transcriptional control can be mediated. BACE1 showed increased activity towards its substrate, APP, leading to increased levels of Aβ40 and 42 in p25 mice, similar to that reported by Cruz et al., (2006) for an inducible p25 mouse line. Pharmacological inhibition of cdk5 reduced the level of BACE1, and Aβ peptides making inhibition of cdk5 a valid target for AD therapeutics.
The cdk5-reponsive region of the BACE1 promoter contains numerous sites, including sites for STAT1/3 and MEF2 that are both transcriptional factors known to be physiological substrates of p25/cdk5 in vivo and in cell culture systems (Fu et al., 2004; Ge et al., 2004; Gong et al., 2003). In addition to a previously reported STAT6 binding site at −931 (Sambamurti et al., 2004), STAT1/3 sites were identified at positions −932, −931, −865, −845, −844, −829 and −439, and a cluster of five sites were identified between −901 to −897. Furthermore, MEF2 sites were identified at −901, −899, −898, and −442 (Ge et al., 2004). These sites are all present in the BACE1P6 promoter region, but absent from BACE1P8 suggesting that cdk5 may be mediating its effect on BACE1 transcription through one, or both of these factors (Ge et al., 2004). Between the transcription initiation site and the physiological translation initiation codon of the human BACE1 gene, there are six upstream ATGs that could serve as the translation initiation codons for five potential ORFs. These μORFs have a negative effect on BACE1 protein translation due to leaky scanning (Lammich et al., 2004; Zhou and Song, 2006). P25/cdk5 activation may also negatively regulate transcription through inhibitory factors such as MEF2, and it is likely that several factors influence the spatial and temporal pattern of target gene transcription during development, and in the diseased brain. STAT1 and 3 sites are also present in many other gene promoters, including the human and mouse APP promoter (Song and Lahiri, 1998). Enhanced amyloidogenic processing could therefore also be the result of APP upregulation. However, the level of full-length APP was not significantly altered by increased p25, either in vitro, or in p25 transgenic mice, suggesting that in the systems studied, p25/cdk5-mediated effects act primarily on BACE1 rather than APP.
Most reports indicate that BACE1 mRNA does not increase in the AD brain (Holsinger et al., 2002; Matsui et al., 2007; Preece et al., 2003; Yasojima et al., 2001) whereas BACE protein/activity does increase (Fukumoto et al., 2002; Harada et al., 2006; Holsinger et al., 2002; Stockley et al., 2006; Tyler et al., 2002; Yang et al., 2003). Several mechanisms have been suggested to account for the increase in BACE1 protein levels including post-transcriptional (Lammich et al., 2004; Zohar et al., 2005) post-translational (Walter et al., 2001), or turnover effects (Qing et al., 2004). BACE1 mRNA levels have, however, been shown to be altered in regions with high plaque density from human AD brain with short post-mortem interval (PMI) (Li et al., 2004) but not longer PMI (Matsui et al., 2007) suggesting that in human brain, post-mortem mRNA degradation may confound interpretation. This does not however explain the increase in BACE1 protein, but not BACE1 mRNA observed in plaque-forming mice (Zhao et al., 2007) leaving the issue of the mechanism by which BACE1 increases in the AD brain unresolved.
The impact of p25/cdk5 on the pathogenesis of Alzheimer's disease is speculative. Although published data suggests that cdk5 activity is increased in AD brain (Lee et al., 1999), the involvement of p25 remains controversial (Patrick et al., 1999; Tandon et al., 2003; Tseng et al., 2002). Cdk5 mediated transcriptional upregulation of BACE could be envisioned to initiate or exacerbate AD pathogenesis in certain situations however. Increased production of p25 and enhancement of cdk5 activity has been shown to occur in rodent models of ischemia (Wang et al., 2003; Wen et al., 2007), and hypoxia due to hypoperfusion is a significant risk factor for long-term susceptibility to AD (Kalaria, 2000; Kalaria and Hedera, 1995). Ischemia in rodents has been shown to increase amyloidogenic processing of APP (Saido et al., 1994; Wen et al., 2004; Yokota et al., 1996), and AD pathology is increased in patients with coexisting evidence of cerebral infarcts (Nagy et al., 1997; Snowdon et al., 1997). Interestingly, pSTAT3 is one of several transcription factors upregulated by ischemia-induced hypoxia (Justicia et al., 2000), and hypoxia has been shown to upregulate BACE1 transcription through hypoxia inducing factor1-alpha (HIF1-α) (Sun et al., 2006). It is thus feasible that hypoxia-induced production of p25 and subsequent enhanced activity of cdk5 leads to local upregulation of BACE1 transcription, Aβ production and initiation or exacerbation of a pathological cascade resulting in AD.
Several substrates in addition to BACE1 are likely to be impacted by enhanced p25/cdk5 activity that could also result in increased amyloidogenic processing in vivo. Systems involved in trafficking that are known to be targets of cdk5 such as cytoskeleton proteins (tau, amphiphysin I and dynamin I), or vesicles regulating endocytosis or exocytosis (Tomizawa et al., 2003; Xin et al., 2004) may alter trafficking and recycling of BACE1 leading to accumulation in early endosomes – a compartment favorable to increased amyloidogenic processing. Phosphorylation of BACE1 itself also results in its redistribution as it has been previously reported that phosphorylation at the S498 site by CK1 alters BACE1 trafficking or sorting (Pastorino et al., 2002; Walter et al., 2001). Although there are two potential cdk5 consensus sites in BACE1 in the ectodomain, the localization (lumen compartment) makes it unlikely that BACE1 is a direct target for cdk5-mediated post-translational modification. Another substrate mediating the cdk5 effect on Aβ production could be APP itself as cdk5-directed phosphorylation of APP at the T668 site has been proposed to alter its accessibility to secretases leading to enhanced Aβ production (Lee et al., 2003). However, recent gene targeting experiments suggest that the T668 site is not involved in Aβ generation in vivo (Sano et al., 2006).
Our data demonstrates that in both young and old mice, elevated p25/cdk5 increases Aβ production through an increase in BACE1, and this effect can be reversed using a pharmacological inhibitor. While STAT3 is a potent inducer of BACE1 transcription, the mechanism of induction is probably not limited to transcriptional regulation via pSTAT3. Overall, given that p25/cdk5 significantly enhances Aβ production in vivo, inhibition of p25/cdk5 may be an appealing therapeutic approach for the treatment, or prevention of AD.
Experimental Procedures
Animals
P25 mice were the generous gift of Pfizer (Ahlijanian et al., 2000) and are available as cryopreserved stock from The Jackson Laboratory, Bar Harbor, ME. The p25 mice used were homozygous, on an FVB background. Ntg mice were age, strain and sex-matched. Some experiments were repeated using hemizygous p25 mice and Ntg littermates. All animals were maintained and killed according to National Institutes of Health and IACUC guidelines. P25 mice were crossed to a mutant APP line (line Tg2576, (Hsiao et al., 1996)). Hemizygous p25/APP and APP littermates at 10−12 months of age were examined. Mice lacking the p727 site in STAT3 (SA mice) were homozygous for the targeted mutation (kind gift of Dr. David Levy, NYU). Controls were age, strain and size matched C57Bl/6 mice.
Tissue culture
P25 transfected SH-SY5Y cells under the control of the tetracycline-regulated mammalian expression T-Rex system (Invitrogen, Carlsbad, CA) were the kind gift of Dr. Luc Buée (Hamdane et al., 2005). For induction of p25 expression, cells were maintained in medium with tetracycline at 1 μg/ml. SH-SY5Y or N2a cells were grown in DMEM supplemented with 10% fetal calf serum and appropriate antibiotics (Invitrogen) in a 5% CO2 humidified incubator at 37°C. Medium was replenished every 3 days. Transfections were performed with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instruction. Flag-GFP was co-transfected to allow normalization in the promoter-deletion experiments.
SH-SY5Y cells were plated at 80% density, then washed twice with PBS prior to incubating them in serum-free media overnight. The following day, cells were washed in PBS, placed in fresh serum-free medium and treated with either vehicle (PBS) or 235 μM STAT3 inhibitor/PBS (EMD Biosciences 573096, Gibstown NJ) for 1 hour (Turkson et al., 2001). After one hour of inhibitor or vehicle pretreatment, 1000 U/mL of IFN-α was added for 30 minutes. Following a 3.5 hour washout period, cells were harvested for biochemical analysis.
Immunoblot analysis
Mice were sacrificed and the brains were immediately removed, dissected and kept on dry ice. Hemi-brains were weighed and homogenized without thawing in 5× RIPA buffer (50 mM Tris-HCl, pH 7.4, 1% NP-40, 0.25% Na-deoxycholate, 150 mM NaCl, 1 mM EDTA, 1 mM PMSF, 1 mM Na3VO4, 1 mM NaF, 10 μl/ml protease inhibitor mix). Cultured cells were similarly lysed using RIPA buffer. Samples were centrifuged for 10 minutes at 20,000 × g at 4°C. A protein assay was performed and the samples were diluted in O+ buffer (62.5 mM Tris-HCl, pH 6.8, 10% (w/v) glycerol, 5% 2-mercaptoethanol, 2.3% SDS, 1 mM EGTA, 1 mM EDTA, 1 mM PMSF, 1 mM Na3VO4, 1 mM NaF, 10 μl/ml protease inhibitor mix) and incubated at 85°C for 10 min. Depending on the primary antibody used, 5−20 μg of protein was analyzed following electorophoresis on SDS-PAGE and transferred onto 0.2 μm nitrocellulose membranes (Whatman, Florham Park, NJ). Immunoblots were blocked, incubated with appropriate primary and secondary antibody dilutions (Jackson Immunoresearch, West Grove PA), and treated using chemiluminescent fluid (Supersignal WestPico, Pierce, Rockford IL). The signal was viewed using the Fujifilm LAS-3000 digital imaging system and Image Gauge software (Fujifilm, Stamford CT).
Aβ ELISA
Soluble murine or human Aβ1−40 and 1−42 from a hemibrain region including cortex and hippocampus was assessed using antibodies Aβx-40 (JRF/cAβ40) and Aβx-42 (JRF/cAβ42/26) for capture; JRF/AβN-25 (human) JRF/rAβ1−15/2 for detection (gifts of Dr. Sonia Jung, Centocor R&D, Radnor PA), as described previously (Schmidt et al., 2005a; Schmidt et al., 2005b). Older p25 or Ntg mice between 18−22 months of age and juvenile mice between 4 and 6 days of age were used. Several sets of mice were assessed for each experiment.
Cdk5 inhibitor treatment
CP-681301 (unpublished, gift of Dr Lit-Fui Lau, Pfizer) at a concentration of 5.8 mg/ml in 30% hydroxypropyl-beta-cyclodextrin (Sigma, St Louis, MO), or vehicle was injected subcutaneously at 100 μl/day once (neonates) or twice, 24 hours apart (adult mice). All mice became lethargic immediately after treatment and were sacrificed 4 hours after final administration.
Antibodies
For cdk5, anti-cdk5 (C-8) and anti-p35/25 (C-19) were used (Santa Cruz Biotechnology, Santa Cruz, CA). Anti human Aβ1−17 for human sAPPα (6E10), and anti-sAPPβ were purchased from Signet Laboratories, Deadham, MA. Monoclonal anti-BACE1 (3D5) was provided by Dr. Robert Vassar; polyclonal anti-BACE1 antibodies 7520 and 7523 were provided by Dr. Christian Haass. Full length APP/CTFs were detected with antibody C1/6.1 that recognizes a C-terminal epitope in APP (provided by Dr. Paul Mathews). Anti-STAT3 (which recognizes all forms of STAT3), pSTAT3(S727), and pSTAT3(Y705) were purchased from Cell Signaling Technology, Danvers, MA. Total tau was detected with either Tau T57120 (monoclonal, BD Transduction Laboratories, San Jose, CA), or Tau A0024 (polyclonal, Dako Cytomation, Carpinteria, CA). MC6 (pTauS235) was provided by Dr. Peter Davies (Albert Einstein, NY). β-actin, α- or β-tubulin were purchased from Sigma.
BACE1 Enzyme Assay
A FRET-based assay, using the substrate, H-RE(EDANS)EVNLDAEFK(DABCYL)R-OH, was performed according to the manufacturer's instruction (EMD Biosciences). Typically, the fluorescent EDANS molecule in the EDANS-XXX-DABCYL complex is quenched by the close proximity of the DABCYL molecule. In the assay, cellular BACE1 specifically cleaves the fluorogenic substrate, which then emits a measurable fluorescent signal. Briefly, cells were plated until they were 70% confluent. Cells were then lysed using the supplied buffer. Cell lysate (10 μL) was combined with the reaction buffer mix containing 10 μM of the fluorescent substrate and was loaded on an opaque plate, maintained at 37°C and monitored every twenty minutes (Ex=350 nm, Em=490nm; Wallac Victor2 1420, Perkin-Elmer Shelton CT). The reaction was carried out in the presence, or absence of a specific BACE1 inhibitor (EMD 565788, 1 μM) (Stachel et al., 2004). HEK 293 cells transfected with BACE1 were used as a positive control for this study.
Quantitative RT-PCR
Reverse transcription was performed on 1 μg of total RNA using the SuperScript III, first-strand supermix (Invitrogen) for qRT-PCR. 10 ng of cDNA was used in each PCR reaction. Quantitative PCR (qPCR) was performed by monitoring in real time, the increase in fluorescence of the iQ SYBR Green dye (Bio-Rad) on a Bio-Rad iQ5 detector system (Bio-Rad) according to the manufacturer's instructions. A standard curve was included in all experiments to monitor PCR efficiency. Each PCR was carried out in triplicate with 25 μl of the iQ SYBR Green Supermix (Bio-Rad). To exclude non-specific contamination by products such as primer dimers, a melting curve analysis was applied to all the final PCR products following the cycling protocols. Values for each sample were normalized to amplification level of mouse β-actin mRNA. The level of human BACE1 mRNA was analyzed with two independent sets of primers, from the 5′ end (5′TGA TCA TTG TGC GGG TGG AGA3′ / 5′TGA TGC GGA AGG ACT GGT TGG TAA3′) and 3′ end (5′ACT CCC TGG AGC CTT TCT TTG3′ / 5′ACT TTC TTG GGC AAA CGA AGG TTG GTG3′) of human BACE1 cDNA. The level of murine BACE1 mRNA level was analyzed with two independent sets of primers: 5′ end (5′ GCT TGC ACC TGT AGG ACA CA3′ / 5′ CTA AAG GAT GCTG GGC AGA G 3′) and the coding sequence from the 3′ end (5′ TCG CTG TCT CAC AGT CAT CC 3′ / 5′ GCA GAG TGG CAA CAT GAA GA 3′). The mRNA level was normalized to actin (5′ GCT CTT TTC CAG CCT TCC TT 3′ / 5′AGT ACT TGC GCT CAG AGG A3′).
Stable transfection of PC12 cells with BACE1 reporter constructs
PC12 cells were transfected with a pGL4.14 (Promega) vector containing a firefly luciferase reporter gene under the control of a 3.3kb fragment of the BACE1 promoter region, spanning from −2975 to +364, and a hygromycin resistance gene. Three days post-transfection, selection with 200 μg/ml hygromycin began and continued for approximately one month until colonies developed in the culture plate. Twenty-four colonies were isolated and grown with continued hygromycin selection until sufficient cells were available for use. One thousand cells derived from each colony were subjected to a firefly luciferase assay (Promega), resulting in a range of baseline activities (data not shown). Based on relative activity, three stable PC12 clones were selected, representing median, low, and high levels of baseline luciferase activity. These cells were then transfected with a p25-GFP expression vector, or a cdk5 expression vector together with a renilla luciferase-containing pRLSV40 plasmid (Promega) as an internal control. Mock transfections contained the renilla plasmid only. Two days following transfection with the expression vectors, activities of both the stably transfected firefly luciferase and the transiently transfected renilla luciferase were measured using a Dual-Glo luciferase detection kit, as per manufacturer's instructions (Promega). The results of p25-GFP and cdk5 over-expression in all three stable PC12 lines were found to be very similar, so these data were combined and analyzed together as a single experiment.
Promoter analysis
Three-six separate wells of N2a cells transfected with BACE1P6 or BACE1P8, with or without p25/GFP or vector were analyzed for CAT activity by ELISA as described previously (Ge et al., 2004). Data were expressed as CAT activity normalized to co-transfected GFP or renilla luciferase expression levels. To identify potential binding sites for the cdk5–associated transcription factors STAT1/3 and MEF2 in the BACE1 5′–flanking region, the TESS and Mat Inspector utilities were used to probe 1kb of BACE1 promoter sequence.
Chromatin immunoprecipitation analysis
ChIP analysis was performed according to manufacturer's instruction using the ChIP-IT Express Kit (Active Motif, Carlsbad, CA). Briefly, SH-SY5Y cells at 80% confluency were treated with vehicle or IFN-α (1000 U/mL for 30 mins), washed with PBS, cross-linked with 1% formaldehyde, and sheared by sonication to less than 500bp fragments. The sheared chromatin was immunoprecipitated with total STAT3 antibody (Cell Signaling Technology). DNA crosslinking was reversed overnight and PCR was then performed with primer pairs recognizing two different regions of the BACE1 promoter. Primer pair BACE-U amplified a region containing multiple STAT3 binding sites (-957/-760) (primer BACE-U: 5′ TTG GTT CAA GGC TTT AAG CTC T3′ / 5′GCC TTG AAC CAA AAG CGT TAG AG3′); whereas primer pair BACE-D amplified a region with a single STAT3 site (581/-393) (primer BACE-D: 5′GCC TTG AAC CAA AAG CGT TAG AG3′ / 5′TCT GGG AAG ACT CAG TGG GAG AA3′). A primer pair recognizing a STAT3 target region in the IL-8 promoter was used as a positive control (5′ GGT TTT CAC AGT GCT TTC AC 3′ / 5′ TTT CCC TCT TTG AGT CAT GC 3′) (Gharavi et al., 2007). PCR products were visualized following electrophoresis on a 2% agarose gel. The level of amplimer generated in each reaction was quantified by real-time PCR using primer pairs BACE1-U and BACE1-D, according to methods described previously for quantitative, mRNA analysis.
Statistical Analysis
All statistical analyses were performed with Prism (Graphpad Software, San Diego, CA), using unpaired Student's t-test between groups. Data shows mean ± SEM. Groups were considered significantly different when p<0.05 (* p<0.05, ** p<0.01 and *** p<0.001). Analysis of the SA STAT3 mice was performed by ANOVA.
Acknowledgements
The authors are grateful to Dr. L. Buée (INSERM U422, Lille, France) for p25 SH-SY5Y cells, to Dr. S. Jung (Centocor R&D, Inc., Radnor, PA) for anti-Aβ40/42 antibodies, Dr. P. Mathews (Nathan Kline Institute, NY) for APP antibodies, Dr. R. Vassar (Northwestern University, IL) for BACE 3D5 antibody and Dr. K. Sambamurti (Medical University of South Carolina, SC) for technical advice. Dr. H. Pant (NIH, MD) is thanked for brain lysate from cdk5 null mice. Dr. D. Levy (New York University, NY) generously supplied STAT3 SA neonates. Dr. S. Barral (Columbia University, NY) provided help with statistical analysis. This study was funded by grants to KD (AG172116, NS48447 and the AA Zenith award) and DKL (AG18379, AG18884 and the AA Zenith Award).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary Material
References
- Ahlijanian MK, Barrezueta NX, Williams RD, Jakowski A, Kowsz KP, McCarthy S, Coskran T, Carlo A, Seymour PA, Burkhardt JE, et al. Hyperphosphorylated tau and neurofilament and cytoskeletal disruptions in mice overexpressing human p25, an activator of cdk5. Proc Natl Acad Sci U S A. 2000;97:2910–2915. doi: 10.1073/pnas.040577797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann K, Mandelkow EM, Biernat J, Piwnica-Worms H, Mandelkow E. Abnormal Alzheimer-like phosphorylation of tau-protein by cyclin-dependent kinases cdk2 and cdk5. FEBS Lett. 1993;336:417–424. doi: 10.1016/0014-5793(93)80849-p. [DOI] [PubMed] [Google Scholar]
- Brierley MM, Fish EN. Stats: multifaceted regulators of transcription. J Interferon Cytokine Res. 2005;25:733–744. doi: 10.1089/jir.2005.25.733. [DOI] [PubMed] [Google Scholar]
- Capell A, Steiner H, Willem M, Kaiser H, Meyer C, Walter J, Lammich S, Multhaup G, Haass C. Maturation and pro-peptide cleavage of beta-secretase. J Biol Chem. 2000;275:30849–30854. doi: 10.1074/jbc.M003202200. [DOI] [PubMed] [Google Scholar]
- Christensen MA, Zhou W, Qing H, Lehman A, Philipsen S, Song W. Transcriptional regulation of BACE1, the beta-amyloid precursor protein beta-secretase. by Sp1. Mol Cell Biol. 2004;24:865–874. doi: 10.1128/MCB.24.2.865-874.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz JC, Kim D, Moy LY, Dobbin MM, Sun X, Bronson RT, Tsai LH. p25/cyclin-dependent kinase 5 induces production and intraneuronal accumulation of amyloid beta in vivo. J Neurosci. 2006;26:10536–10541. doi: 10.1523/JNEUROSCI.3133-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz JC, Tsai LH. A Jekyll and Hyde kinase: roles for Cdk5 in brain development and disease. Curr Opin Neurobiol. 2004;14:390–394. doi: 10.1016/j.conb.2004.05.002. [DOI] [PubMed] [Google Scholar]
- De Strooper B, Saftig P, Craessaerts K, Vanderstichele H, Guhde G, Annaert W, Von Figura K, Van Leuven F. Deficiency of presenilin-1 inhibits the normal cleavage of amyloid precursor protein. Nature. 1998;391:387–390. doi: 10.1038/34910. [DOI] [PubMed] [Google Scholar]
- Dhavan R, Tsai LH. A decade of CDK5. Nat Rev Mol Cell Biol. 2001;2:749–759. doi: 10.1038/35096019. [DOI] [PubMed] [Google Scholar]
- Edbauer D, Winkler E, Regula JT, Pesold B, Steiner H, Haass C. Reconstitution of gamma-secretase activity. Nature cell biology. 2003;5:486–488. doi: 10.1038/ncb960. [DOI] [PubMed] [Google Scholar]
- Fu AK, Fu WY, Cheung J, Tsim KW, Ip FC, Wang JH, Ip NY. Cdk5 is involved in neuregulin-induced AChR expression at the neuromuscular junction. Nat Neurosci. 2001;4:374–381. doi: 10.1038/86019. [DOI] [PubMed] [Google Scholar]
- Fu AK, Fu WY, Ng AK, Chien WW, Ng YP, Wang JH, Ip NY. Cyclin-dependent kinase 5 phosphorylates signal transducer and activator of transcription 3 and regulates its transcriptional activity. Proc Natl Acad Sci U S A. 2004;101:6728–6733. doi: 10.1073/pnas.0307606100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukumoto H, Cheung BS, Hyman BT, Irizarry MC. Beta-secretase protein and activity are increased in the neocortex in Alzheimer disease. Archives of neurology. 2002;59:1381–1389. doi: 10.1001/archneur.59.9.1381. [DOI] [PubMed] [Google Scholar]
- Ge YW, Maloney B, Sambamurti K, Lahiri DK. Functional characterization of the 5' flanking region of the BACE gene: identification of a 91 bp fragment involved in basal level of BACE promoter expression. Faseb J. 2004;18:1037–1039. doi: 10.1096/fj.03-1379fje. [DOI] [PubMed] [Google Scholar]
- Gharavi NM, Alva JA, Mouillesseaux KP, Lai C, Yeh M, Yeung W, Johnson J, Szeto WL, Hong L, Fishbein M, et al. Role of the Jak/STAT pathway in the regulation of interleukin-8 transcription by oxidized phospholipids in vitro and in atherosclerosis in vivo. J Biol Chem. 2007;282:31460–31468. doi: 10.1074/jbc.M704267200. [DOI] [PubMed] [Google Scholar]
- Ghribi O. The role of the endoplasmic reticulum in the accumulation of beta-amyloid peptide in Alzheimer's disease. Current molecular medicine. 2006;6:119–133. doi: 10.2174/156652406775574514. [DOI] [PubMed] [Google Scholar]
- Gong X, Tang X, Wiedmann M, Wang X, Peng J, Zheng D, Blair LA, Marshall J, Mao Z. Cdk5-mediated inhibition of the protective effects of transcription factor MEF2 in neurotoxicity-induced apoptosis. Neuron. 2003;38:33–46. doi: 10.1016/s0896-6273(03)00191-0. [DOI] [PubMed] [Google Scholar]
- Hamdane M, Bretteville A, Sambo AV, Schindowski K, Begard S, Delacourte A, Bertrand P, Buee L. p25/Cdk5-mediated retinoblastoma phosphorylation is an early event in neuronal cell death. J Cell Sci. 2005;118:1291–1298. doi: 10.1242/jcs.01724. [DOI] [PubMed] [Google Scholar]
- Hamdane M, Sambo AV, Delobel P, Begard S, Violleau A, Delacourte A, Bertrand P, Benavides J, Buee L. Mitotic-like tau phosphorylation by p25-Cdk5 kinase complex. J Biol Chem. 2003;278:34026–34034. doi: 10.1074/jbc.M302872200. [DOI] [PubMed] [Google Scholar]
- Harada H, Tamaoka A, Ishii K, Shoji S, Kametaka S, Kametani F, Saito Y, Murayama S. Beta-site APP cleaving enzyme 1 (BACE1) is increased in remaining neurons in Alzheimer's disease brains. Neuroscience research. 2006;54:24–29. doi: 10.1016/j.neures.2005.10.001. [DOI] [PubMed] [Google Scholar]
- Holsinger RM, McLean CA, Beyreuther K, Masters CL, Evin G. Increased expression of the amyloid precursor beta-secretase in Alzheimer's disease. Ann Neurol. 2002;51:783–786. doi: 10.1002/ana.10208. [DOI] [PubMed] [Google Scholar]
- Hong HS, Hwang EM, Sim HJ, Cho HJ, Boo JH, Oh SS, Kim SU, Mook-Jung I. Interferon gamma stimulates beta-secretase expression and sAPPbeta production in astrocytes. Biochemical and biophysical research communications. 2003;307:922–927. doi: 10.1016/s0006-291x(03)01270-1. [DOI] [PubMed] [Google Scholar]
- Hsiao K, Chapman P, Nilsen S, Eckman C, Harigaya Y, Younkin S, Yang F, Cole G. Correlative memory deficits, Abeta elevation, and amyloid plaques in transgenic mice. Science. 1996;274:99–102. doi: 10.1126/science.274.5284.99. [DOI] [PubMed] [Google Scholar]
- Huse JT, Pijak DS, Leslie GJ, Lee VM, Doms RW. Maturation and endosomal targeting of beta-site amyloid precursor protein-cleaving enzyme. The Alzheimer's disease beta-secretase. J Biol Chem. 2000;275:33729–33737. doi: 10.1074/jbc.M004175200. [DOI] [PubMed] [Google Scholar]
- Justicia C, Gabriel C, Planas AM. Activation of the JAK/STAT pathway following transient focal cerebral ischemia: signaling through Jak1 and Stat3 in astrocytes. Glia. 2000;30:253–270. doi: 10.1002/(sici)1098-1136(200005)30:3<253::aid-glia5>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- Kalaria RN. The role of cerebral ischemia in Alzheimer's disease. Neurobiology of aging. 2000;21:321–330. doi: 10.1016/s0197-4580(00)00125-1. [DOI] [PubMed] [Google Scholar]
- Kalaria RN, Hedera P. Differential degeneration of the cerebral microvasculature in Alzheimer's disease. Neuroreport. 1995;6:477–480. doi: 10.1097/00001756-199502000-00018. [DOI] [PubMed] [Google Scholar]
- Kesavapany S, Lau KF, McLoughlin DM, Brownlees J, Ackerley S, Leigh PN, Shaw CE, Miller CC. p35/cdk5 binds and phosphorylates beta-catenin and regulates beta-catenin/presenilin-1 interaction. The European journal of neuroscience. 2001;13:241–247. [PubMed] [Google Scholar]
- Kobayashi S, Ishiguro K, Omori A, Takamatsu M, Arioka M, Imahori K, Uchida T. A cdc2-related kinase PSSALRE/cdk5 is homologous with the 30 kDa subunit of tau protein kinase II, a proline-directed protein kinase associated with microtubule. FEBS Lett. 1993;335:171–175. doi: 10.1016/0014-5793(93)80723-8. [DOI] [PubMed] [Google Scholar]
- Koh YH, von Arnim CA, Hyman BT, Tanzi RE, Tesco G. BACE is degraded via the lysosomal pathway. J Biol Chem. 2005;280:32499–32504. doi: 10.1074/jbc.M506199200. [DOI] [PubMed] [Google Scholar]
- Lahiri DK, Maloney B, Ge YW. Functional domains of the BACE1 and BACE2 promoters and mechanisms of transcriptional suppression of the BACE2 promoter in normal neuronal cells. J Mol Neurosci. 2006;29:65–80. doi: 10.1385/JMN:29:1:65. [DOI] [PubMed] [Google Scholar]
- Lammich S, Schobel S, Zimmer AK, Lichtenthaler SF, Haass C. Expression of the Alzheimer protease BACE1 is suppressed via its 5'-untranslated region. EMBO reports. 2004;5:620–625. doi: 10.1038/sj.embor.7400166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KY, Clark AW, Rosales JL, Chapman K, Fung T, Johnston RN. Elevated neuronal Cdc2-like kinase activity in the Alzheimer disease brain. Neuroscience research. 1999;34:21–29. doi: 10.1016/s0168-0102(99)00026-7. [DOI] [PubMed] [Google Scholar]
- Lee MS, Kao SC, Lemere CA, Xia W, Tseng HC, Zhou Y, Neve R, Ahlijanian MK, Tsai LH. APP processing is regulated by cytoplasmic phosphorylation. J Cell Biol. 2003;163:83–95. doi: 10.1083/jcb.200301115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MS, Kwon YT, Li M, Peng J, Friedlander RM, Tsai LH. Neurotoxicity induces cleavage of p35 to p25 by calpain. Nature. 2000;405:360–364. doi: 10.1038/35012636. [DOI] [PubMed] [Google Scholar]
- Li R, Lindholm K, Yang LB, Yue X, Citron M, Yan R, Beach T, Sue L, Sabbagh M, Cai H, et al. Amyloid beta peptide load is correlated with increased beta-secretase activity in sporadic Alzheimer's disease patients. Proc Natl Acad Sci U S A. 2004;101:3632–3637. doi: 10.1073/pnas.0205689101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Bolon B, Kahn S, Bennett BD, Babu-Khan S, Denis P, Fan W, Kha H, Zhang J, Gong Y, et al. Mice deficient in BACE1, the Alzheimer's beta-secretase, have normal phenotype and abolished beta-amyloid generation. Nat Neurosci. 2001;4:231–232. doi: 10.1038/85059. [DOI] [PubMed] [Google Scholar]
- Maccioni RB, Otth C, Concha, Munoz JP. The protein kinase Cdk5. Structural aspects, roles in neurogenesis and involvement in Alzheimer's pathology. Eur J Biochem. 2001;268:1518–1527. doi: 10.1046/j.1432-1033.2001.02024.x. [DOI] [PubMed] [Google Scholar]
- Maloney B, Ge YW, Greig NH, Lahiri DK. Characterization of the human beta-secretase 2 (BACE2) 5'-flanking region: identification of a 268-bp region as the basal BACE2 promoter. J Mol Neurosci. 2006;29:81–99. doi: 10.1385/JMN:29:1:81. [DOI] [PubMed] [Google Scholar]
- Matsubara M, Kusubata M, Ishiguro K, Uchida T, Titani K, Taniguchi H. Site-specific phosphorylation of synapsin I by mitogen-activated protein kinase and Cdk5 and its effects on physiological functions. J Biol Chem. 1996;271:21108–21113. doi: 10.1074/jbc.271.35.21108. [DOI] [PubMed] [Google Scholar]
- Matsui T, Ingelsson M, Fukumoto H, Ramasamy K, Kowa H, Frosch MP, Irizarry MC, Hyman BT. Expression of APP pathway mRNAs and proteins in Alzheimer's disease. Brain Res. 2007;1161:116–123. doi: 10.1016/j.brainres.2007.05.050. [DOI] [PubMed] [Google Scholar]
- Mawal-Dewan M, Henley J, Van de Voorde A, Trojanowski JQ, Lee VM. The phosphorylation state of tau in the developing rat brain is regulated by phosphoprotein phosphatases. J Biol Chem. 1994;269:30981–30987. [PubMed] [Google Scholar]
- Nagy Z, Esiri MM, Jobst KA, Morris JH, King EM, McDonald B, Joachim C, Litchfield S, Barnetson L, Smith AD. The effects of additional pathology on the cognitive deficit in Alzheimer disease. Journal of neuropathology and experimental neurology. 1997;56:165–170. doi: 10.1097/00005072-199702000-00007. [DOI] [PubMed] [Google Scholar]
- Ohshima T, Ward JM, Huh CG, Longenecker G, Veeranna, Pant HC, Brady RO, Martin LJ, Kulkarni AB. Targeted disruption of the cyclin-dependent kinase 5 gene results in abnormal corticogenesis, neuronal pathology and perinatal death. Proc Natl Acad Sci U S A. 1996;93:11173–11178. doi: 10.1073/pnas.93.20.11173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastorino L, Ikin AF, Nairn AC, Pursnani A, Buxbaum JD. The carboxyl-terminus of BACE contains a sorting signal that regulates BACE trafficking but not the formation of total A(beta). Mol Cell Neurosci. 2002;19:175–185. doi: 10.1006/mcne.2001.1065. [DOI] [PubMed] [Google Scholar]
- Patrick GN, Zukerberg L, Nikolic M, de la Monte S, Dikkes P, Tsai LH. Conversion of p35 to p25 deregulates Cdk5 activity and promotes neurodegeneration. Nature. 1999;402:615–622. doi: 10.1038/45159. [DOI] [PubMed] [Google Scholar]
- Phiel CJ, Wilson CA, Lee VM, Klein PS. GSK-3alpha regulates production of Alzheimer's disease amyloid-beta peptides. Nature. 2003;423:435–439. doi: 10.1038/nature01640. [DOI] [PubMed] [Google Scholar]
- Planel E, Sun X, Takashima A. Role of GSK-3 beta in Alzheimer's disease pathology. Drug Development Research. 2002;56:491–510. [Google Scholar]
- Preece P, Virley DJ, Costandi M, Coombes R, Moss SJ, Mudge AW, Jazin E, Cairns NJ. Beta-secretase (BACE) and GSK-3 mRNA levels in Alzheimer's disease. Brain research. 2003;116:155–158. doi: 10.1016/s0169-328x(03)00233-x. [DOI] [PubMed] [Google Scholar]
- Qing H, Zhou W, Christensen MA, Sun X, Tong Y, Song W. Degradation of BACE by the ubiquitin-proteasome pathway. Faseb J. 2004;18:1571–1573. doi: 10.1096/fj.04-1994fje. [DOI] [PubMed] [Google Scholar]
- Rockenstein E, Torrance M, Adame A, Mante M, Bar-on P, Rose JB, Crews L, Masliah E. Neuroprotective effects of regulators of the glycogen synthase kinase-3beta signaling pathway in a transgenic model of Alzheimer's disease are associated with reduced amyloid precursor protein phosphorylation. J Neurosci. 2007;27:1981–1991. doi: 10.1523/JNEUROSCI.4321-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saido TC, Yokota M, Maruyama K, Yamao-Harigaya W, Tani E, Ihara Y, Kawashima S. Spatial resolution of the primary beta-amyloidogenic process induced in postischemic hippocampus. J Biol Chem. 1994;269:15253–15257. [PubMed] [Google Scholar]
- Sambamurti K, Kinsey R, Maloney B, Ge YW, Lahiri DK. Gene structure and organization of the human beta-secretase (BACE) promoter. Faseb J. 2004;18:1034–1036. doi: 10.1096/fj.03-1378fje. [DOI] [PubMed] [Google Scholar]
- Sano Y, Nakaya T, Pedrini S, Takeda S, Iijima-Ando K, Iijima K, Mathews PM, Itohara S, Gandy S, Suzuki T. Physiological Mouse Brain Abeta Levels Are Not Related to the Phosphorylation State of Threonine-668 of Alzheimer's APP. PLoS ONE. 2006;1:e51. doi: 10.1371/journal.pone.0000051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki S, Shionoya A, Ishida M, Gambello MJ, Yingling J, Wynshaw-Boris A, Hirotsune S. A LIS1/NUDEL/cytoplasmic dynein heavy chain complex in the developing and adult nervous system. Neuron. 2000;28:681–696. doi: 10.1016/s0896-6273(00)00146-x. [DOI] [PubMed] [Google Scholar]
- Schmidt SD, Jiang Y, Nixon RA, Mathews PM. Tissue processing prior to protein analysis and amyloid-beta quantitation. Methods in molecular biology (Clifton, N.J. 2005a;299:267–278. doi: 10.1385/1-59259-874-9:267. [DOI] [PubMed] [Google Scholar]
- Schmidt SD, Nixon RA, Mathews PM. ELISA method for measurement of amyloid-beta levels. Methods in molecular biology (Clifton, N.J. 2005b;299:279–297. doi: 10.1385/1-59259-874-9:279. [DOI] [PubMed] [Google Scholar]
- Shen Y, Schlessinger K, Zhu X, Meffre E, Quimby F, Levy DE, Darnell JE., Jr. Essential role of STAT3 in postnatal survival and growth revealed by mice lacking STAT3 serine 727 phosphorylation. Mol Cell Biol. 2004;24:407–419. doi: 10.1128/MCB.24.1.407-419.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snowdon DA, Greiner LH, Mortimer JA, Riley KP, Greiner PA, Markesbery WR. Brain infarction and the clinical expression of Alzheimer disease. The Nun Study. Jama. 1997;277:813–817. [PubMed] [Google Scholar]
- Song W, Lahiri DK. Isolation of the genomic clone of the rhesus monkey beta-amyloid precursor protein. Biochemistry and molecular biology international. 1998;46:755–764. doi: 10.1080/15216549800204302. [DOI] [PubMed] [Google Scholar]
- Stachel SJ, Coburn CA, Steele TG, Jones KG, Loutzenhiser EF, Gregro AR, Rajapakse HA, Lai MT, Crouthamel MC, Xu M, et al. Structure-based design of potent and selective cell-permeable inhibitors of human beta-secretase (BACE-1). Journal of medicinal chemistry. 2004;47:6447–6450. doi: 10.1021/jm049379g. [DOI] [PubMed] [Google Scholar]
- Stockley JH, Ravid R, O'Neill C. Altered beta-secretase enzyme kinetics and levels of both BACE1 and BACE2 in the Alzheimer's disease brain. FEBS Lett. 2006;580:6550–6560. doi: 10.1016/j.febslet.2006.10.076. [DOI] [PubMed] [Google Scholar]
- Sun X, He G, Qing H, Zhou W, Dobie F, Cai F, Staufenbiel M, Huang LE, Song W. Hypoxia facilitates Alzheimer's disease pathogenesis by up-regulating BACE1 gene expression. Proc Natl Acad Sci U S A. 2006;103:18727–18732. doi: 10.1073/pnas.0606298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamagno E, Parola M, Bardini P, Piccini A, Borghi R, Guglielmotto M, Santoro G, Davit A, Danni O, Smith MA, et al. Beta-site APP cleaving enzyme up-regulation induced by 4-hydroxynonenal is mediated by stress-activated protein kinases pathways. J Neurochem. 2005;92:628–636. doi: 10.1111/j.1471-4159.2004.02895.x. [DOI] [PubMed] [Google Scholar]
- Tandon A, Yu H, Wang L, Rogaeva E, Sato C, Chishti MA, Kawarai T, Hasegawa H, Chen F, Davies P, et al. Brain levels of CDK5 activator p25 are not increased in Alzheimer's or other neurodegenerative diseases with neurofibrillary tangles. J Neurochem. 2003;86:572–581. doi: 10.1046/j.1471-4159.2003.01865.x. [DOI] [PubMed] [Google Scholar]
- Tomizawa K, Sunada S, Lu YF, Oda Y, Kinuta M, Ohshima T, Saito T, Wei FY, Matsushita M, Li ST, et al. Cophosphorylation of amphiphysin I and dynamin I by Cdk5 regulates clathrin-mediated endocytosis of synaptic vesicles. J Cell Biol. 2003;163:813–824. doi: 10.1083/jcb.200308110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai LH, Delalle I, Caviness VS, Jr., Chae T, Harlow E. p35 is a neural-specific regulatory subunit of cyclin-dependent kinase 5. Nature. 1994;371:419–423. doi: 10.1038/371419a0. [DOI] [PubMed] [Google Scholar]
- Tseng HC, Zhou Y, Shen Y, Tsai LH. A survey of Cdk5 activator p35 and p25 levels in Alzheimer's disease brains. FEBS Lett. 2002;523:58–62. doi: 10.1016/s0014-5793(02)02934-4. [DOI] [PubMed] [Google Scholar]
- Turkson J, Ryan D, Kim JS, Zhang Y, Chen Z, Haura E, Laudano A, Sebti S, Hamilton AD, Jove R. Phosphotyrosyl peptides block Stat3-mediated DNA binding activity, gene regulation, and cell transformation. J Biol Chem. 2001;276:45443–45455. doi: 10.1074/jbc.M107527200. [DOI] [PubMed] [Google Scholar]
- Tyler SJ, Dawbarn D, Wilcock GK, Allen SJ. alpha- and beta-secretase: profound changes in Alzheimer's disease. Biochemical and biophysical research communications. 2002;299:373–376. doi: 10.1016/s0006-291x(02)02635-9. [DOI] [PubMed] [Google Scholar]
- Vassar R, Bennett BD, Babu-Khan S, Kahn S, Mendiaz EA, Denis P, Teplow DB, Ross S, Amarante P, Loeloff R, et al. Beta-secretase cleavage of Alzheimer's amyloid precursor protein by the transmembrane aspartic protease BACE. Science. 1999;286:735–741. doi: 10.1126/science.286.5440.735. [DOI] [PubMed] [Google Scholar]
- Walter J, Fluhrer R, Hartung B, Willem M, Kaether C, Capell A, Lammich S, Multhaup G, Haass C. Phosphorylation regulates intracellular trafficking of beta-secretase. J Biol Chem. 2001;276:14634–14641. doi: 10.1074/jbc.M011116200. [DOI] [PubMed] [Google Scholar]
- Wang J, Liu S, Fu Y, Wang JH, Lu Y. Cdk5 activation induces hippocampal CA1 cell death by directly phosphorylating NMDA receptors. Nat Neurosci. 2003;6:1039–1047. doi: 10.1038/nn1119. [DOI] [PubMed] [Google Scholar]
- Wen Y, Onyewuchi O, Yang S, Liu R, Simpkins JW. Increased beta-secretase activity and expression in rats following transient cerebral ischemia. Brain Res. 2004;1009:1–8. doi: 10.1016/j.brainres.2003.09.086. [DOI] [PubMed] [Google Scholar]
- Wen Y, Yang SH, Liu R, Perez EJ, Brun-Zinkernagel AM, Koulen P, Simpkins JW. Cdk5 is involved in NFT-like tauopathy induced by transient cerebral ischemia in female rats. Biochimica et biophysica acta. 2007;1772:473–483. doi: 10.1016/j.bbadis.2006.10.011. [DOI] [PubMed] [Google Scholar]
- Wen Z, Darnell JE., Jr. Mapping of Stat3 serine phosphorylation to a single residue (727) and evidence that serine phosphorylation has no influence on DNA binding of Stat1 and Stat3. Nucleic acids research. 1997;25:2062–2067. doi: 10.1093/nar/25.11.2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Sanada K, Samuels BA, Shih H, Tsai LH. Serine 732 phosphorylation of FAK by Cdk5 is important for microtubule organization, nuclear movement, and neuronal migration. Cell. 2003;114:469–482. doi: 10.1016/s0092-8674(03)00605-6. [DOI] [PubMed] [Google Scholar]
- Xin X, Ferraro F, Back N, Eipper BA, Mains RE. Cdk5 and Trio modulate endocrine cell exocytosis. J Cell Sci. 2004;117:4739–4748. doi: 10.1242/jcs.01333. [DOI] [PubMed] [Google Scholar]
- Yang LB, Lindholm K, Yan R, Citron M, Xia W, Yang XL, Beach T, Sue L, Wong P, Price D, et al. Elevated beta-secretase expression and enzymatic activity detected in sporadic Alzheimer disease. Nature medicine. 2003;9:3–4. doi: 10.1038/nm0103-3. [DOI] [PubMed] [Google Scholar]
- Yasojima K, McGeer EG, McGeer PL. Relationship between beta amyloid peptide generating molecules and neprilysin in Alzheimer disease and normal brain. Brain Res. 2001;919:115–121. doi: 10.1016/s0006-8993(01)03008-6. [DOI] [PubMed] [Google Scholar]
- Yokota M, Saido TC, Tani E, Yamaura I, Minami N. Cytotoxic fragment of amyloid precursor protein accumulates in hippocampus after global forebrain ischemia. J Cereb Blood Flow Metab. 1996;16:1219–1223. doi: 10.1097/00004647-199611000-00016. [DOI] [PubMed] [Google Scholar]
- Zhao J, Fu Y, Yasvoina M, Shao P, Hitt B, O'Connor T, Logan S, Maus E, Citron M, Berry R, et al. Beta-site amyloid precursor protein cleaving enzyme 1 levels become elevated in neurons around amyloid plaques: implications for Alzheimer's disease pathogenesis. J Neurosci. 2007;27:3639–3649. doi: 10.1523/JNEUROSCI.4396-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Song W. Leaky scanning and reinitiation regulate BACE1 gene expression. Mol Cell Biol. 2006;26:3353–3364. doi: 10.1128/MCB.26.9.3353-3364.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zohar O, Pick CG, Cavallaro S, Chapman J, Katzav A, Milman A, Alkon DL. Age-dependent differential expression of BACE splice variants in brain regions of tg2576 mice. Neurobiology of aging. 2005;26:1167–1175. doi: 10.1016/j.neurobiolaging.2004.10.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.