Abstract
Steroid hormone receptors (SHR) belong to a large family of ligand-activated transcription factors that perform their biological functions by enhancing the transcription of specific target genes. The transactivation functions of SHRs are regulated by a specialized group of proteins called coactivators. The SHR coactivators represent a growing class of proteins with various enzymatic activities that serve to modify the chromatin to facilitate the transcription of SHR target genes. The ubiquitin-proteasome pathway enzymes have also been added to the growing list of enzymatic activities that are recruited to the SHR target gene promoters during transcription. One such ubiquitin-proteasome pathway enzyme to be identified and characterized as a SHR coactivator was E6-associated protein (E6-AP). E6-AP is a hect (homologous to E6-associated protein carboxy-terminal domain) domain containing E3 ubiquitin ligase that possesses two independent separable functions; a coactivation function and an ubiquitin-protein ligase activity. Being a component of the ubiquitin-proteasome pathway, it is postulated that E6-AP may orchestrate the dynamics of steroid hormone receptor-mediated transcription by regulating the degradation of the transcriptional complexes. E6-AP has also been shown to be involved in the regulation of various aspects of reproduction such as prostate and mammary gland development. Furthermore, it has been demonstrated that E6-AP expression is down-regulated in breast and prostate tumors and that the expression of E6-AP is inversely associated with that of estrogen and androgen receptors. This review summarizes our current knowledge about the structures, molecular mechanisms, spatiotemporal expression patterns and biological functions of E6-AP.
History
The balance between protein synthesis and degradation is a critical and highly regulated process in a cell. Protein degradation is one of the many vital processes that are essential for cellular homeostasis. One of the most studied and well-understood systems involved in intracellular protein degradation is the ubiquitin proteasome pathway (UPP). In cells, a ubiquitous protein tag known as ubiquitin marks proteins targeted for degradation. Ubiquitin is a 76 amino acid polypeptide that is highly conserved among eukaryotes and is attached to the substrate (protein targeted for degradation) by a process called ubiquitination. Ubiquitination of proteins is carried out by a series of enzymes known as ubiquitin activating enzyme (E1), ubiquitin-conjugating enzyme (E2) and ubiquitin protein ligase (E3). This results in the poly-ubiquitination of proteins that are then recognized by the 26S proteasome and are degraded [Pickart, 2001]. E6-AP is a member of a family of proteins called hect (homologous to E6-associated protein carboxy-terminal domain) domain E3 ligases that share carboxy terminal sequence homology with each other [Salvat et al., 2004; Scheffner et al., 1993]. This growing family of hect domain containing proteins has several members that have been shown to function as E3 ubiquitin ligases through their ability to form thioester bonds with ubiquitin.
The 100 kDa cellular protein E6-AP (UBE3A) was first identified as a protein that interacts with the E6 protein of human papilloma virus (HPV) type 16 and 18 [Scheffner et al., 1993]. E6 is a viral oncogenic protein that forms a complex with E6-AP and then this E6/E6-AP complex binds to p53 and facilitates its ubiquitination and degradation through the UPP [Scheffner et al., 1993]. Although the binding of E6-AP to E6 was required to induce ubiquitination of p53, it was not clear whether E6-AP was actively involved in the ubiquitination of p53. In 1993, Martin et al. demonstrated that the ubiquitination of p53 requires the E6/E6-AP complex and that this complex provides the E3 ligase activity [Scheffner et al., 1993]. Later, the same group also showed that E6-AP has ubiquitin-protein ligase activity in the absence of viral E6 protein. In addition to E6-AP, a variety of other E3-ligases have also been shown to promote p53 degradation including Mdm2 (murine double minute 2), COP1 (constitutively photomorphogenic 1), PIRH2 (p53-induced-RING-H2), etc [Dornan et al., 2004; Leng et al., 2003; Li et al., 2003]. Huibregtse et al. identified two E6-AP homologous; a 100 kDa protein in rat and a 95 kDa protein in yeast (RSP5) with E3-ubiquitin protein ligase activity [Huibregtse et al., 1995].
In 1999, Nawaz et al. demonstrated that E6-AP can also act as a coactivator for steroid hormone receptors (SHRs) such as progesterone receptor (PR), estrogen receptor (ER), androgen receptor (AR), glucocorticoid receptor (GR), retinoic acid receptor-α (RAR-α) and thyroid hormone receptor (TR). It was also shown that the ubiquitin ligase function of E6-AP is not required for the coactivation function of E6-AP and the hect domain of E6-AP is not necessary for the ability of E6-AP to interact with SHR and coactivate SHR function. This indicated that E6-AP possesses two independent, separable functions; a coactivation function and an ubiquitin-protein ligase activity [Nawaz et al., 1999b]. The same year, it was also shown that a genetic disorder, Angelman Syndrome (AS), is caused by the absence of a functional maternal copy of the UBE3A gene that codes for E6-AP [Kishino et al., 1997; Sutcliffe et al., 1997]. It was concluded that the loss of ligase activity, and not the coactivator function, is responsible for the clinical manifestation of this syndrome [Nawaz et al., 1999b].
Structure and organization of the UBE3A/E6-AP gene
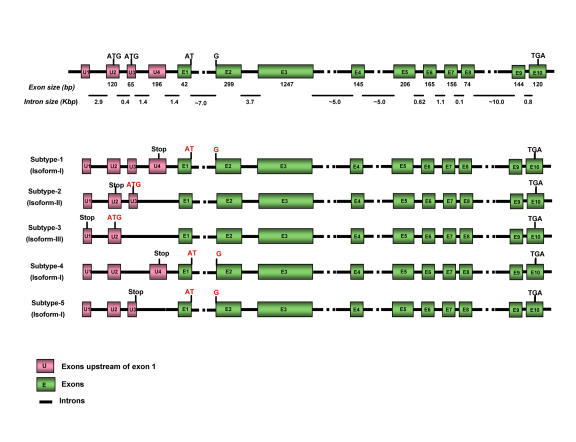
The UBE3A gene that encodes E6-AP is located in the q11-q13 region of chromosome 15 in humans and the UBE3A locus is confined to chromosome 7 in mouse; 7 28.65 cM (http://source.stanford.edu). The E6-AP coding region extends over 60 kilo base pairs (kb) and the length of E6-AP mRNA is approximately 5 kb, with 10 exons. Characterization of the E6-AP gene structure and E6-AP cDNAs has shown that there are at least four different mRNA variants. Sequence analysis revealed five mRNA subtypes 1-5; these subtypes vary in their upstream exons (U1-U4) due to differential splicing. Subtype-1 cDNA contains exons U1, U2, U3 and U4, whereas subtype-2 cDNA lacks exon U4. In subtype-3, exons U3 and U4 are skipped and a potential start codon appears within exon U2. mRNA subtypes 4 and 5 lack exon U4 and exon U3, respectively. These five different cDNA subtypes code for three potential E6-AP protein isoforms (I, II and III), differing in size by as much as 22 amino acids. E6-AP isoform I is coded by cDNA subtypes 1, 4 and 5, while E6-AP isoforms II and III are coded by cDNA subtypes 2 and 3, respectively. Isoform I corresponds to the open reading frame for E6-AP, while isoforms II and III have an additional 20 and 23 amino acids, respectively, at their amino-termini. These additional amino acids do not affect the known functions of E6-AP. The cDNA subtype 2 encodes the longest isoform of the E6-AP protein, isoform II (Figure 1). The physiological function of the different protein isoforms is not yet known. The hect domain is encoded by the last eight exons starting from the 3’ region of exon 3 through exon 10. The E6-binding site is encoded by exon 3 and the active site cysteine residue, which accepts ubiquitin from the E2 ubiquitin-conjugating enzyme, is encoded within exon 10 [Kishino and Wagstaff, 1998; Yamamoto et al., 1997].
Figure 1. Gene structure and exon organization of E6-AP.
(A) The genomic structure of the coding region of E6-AP. The 10 exons that were reported in the first E6-AP gene structure study are indicated by closed boxes in green and numbered E1 to E10. The exons upstream of exon 1 are indicated by closed boxes in pink and are numbered U1 to U4. The three translation initiation sites (ATG) are shown. (B) A set of five E6-AP mRNAs with their potential protein isoforms (isoforms I, II, and III) that differ at their extreme amino-termini. 3’-region of exon 3 and exons 4-10 encode the hect domain. The E6-binding site is encoded within exon 3, and the active site cysteine is encoded within exon 10 (adapted from (Yamamoto et al., 1997)).
Two E6-AP (UBE3A) pseudogenes, UBE3AP1 and UBE3AP2, have also been identified. Both pseudogene sequences revealed a 90-95% nucleotide identity to that of E6-AP. UBE3AP1 and UBE3AP2 were mapped to chromosomes 2 and 21, respectively, but there is no evidence for the expression of these pseudogenes [Kishino and Wagstaff, 1998].
Functional domains of E6-AP and E6-AP interacting proteins
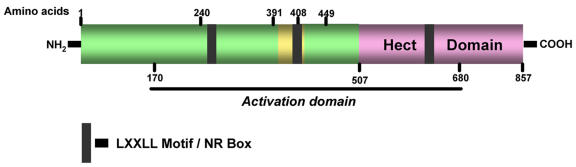
Human E6-AP consists of 865 amino acids, while the murine homolog is slightly longer with 885 amino acids, and these two proteins are 99% similar. E6-AP possesses five well-characterized functional domains: (i) a hect domain, (ii) an E6 binding domain, (iii) a p53 binding domain, (iv) three nuclear receptor interaction domains and (v) an activation domain [Huibregtse et al., 1993; Nawaz et al., 1999b].
The carboxy terminal 350 amino acids form the conserved hect domain of E6-AP, a region of homology shared by several proteins structurally and functionally related to E6-AP [Huibregtse et al., 1995]. The hect domain of E6-AP has a 40 kDa conserved carboxy-terminal catalytic domain that has at least four biochemical functions: 1) it binds to specific E2 ubiquitin-conjugating enzymes like UbcH7, UbcH5B and UbcH8; 2) it accepts ubiquitin from E2 enzyme, forming a ubiquitin-thioester intermediate with its active site cysteine [Kumar et al., 1997]. A conserved cysteine residue at position 833 in the hect domain confers the ubiquitin ligase activity of E6-AP. In vitro studies have shown that the mutation of cysteine 833 residue to alanine or serine renders it unable to form a thioester bond with ubiquitin; 3) it transfers ubiquitin to the ε-amino groups of lysine side chain on the substrate by catalyzing the formation of an isopeptide bond; and 4) it transfers additional ubiquitin molecules to the growing end of the multi-ubiquitin chain [Huang et al., 1999; Scheffner et al., 1995].
E6-AP has been shown to function as an E3 ligase in the ubiquitination of p53 in cooperation with HPV E6 protein [Cooper et al., 2003]. The viral E6 protein acts as an adaptor between p53 and E6-AP [Huibregtse et al., 1993]. This E6-dependent binding of p53 to E6-AP involves amino acids 280-781 of E6-AP, which form the central leucine-rich core that is crucial for the biological function and structural stability of E6-AP. An 18 amino acid region of E6-AP, from 391-408, has been shown to be necessary and sufficient for binding to E6 [Zanier et al., 2005]. This sequence was later characterized as a leucine-rich peptide, LQELL, which is a signature LXXLL motif (a receptor interacting motif) [Cooper et al., 2003; Kishino et al., 1997; Scheffner et al., 1993; Talis et al., 1998]. An essential intermediate step in E6-AP-dependent ubiquitination is the formation of a thioester complex between E6-AP and ubiquitin-conjugating enzymes. In the case of ubiquitin-conjugating enzyme UbcH5B, the implicated binding site corresponding to amino acids 521-679 of E6-AP, falls within the central leucine-rich core of E6-AP (Figure 2). The region of E6-AP involved in complex formation with UbcH7 and UbcH8 was mapped to its hect domain [Huang et al., 1999; Scheffner et al., 1994]. Recently, Mani et al. identified AIB1 (amplified in breast cancer 1)/SRC-3 (steroid receptor coactivator-3) as a substrate for E6-AP-mediated ubiquitination and established that the steady-state level of AIB1 is regulated by E6-AP. The interaction between E6-AP and AIB1 is mediated through the carboxy terminus of AIB1 [Mani et al., 2006].
Figure 2. Schematic representation of the functional domains of E6-AP.
E6-AP possesses five well-characterized functional domains: (i) a hect domain, (ii) an E6 binding domain, (iii) a p53 binding domain, (iv) two nuclear receptor interaction domains and (v) an activation domain, which have been represented with the amino acid numbers they correspond to. The three LXXLL motifs (NR box) are shown dispersed throughout the protein. Two of these motifs are located in the amino terminus and the third one is located within the carboxy terminus of the protein.
Since its identification as a coactivator of SHRs, E6-AP has been shown to interact with a variety of other proteins, which includes SHRs and other coregulators. E6-AP contains 3 consensus receptor-interacting LXXLL motifs. Two of these motifs are located in the amino terminus and the third one is located within the carboxy terminus of the protein. After its characterization as a PR-interacting protein, E6-AP has also been shown to interact with other SHRs, such as AR and ER, and these interactions are enhanced in the presence of appropriate hormone [Dhananjayan et al., 2006; Khan et al., 2006]. Nawaz et al. showed that E6-AP not only interacts with different SHRs, it also contains an intrinsic, transferable activation domain that extends from amino acids 170 to 680 [Nawaz et al., 1999b]. In addition to SHRs, Dhananjayan et al. have recently shown that E6-AP also interacts with a WW-domain binding protein -2 (WBP-2), and further investigation revealed that E6-AP and WBP-2 had additive effect on ER and PR coactivation when both of them were coexpressed [Dhananjayan et al., 2006].
Expression and sub-cellular localization
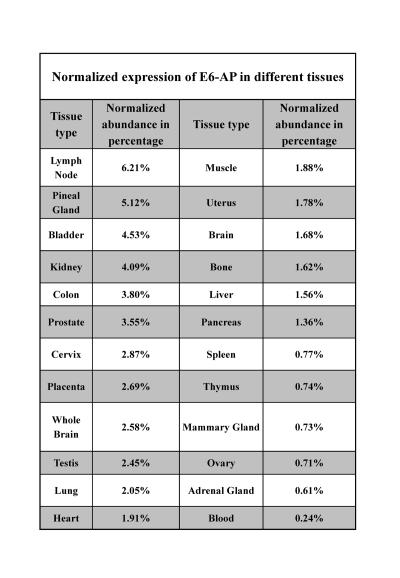
E6-AP is ubiquitously expressed in various tissues (Table 1) and (http://source.stanford.edu). The E6-AP protein was found to be predominantly localized in the cytoplasm, and to a lesser extent in the nucleus, by using fluorescence microscopy [Vaeteewoottacharn et al., 2005]. A recent study using Ube3aYFP knock-in indicated that maternal-derived Ube3a was expressed in most brain regions, including hippocampus, cortex, thalamus, olfactory bulb and cerebellum [Dindot et al., 2008]. The paternal Ube3a is expressed only at a basal level in these brain regions. They also showed that the maternally-derived E6-AP localizes to the nucleus, growth cone and pre- and postsynaptic compartments in the hippocampal neurons, which implies that E6-AP may regulate the development and/or function of synapses [Dindot et al., 2008].
Table 1. Normalized expression of E6-AP in different tissues.
Normalized expression represents the relative expression of E6-AP level in different tissues and is "normalized" for the number of clones from each tissue that are included in UniGene.
Function of E6-AP in the cell
The two well-characterized functions of E6-AP in the cell are 1) regulation of SHR-dependent gene transcription and 2) an E3 ubiquitin ligase activity.
Role of E6-AP in transcription
After its cloning and characterization as a PR-interacting protein, E6-AP has been shown to interact with and coactivate the hormone-dependent transactivation function of several steroid hormone receptors such as ER, AR, PR, GR, TR and RAR. Furthermore, E6-AP reverses the squelching between ER and PR, and it also contains an intrinsic activation function in its amino-terminal domain [Nawaz et al., 1999b]. Additionally, it has been shown that the catalytic function located within the hect domain of E6-AP is not necessary for the ability of E6-AP to interact with and coactivate steroid hormone receptor functions.
Chromatin immunoprecipitation (ChIP) assay is used to reveal the recruitment of transcription factors and coregulators to their target genes and to study the time-dependent recruitment of the transcription machinery to the promoter regions of target genes. E6-AP has been shown to be recruited to the promoter region of the AR-responsive gene, PSA, in a hormone-dependent manner [Khan et al., 2006]. Furthermore, ChIP experiments have indicated that E6-AP is recruited to the estrogen-responsive pS2 promoter and that E6-AP is cyclically associated with the pS2 promoter with a periodicity of 40 min. Re-ChIP analysis of the complex showed that E6-AP is associated with the p300-CBP/SRC-1 complex at the pS2 promoter [Reid et al., 2003]. This strongly suggests that E6-AP is recruited to the hormone-responsive promoters in vivo and regulates hormone-dependent gene expression.
Recent studies have demonstrated that E6-AP is required for transactivation of the human telomerase reverse transcriptase (hTERT) promoter and is recruited to this promoter by the human papilloma virus E6 oncoprotein. In normal cells, telomerase activity is regulated by transcriptional control of the hTERT promoter [Liu et al., 2005]. It has been found that in differentiated epithelial cells that have lost anchorage, hTERT is transcriptionally repressed by the recruitment of histone deacetylase to the hTERT promoter. Inhibition of HDAC1 and induction of histone acetylation is sufficient for activation of hTERT promoter activity in normal human cells. In 2006, James et al. provided evidence that the hTERT promoter is activated by viral E6 protein in association with acetylation of histone H3 at the hTERT promoter. Acetylation of H3 and activation of the hTERT promoter by E6 is dependent on E6-AP and is enhanced by knockdown of p300 (E1A binding protein p300). More importantly, E6-AP knock-out mouse cells and small interfering RNA-mediated knock-down of E6-AP in human cell lines demonstrated that E6-AP is required for E6-mediated hTERT promoter transactivation in both mouse and human cells [James et al., 2006].
Role of E6-AP as an ubiquitin protein ligase
E6-AP was originally discovered as an ubiquitin ligase that interacts with the E6 protein of human papilloma virus and, E6/E6-AP complex targets p53 for ubiquitination and degradation via the UPP [Scheffner et al., 1993]. Since then, numerous other cellular proteins have been reported to interact with E6, and some of these have also been reported to be targeted for degradation in an E6-AP-dependent manner. Some examples include BAK (Bcl-2 homologous antagonist/killer), c-MYC, Mcm-7 (minichromosome maintenance protein 7) and hScrib (the human analogue of the Drosophila Scribble tumor suppressor protein) [Liu et al., 2007; Liu et al., 2005; Oda et al., 1999]. In an attempt to determine if E6-AP could function as an ubiquitin ligase independent of E6, Kumar et al. screened for E6-independent E6-AP substrates that were marked for degradation. In this screen they showed that a DNA repair protein, HHR23A (human homologue of the yeast DNA repair protein RAD23), interacts with E6-AP and is efficiently ubiquitinated in an E6-AP-dependent, but E6-independent manner in vitro. Furthermore, their data also suggested that E6-AP-mediated ubiquitination of HHR23A may be important in regulating its function in DNA repair and cell cycle progression, suggesting a physiological role for E6-AP-mediated ubiquitination [Kumar et al., 1997; Kumar et al., 1999].
Subsequently, it was shown that E6-AP could target itself for degradation in vitro [Nuber et al., 1998]. This was corroborated in vivo by the observation that under conditions of overexpression of E6-AP, it efficiently promoted its own poly-ubiquitination and degradation via the UPP. Such self-ubiquitination has been proposed to result from inter-molecular transfer of ubiquitin on E6-AP molecules that are not bound to substrate proteins. This may be a regulatory mechanism to control the intracellular levels of E6-AP by inactivating and degrading unbound E6-AP molecules [Kao et al., 2000; Nuber et al., 1998].
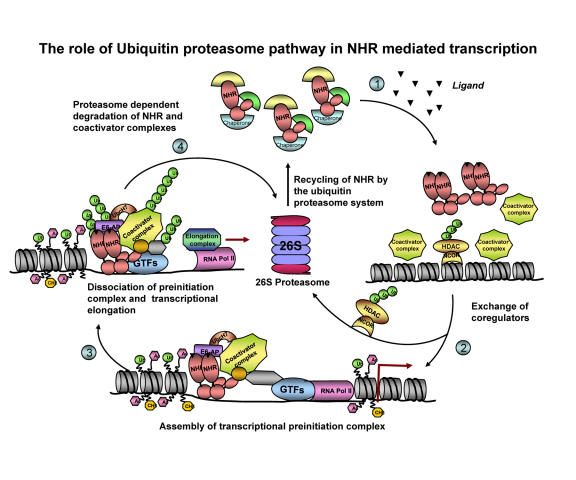
The characterization of E6-AP as a coactivator for SHRs, along with the observations that several nuclear receptors are ubiquitinated and degraded in the course of their transcriptional activities, suggests that ubiquitin proteasome-mediated protein degradation plays a crucial role in eukaryotic transcription (Figure 3) [Alarid, 2006; Ismail and Nawaz, 2005]. The evidence implicating the 26S proteasome in the control of nuclear receptor stability was provided by proteasome inhibition studies using MG132, lactacystin, and other proteasome inhibitors. Proteasome inhibition significantly diminished the ligand-induced transcriptional activity of most of the NRs, which includes AR, ER, PR, RAR-α, TR, peroxisome proliferator-activated receptor (PPAR) and retinoid X receptor (RXR) [Alarid, 2006; Ismail and Nawaz, 2005; Lonard et al., 2000; Nawaz et al., 1999a]. Because of the broad cellular effects of proteasome inhibitors, it is difficult to directly link their functional outcome to the stability of the receptor alone. Thus, considerable effort is being focused on determining the specific E3 ligases involved in targeting the receptors for proteolysis. Although the identity of the E3 ligase(s) involved in ER-α proteolysis has remained controversial and elusive, there is evidence correlating ER-α and AR protein stability to the level of E6-AP expression [Gao et al., 2005; Khan et al., 2006]. The interaction of E6-AP with the ER-α and AR and its recruitment to their target gene promoters supports the idea that E6-AP might be one of the E3 ligases responsible for the ubiquitination and degradation of ER-α and AR [Dhananjayan et al., 2006; Khan et al., 2006; Reid et al., 2003]. This notion was confirmed in the prostate glands of E6-AP null mice, where, although the level of AR protein was elevated, the level of an AR target protein, Probasin, was decreased. Despite the identification of E6-AP as a modulator of receptor transcription function, its activity in relation to ubiquitination and proteolysis of receptors is yet to be determined.
Figure 3. Proposed mechanism for the dynamic assembly of nuclear receptors and their coregulators during transcriptional activation.
Step 1: In the absence of hormone, the NRs are in their inactive form, bound to chaperone proteins. In the presence of hormone, the NRs undergo conformational changes that result in the release of chaperones, which facilitate receptor dimerization and their recruitment to the promoter. Step 2: The negative regulators are cleared of the promoter via the ubiquitin proteasome pathway, followed by recruitment of coactivator complexes, general transcription factors (GTFs) and RNA polymerase II (Pol II) to the target gene promoter (Perissi et al., 2004). Some of the coactivators recruited to the promoter include ubiquitin proteasome enzymes like E6-AP and UBCH7 (Verma et al., 2004). Step 3: The preinitiation complex disassembles and the RNA Pol II escapes from the promoter, leading to transcriptional elongation. Step 4: Then the ubiquitin proteasome pathway degrades the nuclear receptor-transcription complex. According to this model, the active transcription depends on continuous reloading and degradation of the nuclear receptor-transcription complex, thus the ubiquitin-proteasome-dependent degradation and receptor-dependent transactivation are linked.
In 2006, Mani et al. reported the requirement of the E6-AP for the degradation of a nuclear receptor coactivator, AIB1 [Mani et al., 2006]. This study raises the intriguing possibility that the clearance of coactivators from the enhancer/promoter may enable the sequential interaction of an activated receptor with a newly-assembled coactivator complex [Nawaz and O'Malley, 2004].
All these examples show that E6-AP, a component of the ubiquitin-proteasome pathway, could orchestrate the dynamics of NR-mediated transcription by regulating the degradation of the transcriptional complexes [Rochette-Egly, 2005].
Biological role of E6-AP
The physiological role of E6-AP was examined using mice lacking the E6-AP gene (E6-APKO) [Jiang et al., 1998]. E6-APKO mice are viable, but exhibit several interesting phenotypic changes. Postnatal viability is reduced in E6-AP null homozygotes. Heterozygous mice lacking a maternal copy of E6-AP exhibit numerous adverse symptoms such as motor dysfunction and inducible seizures, reminiscent of AS in humans. In addition, in these mice, the cytoplasmic levels of p53 are grossly elevated in Purkinje cells and some hippocampal pyramidal neurons, which may be responsible for the observed deficiencies in context-dependent learning and hippocampal long-term potentiation. These results suggest that E6-AP is required for the proper development of neuronal synapses in the central nervous system [Jiang et al., 1998; Kishino et al., 1997].
Smith et al. have shown that E6-AP KO mice of both sexes are less fertile than wild type mice, due to the reduced size of their gonads. Consistent with this observation, defects in sperm production and ovulation were also seen. Moreover, the hormonal induction of growth of the prostate and uterine glands by testosterone and estradiol was significantly reduced in these mice [Smith et al., 2002]. Recent studies from our laboratory have reported that the deletion of E6-AP in male mice attenuates the growth and development of the prostate gland. This effect was brought about by a partial loss of signaling in the PI3K (phosphoinositide 3-kinase)/Akt mitogenic pathway, which is regulated by E6-AP. The levels of PI3K and both total and phosphorylated Akt are much reduced in E6-AP null prostates. In contrast, the steady state level of RhoA, which negatively regulates AR function and PI3K-Akt signaling, was shown to be elevated in prostate tissue. Furthermore, the levels of p53 and apoptosis-inducing proteins such as Bax, and active caspases are increased in the E6-AP null prostate, accompanied by an increase in apoptosis. Collectively, these data suggest that E6-AP controls the growth of prostate gland by modulating several major cellular pathways, such as AR signaling, PI3K/AKT signaling pathway, RhoA signaling pathway and p53-mediated apoptosis [Khan et al., 2006]. Taken together, these results implicate E6-AP as an important molecule involved in various aspects of reproduction and therefore extend the physiological role of this protein to tissues outside the central nervous system.
Alteration of E6-AP expression in human pathology
Clinical manifestations of mutation in the UBE3A gene locus
The E6-AP (UBE3A) locus is imprinted in certain regions of the brain such that only the maternal copy of the gene is expressed [Sutcliffe et al., 1997]. Angelman Syndrome is caused by maternal deletion of chromosome 15q11-q13, paternal uniparental disomy (pUPD) of chromosome 15, imprinting defects or by loss of function mutations in the UBE3A locus that encodes E6-AP [Kishino et al., 1997; Sutcliffe et al., 1997]. AS is a severe neurological disorder characterized by moderate to severe mental retardation, absence of speech, tremors, ataxia, abnormal gait, inappropriate laughter, sleep disturbance and seizures [Peters et al., 2004]. There are 29 different mutations in E6-AP that are reported to cause AS, including 17 frame shifts, 5 nonsense, 3 missense and one each of single amino acid insertion, single amino acid deletion, splicing and stop codon [Fang et al., 1999]. Subsequent research showed that there is a strong correlation between AS and loss of E3 ubiquitin ligase activity of E6-AP. Since E6-AP is an ubiquitin ligase, it is likely that the improper regulation of E6-AP substrate(s) causes AS, although no disease-relevant E6-AP targets have yet been identified. However, data from E6-AP knockout mice suggests that p53 might be one of the substrates for E6-AP and loss of p53 degradation due to the lack of E6-AP may lead to the development of AS [Jiang et al., 1998].
The paternal deficiency of the same region, ch15q11-q13, causes Prader-Willi Syndrome (PWS). Like AS, PWS is also characterized by defective neurological, developmental and behavioral phenotypes. PWS is characterized by hypotonia, respiratory distress and small stature [Nicholls and Knepper, 2001].
Deregulated expression of E6-AP in tumorigenesis
Recently, the role of coactivators in tumor development and progression has gained increasing attention. In fact, the overexpression of nuclear receptor coactivators, including AIB1, SRA (steroid receptor RNA activator) and AIB3 (amplified in breast cancer 3) has been implicated in human breast cancers [Gao et al., 2002]. Studies from our lab have demonstrated that the expression of E6-AP protein is decreased in human invasive breast and prostate carcinomas compared with their adjacent normal tissues. This down-regulation of E6-AP is accompanied by the elevation of ER in breast and AR in prostate carcinomas [Gao et al., 2005]. Furthermore, in vivo data from E6-AP-knockout animals indicated that the expression levels of ER and AR are increased in E6-AP-null mammary and prostate glands, respectively, when compared with that of normal control animals, suggesting that E6-AP modulates the protein levels of ER in breast and AR in prostate glands [Gao et al., 2005; Khan et al., 2006].
Conclusion
E6-AP was one of the ubiquitin proteasome pathway enzymes that were characterized as a coactivator of NRs. The interaction of E2-ubiquitin conjugating enzyme, E3-ubiquitin ligase enzyme and components of the proteasomal subunit with the transcription machinery and their recruitment to the promoter reinforces the hypothesis that the degradation of nuclear receptors could be linked to transcriptional activation and may be necessary for efficient transcriptional activity. Although a significant amount of work has been done on E6-AP to determine its mechanism of action and to establish its biological relevance, several critical questions still remain unanswered. Further work is warranted to unravel the mechanism of action of E6-AP and to understand its function in SHR signaling and tumorigenesis.
Acknowledgments
We thank Pavan Vaidyanathan, Sarath Dhananjayan and Sathish Srinivasan for critical reading of the manuscript.
Abbreviations
- AIB1
amplified in breast cancer 1
- AR
androgen receptor
- AS
Angelman Syndrome
- ChIP
chromatin immunoprecipitation
- COP1
constitutively photomorphogenic 1
- E6-AP
E6-associated protein
- ER
estrogen receptor
- GR
glucocorticoid receptor
- HDAC
histone deacetylase
- hect
homologous to E6-associated protein carboxy-terminal domain
- HHR23A
human homologue of the yeast DNA repair protein RAD23
- HPV
human papilloma virus
- hTERT
human telomerase reverse transcriptase
- Mdm2
murine double minute 2
- NR
nuclear receptor
- PIRH2
p53-induced-RING-H2
- PR
progesterone receptor
- RAR
retinoic acid receptor
- SHR
steroid hormone receptor
- SRC-3
steroid receptor coactivator 3
- TR
thyroid hormone receptor
- UPP
ubiquitin proteasome pathway
- WBP-2
WW domain binding protein-2;
References
- Alarid E. T. Lives and times of nuclear receptors. Mol Endocrinol. 2006;20:1972–81. doi: 10.1210/me.2005-0481. [DOI] [PubMed] [Google Scholar]
- Cooper B., Schneider S., Bohl J., Jiang Y., Beaudet A., Vande Pol S. Requirement of E6AP and the features of human papillomavirus E6 necessary to support degradation of p53. Virology. 2003;306:87–99. doi: 10.1016/s0042-6822(02)00012-0. [DOI] [PubMed] [Google Scholar]
- Dhananjayan S. C., Ramamoorthy S., Khan O. Y., Ismail A., Sun J., Slingerland J., O'Malley B. W., Nawaz Z. WW domain binding protein-2, an E6-associated protein interacting protein, acts as a coactivator of estrogen and progesterone receptors. Mol Endocrinol. 2006;20:2343–54. doi: 10.1210/me.2005-0533. [DOI] [PubMed] [Google Scholar]
- Dindot S. V., Antalffy B. A., Bhattacharjee M. B., Beaudet A. L. The Angelman syndrome ubiquitin ligase localizes to the synapse and nucleus, and maternal deficiency results in abnormal dendritic spine morphology. Hum Mol Genet. 2008;17:111–8. doi: 10.1093/hmg/ddm288. [DOI] [PubMed] [Google Scholar]
- Dornan D., Wertz I., Shimizu H., Arnott D., Frantz G. D., Dowd P., O'Rourke K., Koeppen H., Dixit V. M. The ubiquitin ligase COP1 is a critical negative regulator of p53. Nature. 2004;429:86–92. doi: 10.1038/nature02514. [DOI] [PubMed] [Google Scholar]
- Fang P., Lev-Lehman E., Tsai T. F., Matsuura T., Benton C. S., Sutcliffe J. S., Christian S. L., Kubota T., Halley D. J., Meijers-Heijboer H., Langlois S., Graham J. M., Jr., Beuten J., Willems P. J., Ledbetter D. H., Beaudet A. L. The spectrum of mutations in UBE3A causing Angelman syndrome. Hum Mol Genet. 1999;8:129–35. doi: 10.1093/hmg/8.1.129. [DOI] [PubMed] [Google Scholar]
- Gao X., Mohsin S. K., Gatalica Z., Fu G., Sharma P., Nawaz Z. Decreased expression of e6-associated protein in breast and prostate carcinomas. Endocrinology. 2005;146:1707–12. doi: 10.1210/en.2004-1198. [DOI] [PubMed] [Google Scholar]
- Gao X., Loggie B. W., Nawaz Z. The roles of sex steroid receptor coregulators in cancer. Mol Cancer. 2002;1:7. doi: 10.1186/1476-4598-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L., Kinnucan E., Wang G., Beaudenon S., Howley P. M., Huibregtse J. M., Pavletich N. P. Structure of an E6AP-UbcH7 complex: insights into ubiquitination by the E2-E3 enzyme cascade. Science. 1999;286:1321–6. doi: 10.1126/science.286.5443.1321. [DOI] [PubMed] [Google Scholar]
- Huibregtse J. M., Scheffner M., Beaudenon S., Howley P. M. A family of proteins structurally and functionally related to the E6-AP ubiquitin-protein ligase. Proc Natl Acad Sci U S A. 1995;92:2563–7. doi: 10.1073/pnas.92.7.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huibregtse J. M., Scheffner M., Howley P. M. Localization of the E6-AP regions that direct human papillomavirus E6 binding, association with p53, and ubiquitination of associated proteins. Mol Cell Biol. 1993;13:4918–27. doi: 10.1128/mcb.13.8.4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail A., Nawaz Z. Nuclear hormone receptor degradation and gene transcription: an update. IUBMB Life. 2005;57:483–90. doi: 10.1080/15216540500147163. [DOI] [PubMed] [Google Scholar]
- James M. A., Lee J. H., Klingelhutz A. J. HPV16-E6 associated hTERT promoter acetylation is E6AP dependent, increased in later passage cells and enhanced by loss of p300. Int J Cancer. 2006;119:1878–85. doi: 10.1002/ijc.22064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y. H., Armstrong D., Albrecht U., Atkins C. M., Noebels J. L., Eichele G., Sweatt J. D., Beaudet A. L. Mutation of the Angelman ubiquitin ligase in mice causes increased cytoplasmic p53 and deficits of contextual learning and long-term potentiation. Neuron. 1998;21:799–811. doi: 10.1016/s0896-6273(00)80596-6. [DOI] [PubMed] [Google Scholar]
- Kao W. H., Beaudenon S. L., Talis A. L., Huibregtse J. M., Howley P. M. Human papillomavirus type 16 E6 induces self-ubiquitination of the E6AP ubiquitin-protein ligase. J Virol. 2000;74:6408–17. doi: 10.1128/jvi.74.14.6408-6417.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan O. Y., Fu G., Ismail A., Srinivasan S., Cao X., Tu Y., Lu S., Nawaz Z. Multifunction steroid receptor coactivator, E6-associated protein, is involved in development of the prostate gland. Mol Endocrinol. 2006;20:544–59. doi: 10.1210/me.2005-0110. [DOI] [PubMed] [Google Scholar]
- Kishino T., Wagstaff J. Genomic organization of the UBE3A/E6-AP gene and related pseudogenes. Genomics. 1998;47:101–7. doi: 10.1006/geno.1997.5093. [DOI] [PubMed] [Google Scholar]
- Kishino T., Lalande M., Wagstaff J. UBE3A/E6-AP mutations cause Angelman syndrome. Nat Genet. 1997;15:70–3. doi: 10.1038/ng0197-70. [DOI] [PubMed] [Google Scholar]
- Kumar S., Talis A. L., Howley P. M. Identification of HHR23A as a substrate for E6-associated protein-mediated ubiquitination. J Biol Chem. 1999;274:18785–92. doi: 10.1074/jbc.274.26.18785. [DOI] [PubMed] [Google Scholar]
- Kumar S., Kao W. H., Howley P. M. Physical interaction between specific E2 and Hect E3 enzymes determines functional cooperativity. J Biol Chem. 1997;272:13548–54. doi: 10.1074/jbc.272.21.13548. [DOI] [PubMed] [Google Scholar]
- Leng R. P., Lin Y., Ma W., Wu H., Lemmers B., Chung S., Parant J. M., Lozano G., Hakem R., Benchimol S. Pirh2, a p53-induced ubiquitin-protein ligase, promotes p53 degradation. Cell. 2003;112:779–91. doi: 10.1016/s0092-8674(03)00193-4. [DOI] [PubMed] [Google Scholar]
- Li M., Brooks C. L., Wu-Baer F., Chen D., Baer R., Gu W. Mono- versus polyubiquitination: differential control of p53 fate by Mdm2. Science. 2003;302:1972–5. doi: 10.1126/science.1091362. [DOI] [PubMed] [Google Scholar]
- Liu X., Disbrow G. L., Yuan H., Tomaic V., Schlegel R. Myc and human papillomavirus type 16 E7 genes cooperate to immortalize human keratinocytes. J Virol. 2007;81:12689–95. doi: 10.1128/JVI.00669-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Yuan H., Fu B., Disbrow G. L., Apolinario T., Tomaic V., Kelley M. L., Baker C. C., Huibregtse J., Schlegel R. The E6AP ubiquitin ligase is required for transactivation of the hTERT promoter by the human papillomavirus E6 oncoprotein. J Biol Chem. 2005;280:10807–16. doi: 10.1074/jbc.M410343200. [DOI] [PubMed] [Google Scholar]
- Lonard D. M., Nawaz Z., Smith C. L., O'Malley B. W. The 26S proteasome is required for estrogen receptor-α and coactivator turnover and for efficient estrogen receptor-α transactivation. Mol Cell. 2000;5:939–48. doi: 10.1016/s1097-2765(00)80259-2. [DOI] [PubMed] [Google Scholar]
- Mani A., Oh A. S., Bowden E. T., Lahusen T., Lorick K. L., Weissman A. M., Schlegel R., Wellstein A., Riegel A. T. E6AP mediates regulated proteasomal degradation of the nuclear receptor coactivator amplified in breast cancer 1 in immortalized cells. Cancer Res. 2006;66:8680–6. doi: 10.1158/0008-5472.CAN-06-0557. [DOI] [PubMed] [Google Scholar]
- Nawaz Z., Lonard D. M., Dennis A. P., Smith C. L., O'Malley B. W. Proteasome-dependent degradation of the human estrogen receptor. Proc Natl Acad Sci U S A. 1999b;96:1858–62. doi: 10.1073/pnas.96.5.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawaz Z., Lonard D. M., Smith C. L., Lev-Lehman E., Tsai S. Y., Tsai M. J., O'Malley B. W. The Angelman syndrome-associated protein, E6-AP, is a coactivator for the nuclear hormone receptor superfamily. Mol Cell Biol. 1999a;19:1182–9. doi: 10.1128/mcb.19.2.1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawaz Z., O'Malley B. W. Urban renewal in the nucleus: is protein turnover by proteasomes absolutely required for nuclear receptor-regulated transcription? Mol Endocrinol. 2004;18:493–9. doi: 10.1210/me.2003-0388. [DOI] [PubMed] [Google Scholar]
- Nicholls R. D., Knepper J. L. Genome organization, function, and imprinting in Prader-Willi and Angelman syndromes. Annu Rev Genomics Hum Genet. 2001;2:153–75. doi: 10.1146/annurev.genom.2.1.153. [DOI] [PubMed] [Google Scholar]
- Nuber U., Schwarz S. E., Scheffner M. The ubiquitin-protein ligase E6-associated protein (E6-AP) serves as its own substrate. Eur J Biochem. 1998;254:643–9. doi: 10.1046/j.1432-1327.1998.2540643.x. [DOI] [PubMed] [Google Scholar]
- Oda H., Kumar S., Howley P. M. Regulation of the Src family tyrosine kinase Blk through E6AP-mediated ubiquitination. Proc Natl Acad Sci U S A. 1999;96:9557–62. doi: 10.1073/pnas.96.17.9557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters S. U., Goddard-Finegold J., Beaudet A. L., Madduri N., Turcich M., Bacino C. A. Cognitive and adaptive behavior profiles of children with Angelman syndrome. Am J Med Genet A. 2004;128:110–3. doi: 10.1002/ajmg.a.30065. [DOI] [PubMed] [Google Scholar]
- Pickart C. M. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001;70:503–33. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- Reid G., Hubner M. R., Metivier R., Brand H., Denger S., Manu D., Beaudouin J., Ellenberg J., Gannon F. Cyclic, proteasome-mediated turnover of unliganded and liganded ERalpha on responsive promoters is an integral feature of estrogen signaling. Mol Cell. 2003;11:695–707. doi: 10.1016/s1097-2765(03)00090-x. [DOI] [PubMed] [Google Scholar]
- Rochette-Egly C. Dynamic combinatorial networks in nuclear receptor-mediated transcription. J Biol Chem. 2005;280:32565–8. doi: 10.1074/jbc.R500008200. [DOI] [PubMed] [Google Scholar]
- Salvat C., Wang G., Dastur A., Lyon N., Huibregtse J. M. The -4 phenylalanine is required for substrate ubiquitination catalyzed by HECT ubiquitin ligases. J Biol Chem. 2004;279:18935–43. doi: 10.1074/jbc.M312201200. [DOI] [PubMed] [Google Scholar]
- Scheffner M., Huibregtse J. M., Howley P. M. Identification of a human ubiquitin-conjugating enzyme that mediates the E6-AP-dependent ubiquitination of p53. Proc Natl Acad Sci U S A. 1994;91:8797–801. doi: 10.1073/pnas.91.19.8797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffner M., Nuber U., Huibregtse J. M. Protein ubiquitination involving an E1-E2-E3 enzyme ubiquitin thioester cascade. Nature. 1995;373:81–3. doi: 10.1038/373081a0. [DOI] [PubMed] [Google Scholar]
- Scheffner M., Huibregtse J. M., Vierstra R. D., Howley P. M. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell. 1993;75:495–505. doi: 10.1016/0092-8674(93)90384-3. [DOI] [PubMed] [Google Scholar]
- Smith C. L., DeVera D. G., Lamb D. J., Nawaz Z., Jiang Y. H., Beaudet A. L., O'Malley B. W. Genetic ablation of the steroid receptor coactivator-ubiquitin ligase, E6-AP, results in tissue-selective steroid hormone resistance and defects in reproduction. Mol Cell Biol. 2002;22:525–35. doi: 10.1128/MCB.22.2.525-535.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe J. S., Jiang Y. H., Galijaard R. J., Matsuura T., Fang P., Kubota T., Christian S. L., Bressler J., Cattanach B., Ledbetter D. H., Beaudet A. L. The E6-Ap ubiquitin-protein ligase (UBE3A) gene is localized within a narrowed Angelman syndrome critical region. Genome Res. 1997;7:368–77. doi: 10.1101/gr.7.4.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talis A. L., Huibregtse J. M., Howley P. M. The role of E6AP in the regulation of p53 protein levels in human papillomavirus (HPV)-positive and HPV-negative cells. J Biol Chem. 1998;273:6439–45. doi: 10.1074/jbc.273.11.6439. [DOI] [PubMed] [Google Scholar]
- Vaeteewoottacharn K., Chamutpong S., Ponglikitmongkol M., Angeletti P. C. Differential localization of HPV16 E6 splice products with E6-associated protein. Virol J. 2005;2:50. doi: 10.1186/1743-422X-2-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y., Huibregtse J. M., Howley P. M. The human E6-AP gene (UBE3A) encodes three potential protein isoforms generated by differential splicing. Genomics. 1997;41:263–6. doi: 10.1006/geno.1997.4617. [DOI] [PubMed] [Google Scholar]
- Zanier K., Charbonnier S., Baltzinger M., Nomine Y., Altschuh D., Trave G. Kinetic analysis of the interactions of human papillomavirus E6 oncoproteins with the ubiquitin ligase E6AP using surface plasmon resonance. J Mol Biol. 2005;349:401–12. doi: 10.1016/j.jmb.2005.03.071. [DOI] [PubMed] [Google Scholar]