Abstract
Macrophage polarization into M1 or M2 phenotypes dictates the nature, duration, and severity of an inflammatory response. The objective of this study was to examine the role of CC chemokine receptor 4 (CCR4) in macrophage polarization during pulmonary oxidative injury in wild-type [WT (CCR4+/+)] and CCR4-deficient (CCR4−/−) mice. Intrapulmonary administration of bleomycin sulfate provoked lethal inflammatory and fibrotic responses in WT (CCR4+/+) mice, but such responses were absent in CCR4−/− mice. Transcript and protein analyses of alveolar and bone marrow-derived macrophages showed that cells isolated from CCR4−/− mice did not exhibit CCL17-dependent M1 activation in response to bleomycin. Instead, CCR4−/− macrophages showed an M2 phenotype characterized by significantly elevated expression of arginase 1 and FIZZ1 (found in inflammatory zone 1), particularly during the peak of pulmonary inflammation. Compared with WT (CCR4+/+) mice, CCR4−/− mice exhibited a significant increase in the expression of the nonsignaling CC chemokine scavenging receptor D6 in whole lung samples and isolated macrophages. Thus, these results demonstrate that CCL17-dependent activation of CCR4 in macrophages plays a central role in free radical-induced pulmonary injury and repair.
Idiopathic pulmonary fibrosis (IPF) is a fatal, interstitial lung disease characterized by relentless tissue scarring for which there is no effective therapy. The diagnostic lesion of IPF is the fibroblastic foci comprised of a heterogeneous mix of epithelial cells and fibroblasts, which, it is postulated, forms as a result of an inappropriate wound healing response to an unknown injurious agent.1 Although the importance of inflammation in IPF remains controversial, some component of the inflammatory response appears to be necessary for the development and maintenance of fibrogenesis.2 The importance of inflammation in this process is particularly evident during acute exacerbations of IPF, which is characterized by profound inflammatory cell infiltration and fibroblastic proliferation and differentiation.3 Thus, it is imperative to understand the cellular and molecular events that ensue during the inflammatory phase after an injurious insult because these early events have a dramatic effect on the magnitude and duration of the resolution phase.
Chemokines are soluble protein mediators that link inflammation to fibrogenesis through their ability to attract and modulate the activity of a variety of immune and nonimmune cells to sites of injury in need of tissue repair.4 One consequence of chemokine activity in the fibrotic lung appears to be the persistence of a Th2-type cytokine microenvironment, which favors a number of profibrotic events including the proliferation, differentiation, and synthetic properties of fibroblasts, epithelial cells, T cells, and macrophages.5 Specifically regarding macrophages, bronchoalveolar lavage (BAL)-derived human6 and mouse7 macrophages exhibit an alternative activation (or M2) profile, characterized by the increased expression of arginase I (Arg1). M2 macrophages can be distinguished from classically activated macrophages (M1) by their increased expression of Arg1 and found in inflammatory zone 1 (FIZZ1), and by their expression patterns of inducible nitric oxide or nitric oxide synthase 2 (NOS2).8 The precise nature of the stimuli that regulate M1 and M2 activation within the inflamed and fibrosing lung remain to be determined but it was recently shown that CCL17, a CCR4 ligand, is integral to regulating the activation of both subsets of macrophages.8,9 Several studies of clinical samples and animal models of pulmonary fibrosis have implicated CCR4 and CCL17 in fibrosis of the lung and elsewhere.10,11
Because chemokines are potent regulators of effector cell functions, endogenous strategies for modulating chemokine activity exist in nature.12 Silent chemokine receptors that lack signal-transducing properties appear to serve as scavengers of chemokines by competing with signaling chemokine receptors and dampening the inflammatory response.13 Additionally, it is hypothesized that these receptors associate with cellular transport machinery and participate in the neutralization of chemokines at endothelial barriers.14,15 The chemokine receptor D6 is a nonsignaling receptor that undergoes ligand-independent internalization, selectively binds inflammatory CC chemokines, and targets them for intracellular degradation.16 Although structurally similar to other chemokine receptors, it is most homologous to CCR4 and CXCR3, thus binding their ligands with highest affinity.17 In vitro studies using D6−/− mice have established a role for D6 in the resolution of various inflammatory disease models.18,19,20,21
In the present study we investigated the mechanism by which CCR4 regulates fibrogenesis in a bleomycin model of pulmonary fibrosis. Specifically, our study addressed the role of CCR4 in the early oxidative injury induced by bleomycin, and the later role of this chemokine receptor on the fibrotic remodeling process. Our data demonstrate a role for CCL17 in the inflammatory or M1 activation of lung-associated and bone marrow-derived macrophages leading to NOS2 induction and oxidative injury. Moreover, our data implicates the scavenging receptor D6 as a novel component in the regulation of CCL17-mediated macrophage function in the development of bleomycin-induced pulmonary fibrosis.
Materials and Methods
Mice
Specific pathogen-free male C57BL/6 [wild-type (WT), WT (CCR4+/+)] mice (6 to 8 weeks of age) were purchased from Taconic (Germantown, NY). CCR4−/− mice were generated as previously described in detail,22 bred, and housed under specific pathogen-free conditions. The Animal Use Committee at the University of Michigan (Ann Arbor, MI) approved all protocols and experiments described herein.
Bleomycin Model of Pulmonary Inflammation and Fibrosis
WT [WT (CCR4+/+)] and CCR4−/− mice received 0.05 U of bleomycin (Blenoxane, sterile bleomycin sulfate; Bristol-Meyers Pharmaceuticals, Evansville, IN) dissolved in phosphate-buffered saline (∼1.7 U/kg) via an intratracheal injection. Groups of WT (CCR4+/+) and CCR4−/− mice (n = 5 to 10 per time point) were monitored for their survival. Other groups were sacrificed and their lung tissues were analyzed at days 1, 3, 7, and 21 after bleomycin injection. Untreated mice (n = 5) did not receive bleomycin and this time point was designated as day 0.
Hydroxyproline Assay
Left lobe samples from WT (CCR4+/+) and CCR4−/− mice (n = 5/group/time point/experiment) before (ie, day 0) and at day 21 after bleomycin challenge were analyzed for hydroxyproline using a previously described assay.23 Hydroxyproline concentrations were calculated from a hydroxyproline standard curve (0 to 100 μg of hydroxyproline/ml). The hydroxyproline levels in each sample were normalized to the protein (in mg) present in each sample measured by the Bradford protein assay.
Bronchoalveolar Lavage
BAL samples were obtained via the instillation of a total of 5 ml of Hanks’ balanced salt solution plus 5 mmol/L ethylenediaminetetraacetic acid (at 4°C) into each mouse.24 For cytospin preparations, cells were centrifuged at 400 × g for 5 minutes using a Cytospin II (Shandon Scientific, Pittsburgh, PA) and stained with Diff-Quik (Dade Behring Inc., Newark, DE) for viewing under a light microscope at ×40 magnification.
BAL Macrophage Culture
Alveolar macrophages were obtained from the BAL of WT [WT (CCR4+/+)] and CCR4−/− mice before and at various times after intrapulmonary bleomycin challenge. BAL cells were plated in 24-well tissue culture plates at a density of 2.5 × 105 cells/ml in RPMI plus 15% fetal calf serum and incubated for 1 hour at 37°C. Nonadherent cells were discarded and adherent alveolar macrophages were subjected to RNA isolation for quantitative TaqMan polymerase chain reaction (PCR) analysis.
Isolation and Culture of Bone Marrow-Derived Macrophages
Before and at various times after intratracheal bleomycin challenge, macrophages were cultured from the bone marrow cells flushed from femur and tibia bones with cold RPMI 1640. Bone marrow cells from a minimum of five WT (CCR4+/+) or CCR4−/− mice were pooled and cultured in 60-mm3 tissue culture dishes with bone marrow medium containing L-cell supernatant as a source of macrophage colony-stimulating factor. On day 3, fresh bone marrow medium was added to each culture. On days 6 or 7 after the initiation of each bone marrow cell culture, bone marrow-derived macrophages were transferred to 24-well plates at a cell density of 2 × 105 cells/well. After 24 hours, one of the following was added to triplicate wells: treatment media (RPMI 1640 plus 0.5% fetal calf serum), CCL22 (10 ng/ml), or CCL17 (10 ng/ml) and incubated for 24 hours at 37°C. CCL22 and CCL17 (<1.0 EU endotoxin level per 1 μg) were purchased from R&D Systems (Minneapolis, MN). RNA was isolated and purified from each well.
Real-Time TaqMan PCR Analysis
Total RNA was prepared from whole lung samples, alveolar macrophages, and bone marrow-derived macrophages using the TRIzol reagent according to the manufacturer’s directions (Invitrogen, Carlsbad, CA). A total of 1.0 μg (from whole lung) or 2.0 μg (BAL or bone marrow macrophages) RNA was reverse-transcribed into cDNA using M-MLV reverse transcriptase (Invitrogen). The cDNA (1.0 μg) was then amplified by real-time quantitative TaqMan PCR using an ABI Prism 7700 sequence detection system (Applied Biosystems, Foster City, CA). GAPDH was analyzed as an internal control. TaqMan gene expression reagents were used to assay CCL17, CCL22, Arg1, and D6 (Applied Biosystems). SYBR Green Master PCR mix (Applied Biosystems) was used to amplify NOS-2, TLR3, and TLR9. Primers for NOS2 were 5′-CGCAGCTGGGCTGTACCAA-3′ and 5′-TGATGTTTGCTTCGGACATCA-3′, for TLR3 were 5′-CCCAGCTCGATCTTTCCTACA-3′ and 5′-AGGCTTGGGAGATAGGAGAAG-3′, and for TLR9 were 5′-AGCTGAACATGAACGGCATCT-3′ and 5′-TGAGCGTGTACTTGTTGAGCG-3′. Primers used for FIZZ-1 TaqMan analysis were 5′-TCCAGCTAACTATCCCTCCACTGT-3′ and 5′-GGCCCATCTGTTCATAGTCTTGA-3′ and the probe was 6FAM-5′-CGAAGACTCTCTCTTGCT-3′-TAMRA. Whole lung gene expression in WT (CCR4+/+) and CCR4−/− mice is expressed as a fold-increase in transcript expression in bleomycin-challenged lung compared with unchallenged lung. In vitro data are expressed as the fold increase in transcript expression in challenged macrophages compared with macrophages incubated in medium alone. The fold difference in mRNA expression between treatment groups was determined by software developed by Applied Biosystems.
Whole Lung Histological Analysis
Whole lungs from unchallenged (ie, day 0) and bleomycin-challenged mice were fully inflated with 10% formalin, dissected, and placed in fresh formalin for 24 hours. Routine histological techniques were used to paraffin-embed the entire lung, and 5-μm sections of whole lung were stained with hematoxylin and eosin (H&E) or Masson Trichrome.
D6 Immunohistochemistry
Paraffin-embedded whole lung samples were analyzed using routine immunohistochemical techniques for the presence of D6. Goat anti-mouse D6 was obtained from Capralogics (Hardwick, MA) and lungs were stained with the mouse horseradish peroxidase-diaminobenzidine cell and tissue staining kit according to the manufacturer’s instructions (R&D Systems). Other histological samples were immunostained with control antibodies (IgG isotype controls and horseradish peroxidase substrate).
Cytokine and Chemokine Enzyme-Linked Immunosorbent Assay (ELISA) Analysis
CCL2, CCL3, CCL5, CCL6, CCL17, CCL22, CXCL1, CXCL2, CXCL9, CXCL10, interferon-γ, C10, interleukin (IL)-10, IL-12, IP-10, tumor necrosis factor, IL-4, IL-13, and transforming growth factor-β were detected in 50-μl samples of cell-free supernatants from whole lung homogenates using a standardized sandwich ELISA technique (R&D Systems). Cytokine and chemokine levels in each sample were normalized to the protein present in cell-free preparation of each sample measured by the Bradford protein assay.
Flow Cytometric Analysis
BAL cells from WT (CCR4+/+) and CCR4−/− mice at day 1 after bleomycin challenge were stained with the indicated Abs (BD Pharmingen, San Diego, CA) or with the Fluorokine kit containing mouse CCL17 biotin conjugate and avidin-fluorescein isothiocyanate (FITC) according to the manufacture’s instructions (R&D Systems) and analyzed using a FACSCalibur and Cell Quest software (BD Biosciences, San Jose, CA).
Statistical Analysis
All results are expressed as mean ± SEM. The means between groups at different time points were compared by two-way analysis of variance. Individual differences were further analyzed using the unpaired t-test with Welch correction or Tukey-Kramer multiple comparisons test where indicated. Values of P < 0.1 (*), P < 0.01 (**), and P < 0.001 (***) were considered significant.
Results
CCR4−/− Mice Were Completely Protected against the Lethal Effects of Bleomycin-Induced Pulmonary Fibrosis
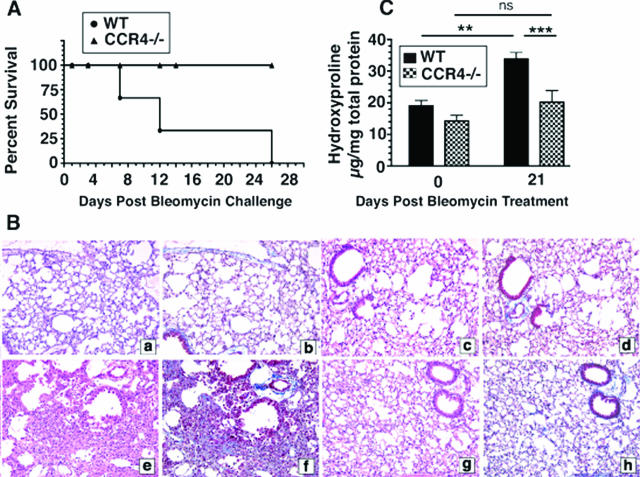
Bleomycin sulfate induces a severe, progressive pulmonary remodeling response that leads to respiratory failure and death. In the present study, age-matched male WT and CCR4−/− mice were challenged intratracheally with 1.7 U/kg of bleomycin dissolved in normal saline.25,26,27 Deaths were first observed at day 7 after bleomycin challenge in the WT group and all of these mice were dead at day 26 after bleomycin. Conversely, no deaths were observed in the CCR4−/− group through to day 26 after bleomycin challenge (Figure 1A). Because all WT mice were dead at day 26, day 21 was selected as the endpoint for an analysis of the pulmonary fibrotic response in both groups of mice. As shown in Figure 1B, WT (Figure 1B, a and b) and CCR4−/− (Figure 1B, c and d) whole lung samples did not differ histologically before bleomycin challenge. In contrast, whole lung samples from WT mice on day 21 after bleomycin exhibited increased inflammation (Figure 1Be) and extracellular matrix deposition (Figure 1Bf) compared with whole lung samples from CCR4−/− mice at the same time after bleomycin (Figure 1B, g and h). These differences in lung histology between WT and CCR4−/− lungs are detectable as early as day 14 after bleomycin, although no striking differences in inflammation were detected on earlier time points (days 1, 3, and 7 after bleomycin; data not shown). Figure 1C summarizes the whole lung hydroxyproline levels detected before and at day 21 after bleomycin challenge. Hydroxyproline levels in whole lung samples from WT mice were significantly increased at day 21 compared with whole lung samples from CCR4−/− mice before bleomycin challenge (Figure 1C). Whole lung samples from CCR4−/− mice exhibited similar hydroxyproline levels to those observed in WT before bleomycin, and levels of hydroxyproline in this group were not elevated above day 0 levels (Figure 1C). More importantly, hydroxyproline levels were significantly lower in CCR4−/− whole lung samples compared with WT whole lung samples at day 21 after bleomycin (Figure 1C). Together, these data demonstrated that CCR4−/− mice were markedly protected from the deleterious effects of systemic bleomycin challenge.
Figure 1.
CCR4−/− mice were protected from the lethal and pulmonary remodeling effects of bleomycin challenge. A: Percent survival of WT versus CCR4−/− mice after bleomycin challenge. B: Histological analysis of lungs and quantification of collagen deposition after bleomycin challenge. Representative H&E-stained histological sections: untreated WT (CCR4+/+) mouse lung (a), untreated CCR4−/− mouse lung (c), WT (CCR4+/+) mouse lung at day 21 after bleomycin (e), CCR4−/− mouse lung at day 21 after bleomycin (g). Representative Masson-trichrome histological sections: untreated WT (CCR4+/+) (b), untreated CCR4−/− mouse lung (d), WT (CCR4+/+) mouse lung at day 21 after bleomycin (f), CCR4−/− mouse lung at day 21 after bleomycin (h). C: Hydroxyproline levels in whole lung homogenates from untreated or bleomycin-challenged mice. Data are mean ± SEM from four to five mice at each time point. ***P < 0.001, **P < 0.01.
CCR4 Deficiency Altered the Transcript and/or Protein Expression of CCL17 and CCL22 in Whole Lung and Macrophages after Bleomycin Challenge
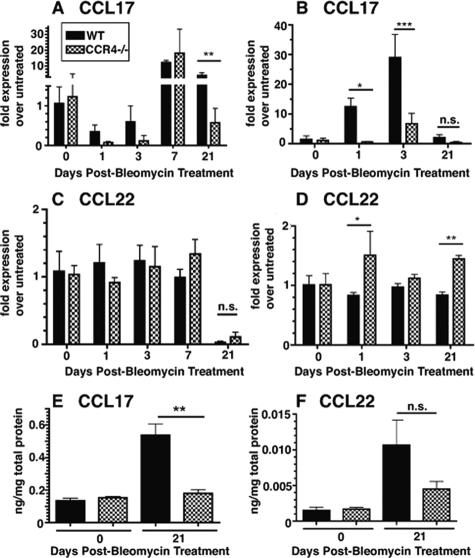
The absence of a chemokine receptor through gene deletion often leads to overt increases in the expression of the chemokine ligand(s) that bind that receptor.28 The CC chemokines, CCL17 and CCL22, share the receptor CCR4. Thus, in the present study, we addressed whether the absence of CCR4 contributed to increased CCL17 and CCL22 transcript and protein expression before and after bleomycin challenge. A quantitative PCR analysis of whole lung (Figure 2A) revealed that CCL17 transcript levels in the CCR4−/− groups did not exceed those detected in the WT groups at any time before and after bleomycin. Furthermore, at day 21 in whole lung samples CCL17 transcript levels were significantly lower in the knockout group compared with the WT group. A similar analysis of CCL22 transcript levels in whole lung (Figure 2C) showed that transcript levels for this CCR4 ligand were not changed in whole lung samples at days 1 and 21 after bleomycin challenge. Using an ELISA analysis of whole lung samples, we observed that CCL17 protein levels were significantly lower at day 21 after bleomycin in the CCR4−/− group compared with the WT group (Figure 2E) but CCL22 levels did not significantly differ between the two groups (Figure 2F).
Figure 2.
Quantitative TaqMan PCR and ELISA analysis of CCL17 and CCL22 expression in whole lung samples after bleomycin challenge. CCL17 transcript expression in whole lung homogenates at days 1, 3, 7, or 21 after bleomycin (A), and in bone marrow-derived macrophages at the same times after bleomycin (B). CCL22 transcript expression in whole lung homogenates at days 1, 3, 7, or 21 after bleomycin (C), and in bone marrow-derived macrophages at the same times after bleomycin (D). For all TaqMan analyses, the data were expressed as the fold-increase in transcript expression above transcript levels measured in whole lung or macrophage samples before bleomycin challenge. ELISA analysis of CCL17 (E) and CCL22 (F) in whole lung homogenates from WT (CCR4+/+) and CCR4−/− mice at day 21 after the bleomycin challenge. Data are mean ± SEM from four to five mice at each time point. **P < 0.01. N.S. = not significant.
Recent data from our laboratory has shown that pulmonary immune responses can affect bone marrow macrophage phenotype.29 Because macrophages are abundant producers of CCL17 and CCL22 in the lung tissue, we next investigated whether the production of CCR4 ligands by these cells was altered in bleomycin-challenged CCR4−/− mice. Similar to results obtained from assessing the whole lung, quantitative PCR analysis of bone marrow-derived macrophages (Figure 2B) also revealed that CCL17 transcript levels in the CCR4−/− groups did not exceed those detected in the WT groups at any time before and after bleomycin. Furthermore, at days 1 and 3 in bone marrow-derived macrophages CCL17 transcript levels were significantly lower in the knockout group compared with the WT group. A similar analysis of CCL22 transcript levels in bone marrow-derived macrophages (Figure 2D) also showed that transcript levels for this CCR4 ligand were significantly elevated in cultured macrophages after bleomycin challenge. Collectively, these data showed that the absence of CCR4 generally resulted in significantly lower CCL17 transcript and protein levels in whole lung and cultured macrophages. In contrast, CCR4 deficiency resulted in unchanged or significantly higher CCL22 transcript, and no difference in protein levels in these same tissues.
CCR4 Deficiency Had a Minor Effect on Inflammatory and Immune Cell Recruitment into the Lung after Bleomycin Treatment
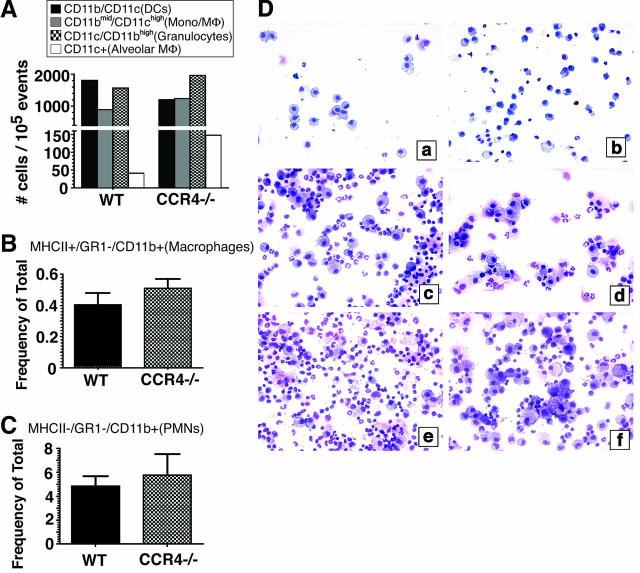
We next determined whether the absence of CCR4 affected the early recruitment of inflammatory leukocytes into the lung after bleomycin challenge. Cells present in the BAL 1 day after bleomycin challenge were analyzed according to their forward scatter and side scatter characteristics by flow cytometry as well as for their expression of CD11b and CD11c as previously described.30 The data demonstrate that the quantity of dendritic cells (Figure 3A), monocytes (Figure 3A), macrophages (Figure 3, A and B), granulocytes (Figure 3, A and C), and alveolar macrophages (Figure 3A) present in BAL samples from WT and CCR4−/− BALF were similar. Interestingly, microscopic analysis of BAL cytospins from both groups of mice at days 1 (Figure 3D, a and b), 3 (Figure 3D, c and d), and 7 (Figure 3D, e and f) after bleomycin suggested that the macrophage population present in the CCR4−/− group were morphologically distinct from the WT group. Specifically, we noted that the macrophages present in CCR4−/− BAL samples were consistently larger and were of a foamy appearance (Figure 3D, b, d, and f) compared with similar cells present in WT BAL samples (Figure 3D, a, c, and e). Together, these data showed that the absence of CCR4 did not alter the movement of inflammatory cells but its absence appeared to alter the morphology of macrophages in the alveolar compartment of bleomycin-challenged mice.
Figure 3.
Cell quantification in BAL samples at days 1, 3, and 7 after bleomycin challenge. Flow cytometric analyses: relative numbers of DCs, monocytes/macrophages, granulocytes, and alveolar macrophages in the BAL at day 1 after bleomycin challenge based on CD11b and CD11c expression (A), and the presence (B, macrophages) and absence of MHC II (C, granulocytes). D: Cytospin analysis. BAL samples were collected at days 1, 3, and 7 after bleomycin challenge, centrifuged onto glass slides, stained with Diff-Quik, and then visualized by light microscopy at ×200 magnification. The representative photomicrographs shown are as follows: a: WT (CCR4+/+) day 1; b: CCR4−/− day 1; c: WT (CCR4+/+) day 3; d: CCR4−/− day 3; e: WT (CCR4+/+) day 7; and f: CCR4−/− day 7.
Macrophages Present in the BAL from Bleomycin-Challenged CCR4−/− Mice Showed a Transient Significant Increase in the Expression of Arg1 and FIZZ-1 Transcripts and No Increase in the Expression of NOS2 Transcript
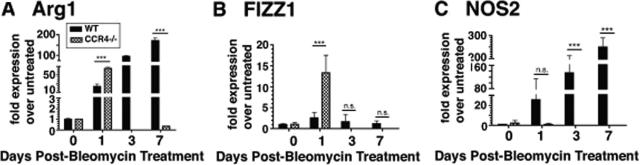
To further explore the nature of the activation state of macrophages in the alveolar compartment during the acute inflammatory phase after bleomycin challenge, the transcript expression of known markers for M2 (ie, Arg1 and FIZZ-1) and M1 macrophages (ie, NOS2) were determined before and at days 1, 3, and 7 after bleomycin treatment in WT and CCR4−/− mice. Baseline (ie, day 0) quantitative analysis of transcript levels of Arg1 (Figure 4A), FIZZ-1 (Figure 4B), and NOS2 (Figure 4C) revealed that all were similar in the WT and CCR4−/− groups. The most striking observation was that BAL-derived WT macrophages maintained a steady increase in the transcript levels of Arg1 and NOS2 through day 7 after bleomycin challenge compared to CCR4−/− macrophages (Figure 4A). Moreover, NO2 production (measured by the Griess reagent) by BAL macrophages was also elevated in WT mice compared to CCR4−/−, which demonstrated undetectable NO2 in macrophage cell supernatants by day 3 (data not shown). Interestingly, FIZZ-1 transcript levels were not increased at any time after bleomycin challenge in BAL-derived WT macrophages (Figure 4B). Another striking observation was that NOS2 was undetectable in CCR4−/− macrophages during the acute inflammatory phase (Figure 4C), whereas Arg1 and FIZZ1 transcript levels were increased at the day 1 time point only in BAL-derived CCR4−/− macrophages (Figure 4, A and B). These increases in Arg1 and FIZZ1 transcript expression at the day 1 time point were significantly greater than those detected in BAL-derived WT macrophages (Figure 4, A and B). These data indicate that on day 1 after bleomycin challenge, BAL-derived WT macrophages in the alveolar compartment exhibited a transient M1 phenotype in contrast to CCR4−/− macrophages, which exhibited a transient M2 phenotype.
Figure 4.
Determination of the BAL macrophage phenotype in WT (CCR4+/+) and CCR4−/− during the acute inflammatory phase. Cells present in the BAL from WT (CCR4+/+) and CCR4−/− mice before and at days 1, 3, and 7 after bleomycin challenge were plated in tissue culture plates at a density of 2.5 × 105 cells/ml in RPMI plus 15% fetal calf serum and incubated for 1 hour at 37°C. Nonadherent cells were discarded and adherent cells were subjected to RNA isolation for quantitative TaqMan PCR analysis of Arg1 (A), FIZZ1 (B), and NOS2 (C) transcript expression. Data are mean ± SEM from BAL macrophages cultured from five mice at each time point. ***P < 0.001. N.S. = not significant.
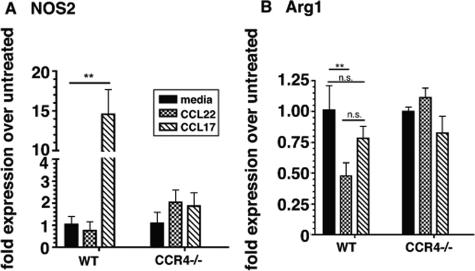
CCL17 Induced NOS2 and CCL22 Suppressed Arg1 Expression in WT Bone Marrow-Derived Macrophages
Because we detected a progressive significant increase in NOS2 transcript expression by BAL-derived WT macrophages but not by BAL-derived CCR4−/− macrophages, we next determined whether CCR4 ligands altered NOS2 transcript expression in cultured macrophages. For these in vitro experiments, we used bone marrow-derived macrophages as our surrogate cell line for the BAL-derived macrophages, because they can be cultured in sufficient quantities for in vitro studies. One day after bleomycin challenge, macrophages were selectively cultured from whole bone marrow cells from WT and CCR4−/− mice. Both groups of bone marrow-derived macrophages were exposed to media alone or media with 10 ng/ml of CCL17 or CCL22 for 24 hours before NOS2 and Arg1 quantitative transcript analysis. Figure 5A shows that exogenous CCL17 was a potent inducer of NOS2 expression in bone marrow-derived WT but not CCR4−/− macrophages. Exogenous CCL22 was a potent suppressor of Arg1 expression in bone marrow-derived WT but not CCR4−/− macrophages (Figure 5B). In control experiments, lipopolysaccharide treatment of WT and CCR4−/− macrophages failed to produce a similar response, excluding the possibility of an lipopolysaccharide contamination artifact (data not shown). Together, these data showed that CCL17 and CCL22 have distinct effects on transcript levels of M1 and M2 markers in macrophages and these effects are dependent on CCR4 expression.
Figure 5.
Effect of CCL17 and CCL22 stimulation on NOS2 and Arg1 transcript expression in bone marrow-derived macrophages. Bone marrow-derived macrophages from WT (CCR4+/+) and CCR4−/− mice at day 1 after bleomycin challenge were stimulated with one of the following: media (RPMI + 0.5% fetal calf serum), 10 ng/ml of CCL22, or 10 ng/ml CCL17 for 24 hours at 37°C. NOS2 (A) and Arg1 (B) transcript expression was measured using quantitative TaqMan PCR analysis. For all TaqMan analyses, the data were expressed as the fold-increase in transcript expression above transcript levels measured in untreated macrophage samples at day 1 after bleomycin challenge. Data are mean ± SEM from bone marrow-derived macrophages cultured from five mice at each time point. **P < 0.01. N.S. = not significant.
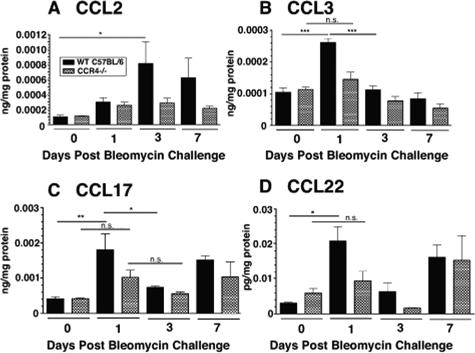
Whole Lung Levels CCL2, CCL3, CCL17, and CCL22 Analyzed by ELISA Were Significantly Increased in WT Mice 1 Day after Bleomycin
A number of chemokines have been implicated in experimental bleomycin-induced fibrosis and these include CCL2, CCL3, CCL5, CCL17, and CCL22.10,28,31,32,33,34 In the present study, we analyzed protein levels of these and several other CXC chemokine ligands and cytokines in the lungs at days 1, 3, 7, and 21 after bleomycin and compared these to untreated day 0 controls. Of the CC chemokines we measured, we detected a significant increase in CCL2 (Figure 6A), CCL3 (Figure 6B), CCL17 (Figure 6C), and CCL22 (Figure 6D) during the early inflammatory phase in the lungs of day 1 WT mice compared to those from the untreated (day 0) mice. In contrast, the CCL2, CCL3, CCL17, and CCL22 levels in the whole lungs of CCR4−/− mice were not significantly altered on days 1, 3, or 7 after bleomycin injection (Figure 6, A–D). Interestingly, only CCL17 remained significantly decreased on day 21 in the lungs of CCR4−/− mice compared to those of WT mice (Figure 2E). Thus, the decreased levels of CCL17 in whole lung samples from CCR4−/− mice were consistent with the decreased pulmonary remodeling response evoked by intratracheal bleomycin.
Figure 6.
Immunoreactive CC chemokine ligand levels in whole lung homogenates from WT (CCR4+/+) and CCR4−/− mice before and at days 1, 3, and 7 days after bleomycin challenge. Significantly elevated levels of CCL2 (A), CCL3 (B), CCL17 (C), and CCL22 (D) were measured in whole lung homogenates from WT (CCR4+/+) mice at day 1 after bleomycin challenge compared to levels of these chemokines in whole lung samples from CCR4−/− mice before the bleomycin challenge. Data are mean ± SEM from four to five mice at each time point. ***P < 0.001, **P < 0.01, *P < 0.05. N.S. = not significant.
CCR4−/− Macrophages Express Increased Levels of the D6 Scavenger Receptor
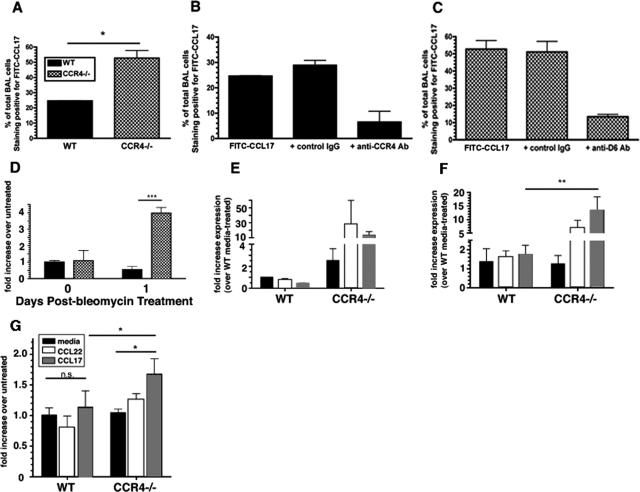
A previous report from our laboratory demonstrated that in the absence of CCR4, both CCL17 and CCL22 levels were unaffected in response to Aspergillus fumigatus conidia challenge.35 This observation is in contrast with other data from receptor knockout mice that show enhancement of the ligand in the absence of its cognate receptor.28 This unique observation in our model (Figure 2) directed us toward analyzing the binding of CCL17 and CCL22 by BAL-derived macrophages from WT and CCR4−/− mice. CCL17 and CCL22 are both ligands for the scavenging receptor, D6. However, CCL22 is rapidly inactivated in circulation by dipeptidyl peptidase IV and this inactive form is not recognized and scavenged by D6.21,36 Thus, we analyzed the binding of CCL17 on BAL-derived WT and CCR4−/− BAL cells both before and at day 1 after bleomycin. We observed more than twice as many CCR4−/− BAL cells stained positive for FITC-conjugated CCL17 compared to WT mice, suggesting that CCR4−/− BAL cells bind significantly greater amounts of CCL17 (Figure 7A). This was particularly surprising because we did not expect to observe any binding of CCL17 by CCR4−/− BAL cells. Control experiments using an irrelevant protein biotinylated to the same degree as CCL17 confirmed the binding specificity of the FITC-conjugated CCL17 (data not shown). An explanation for this increased binding of CCL17 was found in the observation that CCR4−/− BAL macrophages isolated at day 1 after bleomycin challenge expressed significantly greater transcript levels of the chemokine scavenging receptor D6 compared with their WT counterparts (Figure 7B). This hypothesis was strengthened by the observation that treatment of WT BAL cells with a blocking antibody against CCR4 reduced FITC-CCL17 binding (Figure 7C). More importantly, treatment of CCR4−/− BAL cells with a blocking antibody against D6 also reduced FITC-CCL17 binding (Figure 7D), indicating that the binding of FITC-CCL17 to CCR4−/− BAL cells is attributable to D6 expression.
Figure 7.
D6 transcript expression by BAL macrophages and bone marrow macrophages before and at day 1 after bleomycin challenge. A: FITC-CCL17 binding to WT (CCR4+/+) and CCR4−/− BAL macrophages. B: D6 transcript expression in BAL WT and CCR4−/− macrophages before and at day 1 after bleomycin was measured using TaqMan PCR analysis. C: FITC-CCL17 binding to WT (CCR4+/+) in the presence of control IgG or a blocking antibody against CCR4. D: FITC-CCL17 binding to CCR4−/− BAL macrophages in the presence of control IgG or a blocking antibody against D6. BAL macrophages before (E) and at day 1 (F) after bleomycin were cultured with media alone, or with media containing 10 ng/ml of CCL22 or 10 ng/ml of CCL17 for 24 hours and D6 transcript expression was measured. G: Bone marrow macrophages isolated day 1 after bleomycin were cultured and treated with media alone, or with media containing 10 ng/ml of CCL22 or 10 ng/ml of CCL17 for 24 hours and D6 transcript expression was measured. D6 transcript expression was measured using TaqMan and data were expressed as the fold-increase in transcript expression above transcript levels measured in untreated macrophage samples before or at day 1 after bleomycin. Data are mean ± SEM from bone marrow-derived macrophages cultured from five mice at each time point. ***P < 0.001, **P < 0.01, *P < 0.1.
To investigate whether CCR4 ligands affect D6 expression, bone marrow-derived and BAL macrophages isolated from CCR4−/− mice 1 day after bleomycin treatment were treated in vitro with CCR4 ligands. Although the levels of D6 expression in BAL macrophages from untreated (day 0) WT mice that were stimulated with CCL17 and CCL22 (Figure 7E) did not reach statistical significance, BAL macrophages from bleomycin-challenged CCR4−/− mice versus WT mice indeed displayed a significant increase in D6 expression when stimulated in vitro with CCL17 (Figure 7F). Moreover, bone marrow macrophages cultured from WT and CCR4−/− mice 1 day after bleomycin treatment demonstrated that CCL17 stimulation significantly enhanced D6 expression only in CCR4−/− mice (Figure 7G). Thus, these data showed that macrophages from CCR4−/− mice expressed markedly greater levels of the chemokine scavenging receptor D6, especially after bleomycin challenge. Moreover, these data also suggest the possibility that CCL17 can signal through another receptor to induce D6 expression when CCR4 is absent.
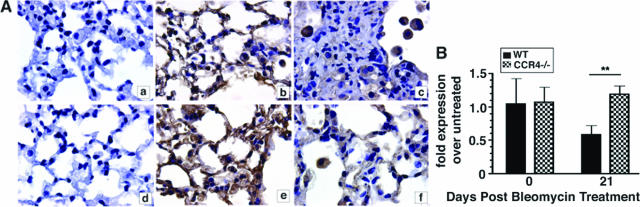
Whole Lung D6 Protein Expression Was Markedly Enhanced in CCR4−/− Mice Compared with WT Mice after Bleomycin
We next analyzed the protein expression of D6 in whole lung samples from both groups of mice at days 1 and 21 after bleomycin challenge. As shown in Figure 8A, whole lung D6 protein staining was greater in CCR4−/− mice at day 1 (Figure 8Ae) and 21 (Figure 8Af) after bleomycin compared with WT mice at the same times after bleomycin treatment (Figure 8A, b and c, respectively). Negative controls for WT and CCR4−/− whole lung samples are shown in Figure 8A, a and d, respectively. It is noteworthy that D6 protein expression was prominently expressed by alveolar macrophages and in interstitial areas of the lungs of both WT and CCR4−/− mice at day 1 after bleomycin (Figure 8A, b and e) but the interstitial expression of D6 appeared to be lost in WT mice (Figure 8Ac). The overall loss of whole lung D6 protein expression in the WT group mirrored the significant decrease in the transcript levels of this scavenging receptor at day 21 after bleomycin (Figure 8B). Thus, D6 expression was markedly enhanced in the absence of CCR4, and this increased expression may explain the absence of increased chemokine levels in whole lung samples from CCR4−/− mice after bleomycin challenge.
Figure 8.
Immunolocalization and quantitative TaqMan analysis of D6 in bleomycin-treated lung. A: Representative histological lung sections from WT (CCR4+/+) and CCR4−/− mice at days 1 [WT (CCR4+/+) (b), CCR4−/− (e)] and 21 [WT (CCR4+/+) (c), CCR4−/− (f)] after bleomycin administration were stained with a goat anti-mouse D6 antibody and processed using routine immunohistochemistry techniques. Control antibody staining in representative whole lung samples WT (CCR4+/+) (a) and CCR4−/− (d) mice at day 1 after bleomycin. B: D6 transcript expression in whole lungs from WT (CCR4+/+) and CCR4−/− before and at day 21 after bleomycin challenge was measured using quantitative TaqMan PCR analysis. The data were expressed as the fold-increase in transcript expression above transcript levels measured in untreated macrophage samples before bleomycin challenge. Data are mean ± SEM from bone marrow-derived macrophages cultured from five mice at each time point. **P < 0.01.
Discussion
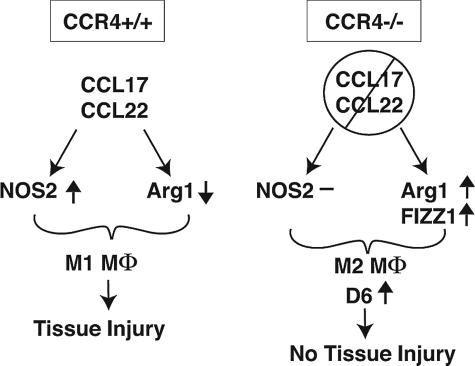
A number of recent clinical studies show that CCR4-positive cells are found in greater abundance in pulmonary diseases associated with increased extracellular matrix deposition including IPF24,37 and collagen vascular disease.38 The present study addressed the role of CCR4 and its ligands, CCL17 and CCL22, in the acute inflammatory and chronic fibrosing effects of bleomycin sulfate in the lung. Mice lacking CCR4 because of gene deletion were significantly protected from the lethal inflammatory and lung-remodeling effects of bleomycin, and data presented herein show that a major contribution for this protective effect is the regulatory role of CCR4 on the phenotype of macrophages during the acute inflammatory phase. Macrophages from bleomycin-challenged WT mice exhibited a time-dependent increase in Arg1 and NOS2 expression. In contrast, CCR4−/− macrophages exhibited a transient increase in Arg1 and FIZZ1 at day 1 after bleomycin, and NOS2 was not induced in these cells at any time after bleomycin. CCL17 was found to induce NOS2 expression in cultured macrophages from WT mice, whereas CCL22 inhibited the expression of Arg1. These effects were absent in CCR4−/− macrophages. BAL macrophages from CCR4−/− mice bound greater amounts of CCL17 compared to WT macrophages, and this increased binding may be attributable to the increased expression of D6, a scavenging chemokine receptor. Surprisingly, CCL17 significantly induced the expression of D6 in CCR4−/− but not WT macrophages. Collectively, this study provides a macrophage-specific mechanism through which the absence of CCR4 provides a protective effect during the oxidative lung injury induced by bleomycin (Figure 9).
Figure 9.
Proposed model for the role of CCR4 in the activation of macrophages in bleomycin-induced pulmonary fibrosis. CCR4+/+: In the presence of CCR4, CCL17 and CCL22 promote tissue injury through the induction of the M1 macrophage phenotype (increased NOS2 and decreased Arg1 expression). CCR4−/−: In the absence of CCR4, the M1 macrophage activation occurs through the suppression of NOS2 and increased Arg1 and FIZZ1 expression. The M2 macrophage phenotype favors the up-regulation of D6, which attenuates inflammation and tissue injury.
Although CCR4 is expressed by a variety of immune cells including Th2 cells, dendritic cells, NK cells, monocytes, platelets, and basophils,39,40 the present study addressed the role of this receptor on the macrophage since previous data provided by Belperio and colleagues31 emphasized its importance in fibrosis. This group demonstrated that CCR4 was expressed predominately by macrophages in bleomycin-treated mice, and the immunoneutralization of CCL17 significantly reduced the numbers of intrapulmonary macrophages and the pulmonary fibrotic effect of bleomycin sulfate in mice.31 Recently, macrophages have gained considerable appreciation for their role in the initiation and persistence of pulmonary fibrosis because of their functional plasticity within a stimulatory microenvironment.41,42 The heterogeneity of the macrophage is reflected by the immunological properties these cells exhibit after defined activation stimuli. Classically activated, or M1 macrophages induced by Th1 signals (interferon-γ, lipopolysaccharide, tumor necrosis factor), have a capacity to present antigen and provide protection against intracellular pathogens by means of increased oxidative burst and nitric oxide (NO) release.43 M1 macrophages also exert cyto-toxic activities, resulting partly from their ability to secrete NO and proinflammatory cytokines such as tumor necrosis factor, IL-1, and IL-6.44,45 M1 macrophages are specifically equipped to scavenge foreign antigen and cellular debris, promote angiogenesis, tissue remodeling, and repair.8,46 Conversely, M2 macrophages are induced by Th2 cytokines (IL-4, IL-13) and are generally characterized by low production of proinflammatory cytokines. These macrophages display up-regulation of FIZZ1, a molecule recently shown to activate fibroblast and myofibroblast differentiation in bleomycin-induced pulmonary fibrosis.47,48 The functions of the M2 macrophage are dependent on the Arg1 pathway, which leads to polyamine and proline synthesis that contribute to cell growth, collagen deposition, and synthesis of extracellular matrix.49
Although the persistence of M2 cells in tissues has been linked to clinical43 and experimental7 fibrosis, the transient presence of these cells protects tissues from oxidative injury. For example, Arg1 expression in macrophages has been shown to diminish the tissue injury and fibrosis associated with the intratracheal instillation of silica.41 In the present study, we observed that lung and bone marrow-derived macrophages from CCR4−/− mice exhibited transient M2 activation, as evidenced by increased Arg1 and FIZZ1, and these macrophages did not show an increase in NOS2 expression at any time after the in vivo bleomycin challenge. The absence of NOS2 expression in CCR4−/− macrophages was intriguing and prompted further examination of the effect of CCL17 on the expression of this enzyme. We observed that CCL17 was a potent inducer of NOS2 in WT but not CCR4−/− macrophages. CCL22 had no effect on the expression of NOS2 in either macrophage group. At present the mechanism through which CCL17 induced NOS2 is not evident but these findings are in agreement with a previous clinical study showing that therapeutic improvement in pediatric asthma was associated with decreased NO and CCL17.50 The present study also demonstrated that CCL22 inhibited the expression of Arg1 in WT but not CCR4−/− macrophages. The inhibitory effect of CCL22 on Arg1 expression is not readily explained by previously published observations. Another question arising from the present study pertains to the discrepant effects of these chemokine ligands on the l-arginine-using enzymes. To date, CCR4 is a functional receptor for both ligands51 and yet we11 and others31 have shown that CCL17 and CCL22 have divergent roles in pulmonary disease. This discrepancy may be explained by the fact that CCL17 and/or CCL22 can bind to molecules other than CCR4 but little evidence has been presented to show that these additional interactions involve functional chemokine receptors.
Data from the present study showed that significantly higher levels of FITC-labeled CCL17 bound to CCR4−/− macrophages compared with WT macrophages. These data suggested that CCL17 was binding another receptor on macrophages and further investigation revealed that this binding receptor was D6. D6 is a decoy receptor because it lacks the DRY motif in the second intracellular loop as well as the TXP motif in the second transmembrane domain, which are critical for the G-protein coupling and signaling functions of chemokine receptors.16 D6 binds inflammatory CC chemokines ligands that normally interact with CCR1, CCR2, CCR3, CCR4, and CCR5. However, CCL22 is rapidly deactivated in circulation by enzymatic cleavage, and this processed form is not recognized by D6.36 D6 is highly expressed by endothelial cells of lymphatic afferent vessels in the skin, gut, and lung20,21,36,52,53 and on macrophages and trophoblasts14 and we have also detected D6 expression in human lung fibroblasts from healthy and fibrotic patients (G.T. and C.M.H., unpublished data). D6 functions to negatively regulate its chemokine ligands by constitutively binding and internalizing them for intracellular degradation. This chemokine-scavenging function has been reported to be completely dependent on D6 interaction with the cytoplasmic adaptor, β-arrestin, because cells deficient in β-arrestin fail to display surface expression of D6.54 Of interest in the present study was the observation that CCL17 induced the expression of D6 in bone marrow-derived and BAL macrophages from CCR4−/− but not WT mice. Moreover, histological data suggests that treatment of CCR4−/− mice with a neutralizing antibody against D6 before bleomycin challenge results in increased lung injury and inflammation, as well as clusters of shed alveolar epithelial cells and several types of inflammatory cells (including neutrophils and macrophages) present in the BAL (G.T. and C.M.H., unpublished data). The mechanism through which CCL17 augments D6 expression in CCR4−/− macrophages is unknown at present. The data presented herein inspires further analysis of D6 regulation, and studies are presently underway in our laboratory to determine whether CCL17 can signal through D6, and whether this signaling process is β-arrestin-dependent. Together, these data suggest that D6 expression provides protection against bleomycin-induced pulmonary injury.
In conclusion, lung injury attributable to the intrapulmonary introduction of bleomycin is mediated, in part, via a CCR4-dependent mechanism specific to macrophages. CCL17 and CCL22 differentially regulate the l-arginine using enzymes NOS2 and Arg1, respectively. In the absence of CCR4, lung and bone marrow macrophages appeared to transiently revert to a M2 phenotype characterized by Arg1 and FIZZ1 expression, which appears to limit the pulmonary destructive effects of bleomycin and limits the corresponding inflammatory response. Also, the CCR4-deficient macrophage expressed significantly greater amounts of the scavenging chemokine receptor D6, which was positively regulated by CCL17. Thus, these data support therapeutically targeting CCR4 during the inflammatory phase that predispose the lung to progressive and lethal fibrosis. Pignatti and colleagues24 have previously reported that CCR4 and CCL17 are elevated in BAL from patients with IPF compared to healthy control patients or patients with sarcoidosis. Therefore, the targeting of CCR4 may be a particularly relevant approach to the recent clinical evidence that describes acute exacerbations in IPF patients.55 Studies that explore the role of the macrophage in acute exacerbations of IPF in human cells are currently underway in our laboratory.
Acknowledgments
We thank Ms. Robin G. Kunkel for her artistic assistance in the preparation of the manuscript, and Drs. Judith Connett and Matthew Schaller for critical reading of the manuscript.
Footnotes
Address reprint requests to Glenda Trujillo, Ph.D., Immunology Program, Department of Pathology, University of Michigan, 4071 BSRB, 109 Zina Pitcher Pl., Ann Arbor, MI 48109-2200. E-mail: glendat@med.umich.edu.
Supported by the National Institutes of Health (grant R01-HL069865 to C.M.H., grants PO1-HL031963 and R01-HL035276 to S.L.K., and training grant T32 HL07749 to G.T.).
References
- King TE, Jr, Tooze JA, Schwarz MI, Brown KR, Cherniack RM. Predicting survival in idiopathic pulmonary fibrosis: scoring system and survival model. Am J Respir Crit Care Med. 2001;164:1171–1181. doi: 10.1164/ajrccm.164.7.2003140. [DOI] [PubMed] [Google Scholar]
- Strieter RM, Keane MP, Burdick MD, Sakkour A, Murray LA, Belperio JA. The role of CXCR2/CXCR2 ligands in acute lung injury. Curr Drug Targets Inflamm Allergy. 2005;4:299–303. doi: 10.2174/1568010054022178. [DOI] [PubMed] [Google Scholar]
- Tajima S, Oshikawa K, Tominaga S, Sugiyama Y. The increase in serum soluble ST2 protein upon acute exacerbation of idiopathic pulmonary fibrosis. Chest. 2003;124:1206–1214. doi: 10.1378/chest.124.4.1206. [DOI] [PubMed] [Google Scholar]
- Charo IF, Ransohoff RM. The many roles of chemokines and chemokine receptors in inflammation. N Engl J Med. 2006;354:610–621. doi: 10.1056/NEJMra052723. [DOI] [PubMed] [Google Scholar]
- Wynn TA. Fibrotic disease and the T(H)1/T(H)2 paradigm. Nat Rev Immunol. 2004;4:583–594. doi: 10.1038/nri1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasse A, Germann M, Pechkovsky DV, Markert A, Verres T, Stahl M, Melchers I, Luttmann W, Muller-Quernheim J, Zissel G. IL-10-producing monocytes differentiate to alternatively activated macrophages and are increased in atopic patients. J Allergy Clin Immunol. 2007;119:464–471. doi: 10.1016/j.jaci.2006.09.030. [DOI] [PubMed] [Google Scholar]
- Mora AL, Torres-Gonzalez E, Rojas M, Corredor C, Ritzenthaler J, Xu J, Roman J, Brigham K, Stecenko A. Activation of alveolar macrophages via the alternative pathway in herpesvirus-induced lung fibrosis. Am J Respir Cell Mol Biol. 2006;35:466–473. doi: 10.1165/rcmb.2006-0121OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani A, Sica A, Sozzani S, Allavena P, Vecchi A, Locati M. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. 2004;25:677–686. doi: 10.1016/j.it.2004.09.015. [DOI] [PubMed] [Google Scholar]
- Katakura T, Miyazaki M, Kobayashi M, Herndon DN, Suzuki F. CCL17 and IL-10 as effectors that enable alternatively activated macrophages to inhibit the generation of classically activated macrophages. J Immunol. 2004;172:1407–1413. doi: 10.4049/jimmunol.172.3.1407. [DOI] [PubMed] [Google Scholar]
- Inoue T, Fujishima S, Ikeda E, Yoshie O, Tsukamoto N, Aiso S, Aikawa N, Kubo A, Matsushima K, Yamaguchi K. CCL22 and CCL17 in rat radiation pneumonitis and in human idiopathic pulmonary fibrosis. Eur Respir J. 2004;24:49–56. doi: 10.1183/09031936.04.00110203. [DOI] [PubMed] [Google Scholar]
- Jakubzick C, Wen H, Matsukawa A, Keller M, Kunkel SL, Hogaboam CM. Role of CCR4 ligands. CCL17 and CCL22, during Schistosoma mansoni egg-induced pulmonary granuloma formation in mice. Am J Pathol. 2004;165:1211–1221. doi: 10.1016/S0002-9440(10)63381-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locati M, Torre YM, Galliera E, Bonecchi R, Bodduluri H, Vago G, Vecchi A, Mantovani A. Silent chemoattractant receptors: D6 as a decoy and scavenger receptor for inflammatory CC chemokines. Cytokine Growth Factor Rev. 2005;16:679–686. doi: 10.1016/j.cytogfr.2005.05.003. [DOI] [PubMed] [Google Scholar]
- Comerford I, Litchfield W, Harata-Lee Y, Nibbs RJ, McColl SR. Regulation of chemotactic networks by ‘atypical’ receptors. Bioessays. 2007;29:237–247. doi: 10.1002/bies.20537. [DOI] [PubMed] [Google Scholar]
- Borroni EM, Buracchi C, de la Torre YM, Galliera E, Vecchi A, Bonecchi R, Mantovani A, Locati M. The chemoattractant decoy receptor D6 as a negative regulator of inflammatory responses. Biochem Soc Trans. 2006;34:1014–1017. doi: 10.1042/BST0341014. [DOI] [PubMed] [Google Scholar]
- Mantovani A, Bonecchi R, Locati M. Tuning inflammation and immunity by chemokine sequestration: decoys and more. Nat Rev Immunol. 2006;6:907–918. doi: 10.1038/nri1964. [DOI] [PubMed] [Google Scholar]
- Nibbs RJ, Wylie SM, Yang J, Landau NR, Graham GJ. Cloning and characterization of a novel promiscuous human beta-chemokine receptor D6. J Biol Chem. 1997;272:32078–32083. doi: 10.1074/jbc.272.51.32078. [DOI] [PubMed] [Google Scholar]
- Blackburn PE, Simpson CV, Nibbs RJ, O’Hara M, Booth R, Poulos J, Isaacs NW, Graham GJ. Purification and biochemical characterization of the D6 chemokine receptor. Biochem J. 2004;379:263–272. doi: 10.1042/BJ20031266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamieson T, Cook DN, Nibbs RJ, Rot A, Nixon C, McLean P, Alcami A, Lira SA, Wiekowski M, Graham GJ. The chemokine receptor D6 limits the inflammatory response in vivo. Nat Immunol. 2005;6:403–411. doi: 10.1038/ni1182. [DOI] [PubMed] [Google Scholar]
- Martinez de la Torre Y, Buracchi C, Borroni EM, Dupor J, Bonecchi R, Nebuloni M, Pasqualini F, Doni A, Lauri E, Agostinis C, Bulla R, Cook DN, Haribabu B, Meroni P, Rukavina D, Vago L, Tedesco F, Vecchi A, Lira SA, Locati M, Mantovani A. Protection against inflammation- and autoantibody-caused fetal loss by the chemokine decoy receptor D6. Proc Natl Acad Sci USA. 2007;104:2319–2324. doi: 10.1073/pnas.0607514104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Graham GJ, Damodaran A, Hu T, Lira SA, Sasse M, Canasto-Chibuque C, Cook DN, Ransohoff RM. Cutting edge: the silent chemokine receptor D6 is required for generating T cell responses that mediate experimental autoimmune encephalomyelitis. J Immunol. 2006;177:17–21. doi: 10.4049/jimmunol.177.1.17. [DOI] [PubMed] [Google Scholar]
- Whitehead GS, Wang T, DeGraff LM, Card JW, Lira SA, Graham GJ, Cook DN. The chemokine receptor D6 has opposing effects on allergic inflammation and airway reactivity. Am J Respir Crit Care Med. 2007;175:243–249. doi: 10.1164/rccm.200606-839OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chvatchko Y, Hoogewerf AJ, Meyer A, Alouani S, Juillard P, Buser R, Conquet F, Proudfoot AE, Wells TN, Power CA. A key role for CC chemokine receptor 4 in lipopolysaccharide-induced endotoxic shock. J Exp Med. 2000;191:1755–1764. doi: 10.1084/jem.191.10.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogaboam CM, Blease K, Mehrad B, Steinhauser ML, Standiford TJ, Kunkel SL, Lukacs NW. Chronic airway hyperreactivity, goblet cell hyperplasia, and peribronchial fibrosis during allergic airway disease induced by Aspergillus fumigatus. Am J Pathol. 2000;156:723–732. doi: 10.1016/S0002-9440(10)64775-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pignatti P, Brunetti G, Moretto D, Yacoub MR, Fiori M, Balbi B, Balestrino A, Cervio G, Nava S, Moscato G. Role of the chemokine receptors CXCR3 and CCR4 in human pulmonary fibrosis. Am J Respir Crit Care Med. 2006;173:310–317. doi: 10.1164/rccm.200502-244OC. [DOI] [PubMed] [Google Scholar]
- Liu T, Chung MJ, Ullenbruch M, Yu H, Jin H, Hu B, Choi YY, Ishikawa F, Phan SH. Telomerase activity is required for bleomycin-induced pulmonary fibrosis in mice. J Clin Invest. 2007;117:3800–3809. doi: 10.1172/JCI32369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tager AM, LaCamera P, Shea BS, Campanella GS, Selman M, Zhao Z, Polosukhin V, Wain J, Karimi-Shah BA, Kim ND, Hart WK, Pardo A, Blackwell TS, Xu Y, Chun J, Luster AD. The lysophosphatidic acid receptor LPA(1) links pulmonary fibrosis to lung injury by mediating fibroblast recruitment and vascular leak. Nat Med. 2008;14:45–54. doi: 10.1038/nm1685. [DOI] [PubMed] [Google Scholar]
- Pilling D, Roife D, Wang M, Ronkainen SD, Crawford JR, Travis EL, Gomer RH. Reduction of bleomycin-induced pulmonary fibrosis by serum amyloid P. J Immunol. 2007;179:4035–4044. doi: 10.4049/jimmunol.179.6.4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blease K, Mehrad B, Standiford TJ, Lukacs NW, Gosling J, Boring L, Charo IF, Kunkel SL, Hogaboam CM. Enhanced pulmonary allergic responses to Aspergillus in CCR2−/− mice. J Immunol. 2000;165:2603–2611. doi: 10.4049/jimmunol.165.5.2603. [DOI] [PubMed] [Google Scholar]
- Joshi AD, Raymond T, Coelho AL, Kunkel SL, Hogaboam CM. A systemic granulomatous response to Schistosoma mansoni eggs alters responsiveness of bone marrow-derived macrophages to Toll-like receptor agonists. J Leukoc Biol. 2008;83:314–324. doi: 10.1189/jlb.1007689. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Juarrero M, Shim TS, Kipnis A, Junqueira-Kipnis AP, Orme IM. Dynamics of macrophage cell populations during murine pulmonary tuberculosis. J Immunol. 2003;171:3128–3135. doi: 10.4049/jimmunol.171.6.3128. [DOI] [PubMed] [Google Scholar]
- Belperio JA, Dy M, Murray L, Burdick MD, Xue YY, Strieter RM, Keane MP. The role of the Th2 CC chemokine ligand CCL17 in pulmonary fibrosis. J Immunol. 2004;173:4692–4698. doi: 10.4049/jimmunol.173.7.4692. [DOI] [PubMed] [Google Scholar]
- Capelli A, Di Stefano A, Gnemmi I, Donner CF. CCR5 expression and CC chemokine levels in idiopathic pulmonary fibrosis. Eur Respir J. 2005;25:701–707. doi: 10.1183/09031936.05.00082604. [DOI] [PubMed] [Google Scholar]
- Moore BB, Kolodsick JE, Thannickal VJ, Cooke K, Moore TA, Hogaboam C, Wilke CA, Toews GB. CCR2-mediated recruitment of fibrocytes to the alveolar space after fibrotic injury. Am J Pathol. 2005;166:675–684. doi: 10.1016/S0002-9440(10)62289-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuma T, Terasaki Y, Kaikita K, Kobayashi H, Kuziel WA, Kawasuji M, Takeya M. C-C chemokine receptor 2 (CCR2) deficiency improves bleomycin-induced pulmonary fibrosis by attenuation of both macrophage infiltration and production of macrophage-derived matrix metalloproteinases. J Pathol. 2004;204:594–604. doi: 10.1002/path.1667. [DOI] [PubMed] [Google Scholar]
- Schuh JM, Power CA, Proudfoot AE, Kunkel SL, Lukacs NW, Hogaboam CM. Airway hyperresponsiveness, but not airway remodeling, is attenuated during chronic pulmonary allergic responses to Aspergillus in CCR4−/− mice. FASEB J. 2002;16:1313–1315. doi: 10.1096/fj.02-0193fje. [DOI] [PubMed] [Google Scholar]
- Bonecchi R, Locati M, Galliera E, Vulcano M, Sironi M, Fra AM, Gobbi M, Vecchi A, Sozzani S, Haribabu B, Van Damme J, Mantovani A. Differential recognition and scavenging of native and truncated macrophage-derived chemokine (macrophage-derived chemokine/CC chemokine ligand 22) by the D6 decoy receptor. J Immunol. 2004;172:4972–4976. doi: 10.4049/jimmunol.172.8.4972. [DOI] [PubMed] [Google Scholar]
- Yoshinouchi T, Naniwa T, Shimizu S, Ohtsuki Y, Fujita J, Sato S, Eimoto T, Ueda R. Expression of chemokine receptors CXCR3 and CCR4 in lymphocytes of idiopathic nonspecific interstitial pneumonia. Respir Med. 2007;101:1258–1264. doi: 10.1016/j.rmed.2006.10.019. [DOI] [PubMed] [Google Scholar]
- Shimizu S, Yoshinouchi T, Niimi T, Ohtsuki Y, Fujita J, Maeda H, Sato S, Yamadori I, Eimoto T, Ueda R. Differing distributions of CXCR3- and CCR4-positive cells among types of interstitial pneumonia associated with collagen vascular diseases. Virchows Arch. 2007;450:51–58. doi: 10.1007/s00428-006-0330-2. [DOI] [PubMed] [Google Scholar]
- D’Ambrosio D, Iellem A, Bonecchi R, Mazzeo D, Sozzani S, Mantovani A, Sinigaglia F. Selective up-regulation of chemokine receptors CCR4 and CCR8 upon activation of polarized human type 2 Th cells. J Immunol. 1998;161:5111–5115. [PubMed] [Google Scholar]
- Bonecchi R, Bianchi G, Bordignon PP, D’Ambrosio D, Lang R, Borsatti A, Sozzani S, Allavena P, Gray PA, Mantovani A, Sinigaglia F. Differential expression of chemokine receptors and chemotactic responsiveness of type 1 T helper cells (Th1s) and Th2s. J Exp Med. 1998;187:129–134. doi: 10.1084/jem.187.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misson P, van den Brule S, Barbarin V, Lison D, Huaux F. Markers of macrophage differentiation in experimental silicosis. J Leukoc Biol. 2004;76:926–932. doi: 10.1189/jlb.0104019. [DOI] [PubMed] [Google Scholar]
- Prasse A, Pechkovsky DV, Toews GB, Jungraithmayr W, Kollert F, Goldmann T, Vollmer E, Muller-Quernheim J, Zissel G. A vicious circle of alveolar macrophages and fibroblasts perpetuates pulmonary fibrosis via CCL18. Am J Respir Crit Care Med. 2006;173:781–792. doi: 10.1164/rccm.200509-1518OC. [DOI] [PubMed] [Google Scholar]
- MacMicking J, Xie QW, Nathan C. Nitric oxide and macrophage function. Annu Rev Immunol. 1997;15:323–350. doi: 10.1146/annurev.immunol.15.1.323. [DOI] [PubMed] [Google Scholar]
- Okamoto T, Gohil K, Finkelstein EI, Bove P, Akaike T, van der Vliet A. Multiple contributing roles for NOS2 in LPS-induced acute airway inflammation in mice. Am J Physiol. 2004;286:L198–L209. doi: 10.1152/ajplung.00136.2003. [DOI] [PubMed] [Google Scholar]
- Speyer CL, Neff TA, Warner RL, Guo RF, Sarma JV, Riedemann NC, Murphy ME, Murphy HS, Ward PA. Regulatory effects of iNOS on acute lung inflammatory responses in mice. Am J Pathol. 2003;163:2319–2328. doi: 10.1016/S0002-9440(10)63588-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani A, Sica A, Locati M. New vistas on macrophage differentiation and activation. Eur J Immunol. 2007;37:14–16. doi: 10.1002/eji.200636910. [DOI] [PubMed] [Google Scholar]
- Liu T, Dhanasekaran SM, Jin H, Hu B, Tomlins SA, Chinnaiyan AM, Phan SH. FIZZ1 stimulation of myofibroblast differentiation. Am J Pathol. 2004;164:1315–1326. doi: 10.1016/S0002-9440(10)63218-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Jin H, Ullenbruch M, Hu B, Hashimoto N, Moore B, McKenzie A, Lukacs NW, Phan SH. Regulation of found in inflammatory zone 1 expression in bleomycin-induced lung fibrosis: role of IL-4/IL-13 and mediation via STAT-6. J Immunol. 2004;173:3425–3431. doi: 10.4049/jimmunol.173.5.3425. [DOI] [PubMed] [Google Scholar]
- Shearer JD, Richards JR, Mills CD, Caldwell MD. Differential regulation of macrophage arginine metabolism: a proposed role in wound healing. Am J Physiol. 1997;272:E181–E190. doi: 10.1152/ajpendo.1997.272.2.E181. [DOI] [PubMed] [Google Scholar]
- Hung CH, Li CY, Lai YS, Hsu PC, Hua YM, Yang KD. Discrepant clinical responses and blood chemokine profiles between two non-steroidal anti-inflammatory medications for children with mild persistent asthma. Pediatr Allergy Immunol. 2005;16:306–309. doi: 10.1111/j.1399-3038.2005.00273.x. [DOI] [PubMed] [Google Scholar]
- Imai T, Nagira M, Takagi S, Kakizaki M, Nishimura M, Wang J, Gray PW, Matsushima K, Yoshie O. Selective recruitment of CCR4-bearing Th2 cells toward antigen-presenting cells by the CC chemokines thymus and activation-regulated chemokine and macrophage-derived chemokine. Int Immunol. 1999;11:81–88. doi: 10.1093/intimm/11.1.81. [DOI] [PubMed] [Google Scholar]
- Nibbs RJ, Kriehuber E, Ponath PD, Parent D, Qin S, Campbell JD, Henderson A, Kerjaschki D, Maurer D, Graham GJ, Rot A. The beta-chemokine receptor D6 is expressed by lymphatic endothelium and a subset of vascular tumors. Am J Pathol. 2001;158:867–877. doi: 10.1016/s0002-9440(10)64035-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez de la Torre Y, Locati M, Buracchi C, Dupor J, Cook DN, Bonecchi R, Nebuloni M, Rukavina D, Vago L, Vecchi A, Lira SA, Mantovani A. Increased inflammation in mice deficient for the chemokine decoy receptor D6. Eur J Immunol. 2005;35:1342–1346. doi: 10.1002/eji.200526114. [DOI] [PubMed] [Google Scholar]
- Galliera E, Jala VR, Trent JO, Bonecchi R, Signorelli P, Lefkowitz RJ, Mantovani A, Locati M, Haribabu B. Beta-arrestin-dependent constitutive internalization of the human chemokine decoy receptor D6. J Biol Chem. 2004;279:25590–25597. doi: 10.1074/jbc.M400363200. [DOI] [PubMed] [Google Scholar]
- Collard HR, Moore BB, Flaherty KR, Brown KK, Kaner RJ, King TE, Jr, Lasky JA, Lloyd JE, Noth I, Olman MA, Raghu G, Roman J, Ryu JH, Zisman DA, Hunnighake GW, Colby TV, Egan JJ, Hansell DM, Johkoh T, Kaminski N, Kim DS, Kondoh Y, Lynch DA, Muller-Quernheim J, Myers JL, Nicholson AG, Selman M, Toews GB, Wells AU, Martinez FJ, Idiopathic Pulmonary Fibrosis Clinical Research Network Investigators Acute exacerbations of idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2007;176:636–643. doi: 10.1164/rccm.200703-463PP. [DOI] [PMC free article] [PubMed] [Google Scholar]