Abstract
The hepatocyte growth factor (HGF)/Met signaling system is essential for liver development, homeostasis, and function. In this study, we took advantage of a liver-specific, Met-conditional knockout mouse generated in our laboratory to address the molecular mechanisms of HGF/Met signaling in adult liver progenitor cell (oval cell) biology. For this purpose, we isolated oval cells from 3,5-diethoxycarbonyl-1,4-dihydro-collidine-treated Metflx/flx mice and established oval cell-derived cell lines that carried either functional (Metflx/flx) or a nonfunctional (Met−/−) met gene using virus-mediated Cre-loxP recombination. Oval cells lacking Met tyrosine kinase activity displayed neither Met phosphorylation nor activation of downstream targets and were refractory to HGF stimulation. Although Met−/− and Metflx/flx cells proliferated at similar rates under 10% serum, Met-deficient cells demonstrated decreased cell viability and were more prone to apoptosis when challenged with either serum starvation or the pro-apoptotic cytokine transforming growth factor-β. Treatment with HGF reduced transforming growth factor-β-mediated cell death in Metflx/flx but not Met−/− cells. Importantly, Metflx/flx and Met−/− cells both constitutively expressed hgf, and conditioned medium from serum-starved oval cells exhibited anti-apoptotic activity in Metflx/flx cells. Furthermore, serum-starved Metflx/flx cells showed persistent activation of the Met tyrosine kinase, suggesting HGF/Met autocrine regulation. In conclusion, these data reveal a critical, functional role for Met in oval cell survival through an autocrine mechanism.
Oval cells are the progenies of the hepatic stem cells that are thought to reside in the terminal branches of the biliary tree termed the canals of Hering. Oval cells have phenotypic features intermediate between hepatocytes and biliary epithelial cells, and co-express some markers of hematopoietic stem cells. These cells are normally quiescent but become activated under certain conditions, for example, following administration of a variety of hepatotoxins, and are responsible for the regenerative growth after liver injuries that impede the proliferation or function of existing hepatocytes.1,2
Besides the beneficial effects, oval cells may also play a role in neoplasia. In fact, oval cell proliferation is often associated with exposure to hepatocarcinogens. Moreover, cells with progenitor cell features have been identified in a number of human chronic liver diseases including viral hepatitis, cirrhosis, focal nodular hyperplasia, and hepatocellular adenoma, as well as in hepatocellular carcinomas.3,4 The molecular events mediating oval cell expansion and differentiation are now beginning to be elucidated. Interestingly, the same regulatory signals operating during hepatocyte-mediated regeneration represent the important components of the oval cell-triggered regenerative response.5 One of the potentially important regulatory signaling networks is the hepatocyte growth factor (HGF)/Met system. HGF is a pleiotropic growth factor that promotes mitogenesis, motogenesis, morphogenesis, and survival in a variety of cell types,6,7 and plays a major role in tissue formation and homeostasis. Responses to HGF are mediated by binding to its tyrosine kinase receptor, Met. Ligand-receptor binding results in phosphorylation of specific tyrosine residues located in the C-terminal domain of the receptor, and subsequent activation of an array of adapter and signal transducing proteins, including growth factor receptor-bound protein 2/son-of-sevenless, Ras-mitogen-activated protein kinase, growth factor receptor-bound protein 2-associated binder 1, phosphoinositide-3 kinase, phospholipase C-γ, and signal transducer and activator of transcription, which mediate the biological effects of HGF/Met.8 In the liver, HGF is best known as a potent mitogen for hepatocytes and an inducer of regeneration9,10,11 but it also promotes survival of embryonic and adult hepatocytes.12,13 A requirement for HGF and its receptor Met in liver development has been demonstrated by gene ablation studies.14,15,16 Recently, we and others have provided direct genetic evidence for the essential role of HGF/Met in liver regeneration using liver specific Met conditional knock-out mouse models.17,18,19
The importance of HGF in liver physiology is not limited to its direct action on hepatocytes. Oval cells express Met among other growth factor receptors.20 Furthermore, HGF infusion during liver regeneration induced by N-acetyl-2-amino-fluorene/partial hepatectomy protocol accelerated oval cell proliferation in vivo.21 HGF-elicited mitogenic and/or morphogenetic responses have been also documented in liver progenitor cells in vitro.22,23,24,25
Based on these data, we hypothesized that HGF/Met signaling may play a fundamental role during the oval cell-driven regenerative response. In this study, we examined the effects of inactivating Met on the behavior of oval cells. We have established oval cell-derived lines from the Metflx/flx mice,18 and deleted the floxed allele by virus-mediated Cre-loxP recombination. We found that the Met signaling mutants displayed an amplified apoptotic response to both serum deprivation and treatment with transforming growth factor-beta (TGF-β) but apparently intact proliferative function in normal culture conditions. Our results demonstrate that Met-supported signaling pathway plays an essential role in promoting oval cell survival. We also provide evidence for an autocrine mechanism involved in the Met-mediated pro-survival activity in liver progenitor cells.
Materials and Methods
Development of Oval Cell Lines and Culture Conditions
Generation of mice homozygous for the floxed c-met allele has been described previously.18,26 To induce oval cells, nine-week-old male Metflx/flx mice were maintained on 1% 3,5-diethoxycarbonyl-1,4-dihydro-collidine-supplemented diet for 13 days.27 Oval cell-enriched nonparenchymal cell fraction was isolated as described elsewhere.28 Briefly, the liver was washed from blood with HBBS without Ca2+ and Mg2+ and then perfused with Williams E medium containing 0.1% w/v pronase E (Sigma, Madrid, Spain) and 0.1% collagenase type 1 (Worthington Biomedical Corporation, Lakewood, NJ). Then, liver was excised, minced in the Williams E medium containing 0.1% w/v pronase E (Sigma), 0.1% collagenase type 1 (Worthington Biomedical Corporation) and 0.005% DNase I (Worthington Biomedical Corporation), and incubated in the same solution for an additional 30 to 45 minutes at 37°C in a waterbath with shaking. The digest was diluted with Williams E medium containing 10% fetal bovine serum, passed through a 40 μm nylon mesh filter and sedimented by centrifugation. After three consecutive washes in Williams E medium containing 10% fetal bovine serum, cells were purified on a Percoll gradient (30% and 70% Percoll). The cell fraction localized at the interphase between two Percoll phases was collected, washed twice in Williams E medium containing 10% fetal bovine serum, and sedimented by centrifugation. Cells were plated in plastic dishes (BD Falcon Cell Culture Dishes) in the attachment/growth medium containing high-glucose Dulbecco’s modified Eagle’s medium/Ham’s F12 medium with glutamine (1:1 mix) supplemented with 1g/L insulin-transferrin-selenium+, 1g/L D-galactose, 0.3 g/L proline, 1.5 mmol/L Na Pyruvate, 0.018M HEPES, penicillin/streptomycin, and 10% fetal bovine serum. The following day, the non-attached cells were removed, and a mixed culture containing different cell types was maintained for a few days until small colonies of cells displaying epithelial morphology became apparent. Individual colonies were picked up using cloning cylinders, and subcultured at limiting dilution. Several single cell epithelial clones were selected and further subcultured for expansion and characterization. Once established, cell lines were routinely maintained in Dulbecco’s modified Eagle’s medium (Invitrogen-Gibco, Barcelona, Spain) in a humidified incubator at 37°C and a 5% CO2 atmosphere. Medium was replaced every three days, and cells were harvested at 80% to 90% confluence using trypsin-EDTA and replated at 1:10 dilution for maintenance.
In vitro inactivation of Met was achieved by infecting the parental oval cell lines with adenovirus expressing the Cre recombinase under the cytomegalovirus promoter (Ad-CMV-Cre) (Vector Biolabs, Philadelphia, PA). Cells were plated 24 hours before infection at a density that guaranteed 80% confluence at the day of infection. Virus was diluted in infection medium (complete growth medium supplemented with 2.5 μg/ml polybrene) at a multiplicity of infection = 20. Original cell culture medium was replaced with the virus-containing medium (0.5 ml/well in a 12-well plate), cells were incubated for 1 hour in the incubator with occasional shacking, and then fresh medium was added to complete volume. After 48 hours of infection, cells were trypsinized and replated following the limiting dilution protocol. An aliquot of the infected cells was used to isolate genomic DNA following standard procedures to identify the deleted allele by PCR using specific oligonucleotides.18 A total of 52 cell clones were selected, expanded and genotyped, and 10 of them were used for further characterization.
Immunofluorescence Staining for Flow Cytometry and Confocal Microscopy Analysis
For flow cytometry analysis, cells were detached with trypsin-EDTA. For detection of cell surface antigen A6, specific for oval cells,29 freshly isolated cells were stained with rat anti-A6 at 1:25 dilution for 30 minutes at 4°C in 0.1% bovine serum albumin-PBS followed by incubation with an Alexa 488-conjugated anti-rat antibody (Invitrogen). Before analysis, cells were incubated with 0.005% propidium iodide to gate out dead cells. Analysis was performed in a CyAn MLE flow cytometer (Dako, Glostrup, United Kingdom), equipped with three laser lines (365, 488, and 637 nm). For detection of albumin, cells were fixed with 3.5% paraformaldehyde for 5 minutes at room temperature, washed with PBS, incubated with methanol (−20°C) for 30 seconds at room temperature, and washed again. Rabbit polyclonal anti-albumin was used at 1:50 dilution (Nordic Immunological Laboratories, Tilburg, Netherlands) at room temperature for 30 minutes. After washing, cells were incubated with a Cy3-conjugated anti-rabbit antibody (c-2306, Sigma).
To analyze cytokeratin18 and albumin expression by fluorescence microscopy, cells were fixed in methanol (−20°C) for 2 minutes and incubated for 1 hour at 37°C with primary antibodies diluted 1:50 in 1% bovine serum albumin (mouse monoclonal anti-cytokeratin18 from Progen, Heidelberg, Germany; rabbit polyclonal anti-albumin). After washing in PBS, cells were incubated either with Cy3-conjugated anti-rabbit or Green Oregon-conjugated anti-mouse (1:500 and 1:200, respectively) for 1 hour at room temperature in the presence of 4,6-diamidino-2-phenylindole (5 μg/ml, Sigma) for nuclei staining. Cells were examined in a Radiance 2100 confocal microscope (Carl Zeiss, Jena, Germany) and analyzed using Laserpix software (Bio-Rad, Hercules, CA).
Reverse Transcriptase-Polymerase Chain Reaction Analysis
Total cellular RNA was isolated using the RNeasy Kit (Qiagen, Valencia, CA). RNA yield and purity were analyzed using a spectrophotometer (UV-visible recording spectrophotometer UV-160, Shimadzu). Three μg total RNA was reverse-transcribed into complementary DNA using SuperScript III RNase H Reverse Transcriptase (Invitrogen) and oligo-dT (Amersham Biosciences, Piscataway, NJ) as a primer. The PCR primers were as follows: α-fetoprotein (forward 5′-CACTGCTGCAACTCTTCGTA-3′, reverse 5′-CTTTGGACCCTCTTCTGTGA-3′), albumin (forward 5′-CTGCCGATCTGCCCTCAATAGC-3′, reverse 5′-GTGCCCACTCTTCCCAGGTTTCT-3′), cytokeratin19 (forward 5′-GTGCCACCATTGACAACTCC-3′, reverse 5′-AATCCACCTCCACACTGACC-3′), CD34 (forward 5′-TCCTGATGAACCGTCGCAGTT-3, reverse 5′-TGTCAGCCACCACATGTTGTC-3′), thy-1 (forward 5′-AGAAGGTGACCAGCCTGACA-3′, reverse 5′-AATGAAGTCCAGGGCTTGGA-3′), connexin43 (forward 5′-GATGAGGAAGGAAGAGAAGC-3′, reverse 5′-TTGTTTCTGTCACCAGTAAC-3′), hnf-1β (forward 5′-TCAGTCAACAGAACCAGGGCC-3′, reverse 5′-GCCGGGGAGACTTGTTGTAAA-3′), hnf-1α (forward 5′-CGGACTGATTGAAGAGCCCAC-3′, reverse 5′-CTGGTTGAGACCTGGAGACGT-3′), hnf-4 (forward 5′-AGTACATCCCGGCCTTCTGTG-3′, reverse 5′-GACCCTCCAAGCAGCATCTCC-3′), hnf-6 (forward 5′-GCAATGGAAGTAATTCAGGGCAG-3′, reverse 5′-CATGAAGAAGTTGCTGACAGTGC-3′), glyceraldehyde-3-phosphate dehydrogenase (forward 5′-TGAAGGTCGGTGTGAACGGATTTGGC-3′, reverse 5′-CATGTAGGCCATGAGGTCCACCAC-3′), hgf (forward 5′-ATGTCCTCCTGCACCTCCTC-3′, reverse 5′-TTACCGCGATAGCTCGAAG-3′), β-actin (forward 5′-ATGCCATCCTGCGTCTGGACCTGGC-3′, reverse 5′-AGCATTTGCGGTGCACGATGGAGGG-3′).
Cycling parameters were: denaturation at 94°C for 1 minute, annealing temperatures of 57 to 65°C for 1 minute, and extension at 72°C for 1 minute (30 to 35 cycles of amplification). Amplified products were subjected to electrophoresis in 1.2 to 1.5% agarose gels and stained with ethidium bromide for visualization.
DNA Synthesis and Cell Growth Analysis
Cells were plated at a density of 17,500 cells/sq cm in 10% fetal bovine serum-Dulbecco’s modified Eagle’s medium. At different times after plating, cells were trypsinized following routine protocols and counted using a hemacytometer under a Nikon Eclipse, TS-100 microscope. Cell viability was determined by trypan blue dye exclusion. Six hours after plating, time required for 100% cell attachment, was used as a starting reference point.
To evaluate DNA synthesis cells were plated at the same density and incubated for 48 hours in serum-free medium with or without growth factors: 20 ng/ml of epidermal growth factor (Serono Laboratories, Madrid, Spain) or 10, 20, 40 ng/ml of HGF (a kind gift of Dr. T. Nakamura). Incorporation of 3H-thymidine during the last 40 hours of culture was measured in trichloroacetic acid-precipitable material following a previously described protocol.30
Cell Viability and Apoptosis Assays
For apoptosis studies, complete Dulbecco’s modified Eagle’s medium was replaced with serum-free Dulbecco’s modified Eagle’s medium in the presence or absence of 1 ng/ml of TGF-β (Calbiochem, La Jolla, CA). When indicated, HGF (20 ng/ml) was added before the TGF-β treatment for a minimum of 6 hours. Cell viability was analyzed by staining with crystal violet and spectrophotometric reading of the solubilized dye at 560 nm, as described.31 Cells undergoing apoptosis were scored under inverted fluorescence microscope (Eclipse TE300, Nikon) at high magnification (×600) following standard morphological criteria such as chromatin condensation, nuclear pyknosis, and nuclear fragmentation. A cluster of closely packed apoptotic bodies was scored as one. Cells were stained with PI as described,32 and apoptotic indices were calculated after counting 1000 to 2000 cells per treatment in a blinded manner.
Measurement of Caspase-3-Like Enzymatic Activity
To quantify Caspase-3-like activity we used a fluorometric assay in the presence of Ac-DEVD-AMC as fluorogenic Caspase-3 substrate (PharMingen, San Diego, CA).33 Cleavage of the substrate was monitored in a Microplate Fluorescence Reader FL600 (Bio-Tek) (excitation, 380 nm; emission, 440 nm). A unit of caspase activity is the amount of enzyme that will lead to a one unit increase in the fluorescence intensity. Protein concentration was estimated and results are expressed as units of activity per microgram of protein.
Protein Isolation and Immunoblot Analysis
Cells were lysed in a modified radioimmunoprecipitation assay buffer (30 mmol/L Tris, pH7.5; 150 mmol/L NaCl; 1% NP-40; 0.5% sodium deoxycholate; 0.1% SDS; 5 mmol/L EDTA, 10% glycerol) supplemented with 1 mmol/L phenylmethylsulfonyl fluoride, 10 μg/ml aprotinin and leupeptin; 1 mmol/L Na orthovanadate). Forty to 80 μg of protein were separated in 10% acrylamide sodium dodecyl sulfate-polyacrylamide electrophoresis gels and blotted to Immobilon-P membranes (Millipore, Bedford, MA). Membranes were probed with the following primary antibodies: anti-phospho-protein kinase B (AKT) (Ser 473) (CS-9271), anti-AKT (CS-9272), anti-phosphoextracellular signal-regulated kinases (CS-9101), anti-extracellular signal-regulated kinases (CS-9102) (all rabbit polyclonal from Cell Signaling, Beverly, MA, diluted 1:500 to 1:1000 in Tris-buffered saline containing Tween 20-0.5% nonfat dried milk), rabbit polyclonal anti c-Met (sc-162, Santa Cruz Biotechnology, Inc, CA, diluted 1:500), and mouse monoclonal anti-phosphotyrosine (recombinant clone 4G10, Upstate Biotechnology, Inc., Lake Placid, NY, diluted 1:500).
For Met immunoprecipitation, total cell extracts prepared with a lysis buffer containing 10 mmol/L Tris, pH = 7.4; 150 mmol/L NaCl; 1% NP-40; 1% Na deoxycholate; 0.1% SDS; 2 mmol/L EDTA; plus protease and phosphatase inhibitors (as describe above), were immunoprecipitated using a rabbit polyclonal anti-c-Met antibody (sc-162) and protein A-agarose beads. Immunoprecipitates were washed three times with the lysis buffer and heated for 5 minutes at 95°C in Laemmli sample buffer for Western blot analysis.
Statistical Analysis
Statistical analysis was performed by Student’s t-test analysis. The differences were assumed significant at P < 0.05.
Results
Generation of Met−/− Oval Cell Lines
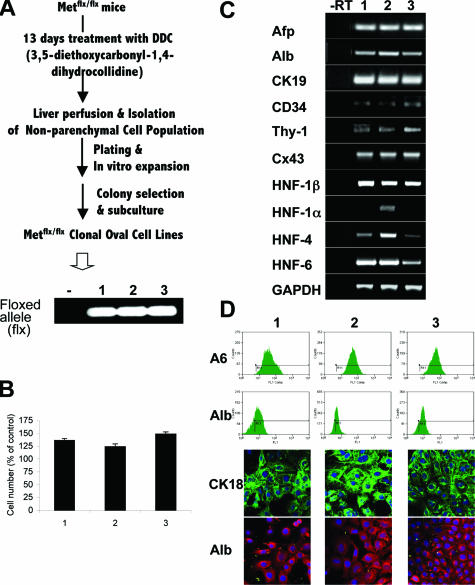
The liver-specific, Met-conditional, knock-out mouse model has been previously generated and used to address the contribution of Met signaling to liver regeneration and carcinogenesis.18,26 In this study, we used this mouse model to directly asses the role of Met in oval cell biology. Nine-week-old mice homozygous for the conditional genetic modification of Met (Metflx/flx) were given a 3,5-diethoxycarbonyl-1,4-dihydro-collidine-supplemented diet, a protocol shown to induce a rapid and massive oval cell proliferation.27 After 13 days of treatment, livers were perfused, and the oval cell-enriched nonparenchymal cell fraction was isolated as described in Material and Methods. The clones composed of small epithelial cells based on morphological and growth properties were designated as oval cells and used for establishment of Metflx/flx single-cell clonal oval cell lines. PCR genotyping confirmed the presence of the floxed allele and absence of the wt allele (Figure 1A). All oval cell lines responded to HGF treatment by increasing cell proliferation indicating that HGF/Met signaling was intact in Metflx/flx cells (Figure 1B). In addition, we examined the expression of several lineage specific markers using semiquantitative RT-PCR, flow cytometry and confocal microscopy. As shown in Figure 1, C and D, all oval cell lines were positive for the surface marker A6, one of the few antigens specifically detected in mouse oval cells, and shared by biliary epithelial cells,29 as well as for connexin 43, a gap junction protein present in oval cells, although they did not express connexin 32 characteristic for mature hepatocytes (data not shown). However, the oval cells expressed α-fetoprotein, albumin, and cytokeratins 18 & 19, consistent with their epithelial origin. The oval cells also expressed liver specific transcription factors such as hnf-1β and hnf-6, low levels of hnf-4, and very low or undetectable levels of hnf-1α. Besides classical epithelial cell markers, the oval cells showed a low expression of CD34 and thy-1, two hematopoietic stem cell markers.34,35 Essentially, no differences in gene expression were observed between the established cell lines, with the exception of slightly higher levels of hnf-4 and a low level of hnf-1α in the line number 2, suggesting that this cell line may be slightly more differentiated as compared to others. Together, these data show that the oval cell-derived cell lines exhibit bipotent liver progenitor cell phenotype.
Figure 1.
Development and phenotypic characterization of oval cell-derived cell lines. A: Scheme used to generate oval cell lines. PCR genotyping confirmed the presence of the floxed allele (flx) in three established parental lines (marked as 1, 2, 3). B: Growth response to HGF. Cells were treated with 20 ng/ml HGF for 48 hours. Cell number was determined by crystal violet staining. Data are shown as percentage of untreated cells and are mean ± SEM of duplicate experiments. Analysis of the expression of stem cell and hepatic lineage markers by semiquantitative RT-PCR (C), flow cytometry and confocal microscopy (D). For C and D, a representative experiment of three is shown.
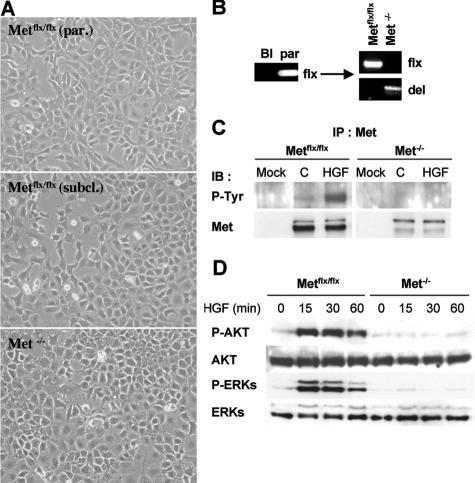
Once the Metflx/flx oval cell lines were established and characterized, we proceeded with the in vitro deletion of Met. For this purpose, cells were infected with adenovirus expressing the Cre recombinase under the cytomegalovirus promoter. Infected cells were then subcultured at a limiting dilution to allow for a clonal expansion and selection. A number of subclones were selected for further characterization. The infection/selection process did not affect the morphological features of oval cells. Both Metflx/flx and Met−/− subclones grew as small polygonal epithelial cells with a high nuclear-cytoplasmic ratio, similar to the parental lines (Figure 2A). The genotype of the oval cell subclones was analyzed by PCR (Figure 2B). As expected, Met−/− oval cell lines did not show tyrosine phosphorylation of Met in response to a short treatment with HGF (Figure 2C), and consequently, they lacked activation of downstream tyrosine kinases, such as AKT and extracellular signal-regulated kinases (Figure 2D). These data serve as a functional validation for generation of Met-deficient oval cell lines.
Figure 2.
Characterization of Metflx/flx and Met−/− oval cell lines. A: Light microscopy photographs of the parental Metflx/flx cell line and the stable Metflx/flx and Met−/− subclones established after infection of the parental cells with AdCMVCre. Magnification: ×100. B: PCR genotyping of parental and subcloned cell lines. C. Met activation in Metflx/flx but not in Met−/− cells in the absence (C) or presence of HGF (20 ng/ml) for 10 minutes. Whole protein extracts were used for Met immunoprecipitation. Phosphorylation was detected by immunoblotting with anti-P-tyrosine antibody. Western blot for Met was used as a loading control. Two bands were detected by Western blot using a Met-specific antibody corresponding to the 170-kDa inactive single-chain precursor form (upper band) and the 145-kDa proteolytically processed mature form (lower band). D: Western blot analysis of AKT and extracellular signal-regulated kinase activation after HGF treatment. HGF induced AKT and extracellular signal-regulated kinase phosphorylation in Metflx/flx but not in Met−/−.
Proliferation Is Not Affected in Met−/− Oval Cells
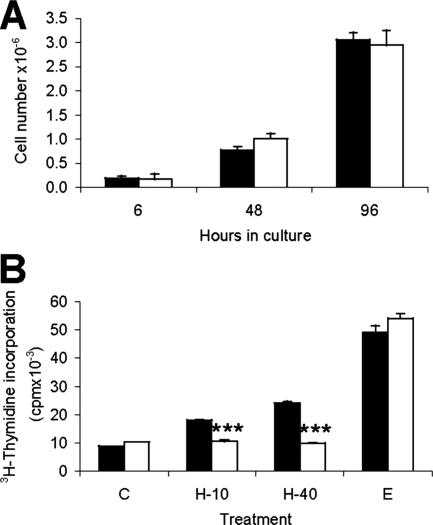
Given that HGF is a well known mitogen for both hepatocytes and liver progenitor cells, we compared the proliferative capacity of the Metflx/flx and Met−/− oval cells in culture using two approaches. First, we analyzed the growth properties of the cells throughout the culture in the presence of 10% serum. As shown in Figure 3A, no significant differences were observed in cell number between Metflx/flx and Met−/− oval cells during 4 days in culture. We then measured the DNA synthesis rate in Metflx/flx and Met−/− cells cultured in the absence or presence of specific mitogenic stimuli, such as HGF and epidermal growth factor. The latter was included as another important mitogen for hepatocytes.36 As expected, HGF increased the rate of DNA synthesis in a dose-dependent fashion only in Metflx/flx but not in Met−/− oval cells (Figure 3B). Interestingly, the proliferative response to epidermal growth factor was much stronger and comparable between two genotypes whereas the basal rate of DNA synthesis (in the absence of exogenous stimuli) was also similar in both cell types.
Figure 3.
Metflx/flx and Met−/− oval cells show similar proliferation rate in the presence of mitogenic stimuli others than HGF. A: Cell growth analysis of oval cells cultured in the presence of 10% serum. Cell number was determined by trypan blue exclusion assay under light microscopy. Black bars, Metflx/flx. White bars, Met−/−. Data are mean ± SEM of triplicates. B: [3H]Thymidine incorporation assay performed in oval cells treated for 48 hours with different concentrations of HGF (10 and 40 ng/ml) and epidermal growth factor (20 ng/ml). Results are expressed as cpm/dish. Data are mean ± SEM of triplicate experiments. ***P < 0.001.
Met−/− Oval Cells Are More Sensitive to Serum Deprivation and TGF-β-Induced Apoptosis
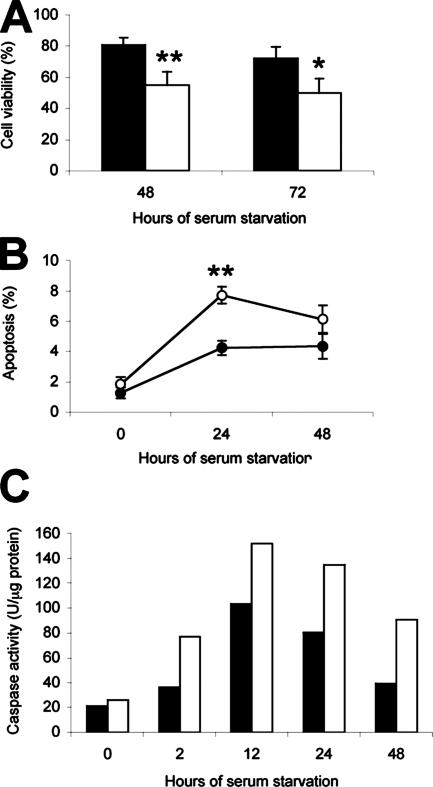
Since there were no apparent differences in the proliferative potential between Met−/− and Metflx/flx oval cells cultured in the presence of serum, we decided to challenge the cells by submitting them to serum deprivation. Under these conditions, Met−/− cells exhibited a significant reduction in cell viability whereas Metflx/flx cells were considerably more resistant to serum deprivation (Figure 4A). To clarify if the reduction in cell number was a reflection of cell death, we measured the rate of apoptosis in serum-starved Metflx/flx and Met−/− oval cells. Although a moderate increase in the apoptotic indexes was observed in both cultures, Met−/− cells died at a considerably higher rate than Metflx/flx cells, particularly during the first 24 hours of starvation (Figure 4B). These data were consistent with kinetics of caspase activation after serum deprivation. Differently from Metflx/flx cells, Met−/− cells displayed a very rapid response starting at 2 hours, a stronger peak, and a longer increase in caspase activity (Figure 4C).
Figure 4.
Met−/− cells show increased sensitivity to serum deprivation. A: Cell viability after 48 hours and 72 hours of culture in the absence of serum. Cell number was determined by crystal violet staining. Data are shown as percent relative to the value at the start of starvation. Black bars, Metflx/flx. White bars, Met−/−. Data are mean ± SEM of six independent experiments. *P < 0.05. **P < 0.01. B: Apoptotic index at different time points after serum withdrawal. Cells were fixed and stained with propidium iodide, and the percentage of apoptotic nuclei was estimated under a fluorescence microscope in a blinded manner. A total of 1000 to 2000 cells were counted per dish in each experiment. Solid circles, Metflx/flx; open circles, Met−/−. Data are mean ± SEM of at least eight experiments. **P < 0.01. C: Analysis of caspase-3 activity in Metflx/flx and Met−/− oval cells at different times after serum withdrawal. Black bars, Metflx/flx. White bars, Met−/−. A representative experiment of three performed is shown.
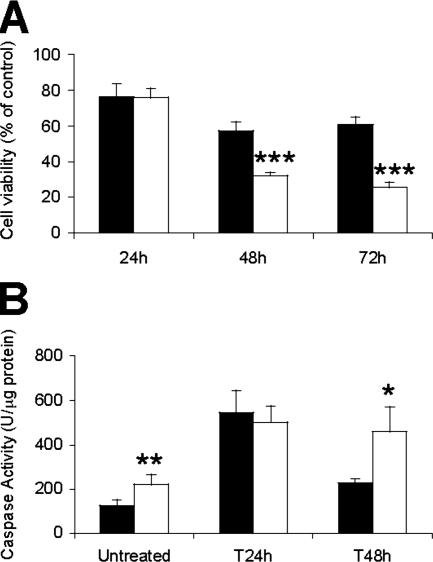
Next we challenged Met−/− cells with TGF-β, an important pro-apoptotic stimuli in fetal and adult hepatocytes.31,37 In general, TGF-β treatment caused a more detrimental effect on oval cell viability than serum deprivation. After 24 hours, the lack of Met function did not seem to affect cell viability. However, by 48 and 72 hours, TGF-β caused significantly more cell death in Met−/− cultures than in control cells (Figure 5, A and B). Consistent with a higher cell loss, Met−/− cells showed a continuous increase in caspase activity differently from Metflx/flx oval cells, which displayed a remarkably decreased caspase activity by 48 hours (Figure 5B). In contrast, the effect of TGF-β on cell proliferation was comparable in cells of both genotypes (see Supplemental Figure S1A at http://ajp.amjpathol.org). Interestingly, low doses of TGF-β (0.1−0.25 ng/ml) resulted in inhibition of cell proliferation without inducing apoptosis (see Supplemental Figure S1B and S1C at http://ajp.amjpathol.org), suggesting that apoptotic response elicited by TGF-β in oval cells is not just the consequence of its growth inhibitory action.
Figure 5.
Met −/− cells are more sensitive to TGF-β-induced apoptosis. A: Cell viability after TGF-β treatment as measured by crystal violet staining. Data are expressed as percentage of untreated cells and represent mean ± SEM of three independent experiments performed in triplicates. Black bars, Metflx/flx cells. White bars, Met−/− cells. ***P < 0.001. B: Caspase-3 activity in Metflx/flx and Met−/− oval cells after treatment with TGF-β for 24 hours and 48 hours. Black bars, Metflx/flx. White bars, Met−/−. Data are mean ± SEM of four independent experiments. *P < 0.05. **P < 0.01.
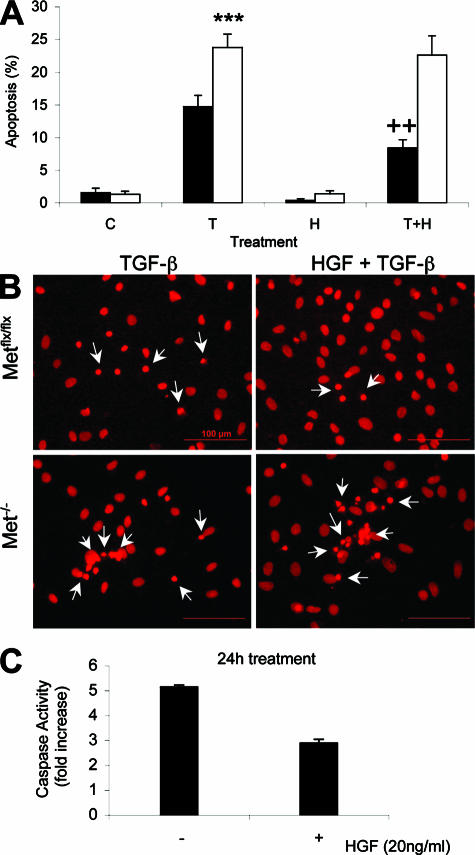
The differences in the rate of apoptosis observed between Metflx/flx and Met−/− in response to TGF-β were more pronounced in the presence of HGF. Pretreatment with HGF was able to reduce the percentage of dying cells by 40% to 50% only in Metflx/flx but not in Met−/− cells (Figure 6, A and B). The HGF-mediated decrease in apoptosis was accompanied by a reduction in the levels of activated caspase-3 (Figure 6C).
Figure 6.
HGF reduces the TGF-β-induced apoptosis in Metflx/flx but not in Met−/− oval cells. A: Apoptotic index in oval cells treated with TGF-β (T; 1 ng/ml) for 48 hours in the absence or presence of HGF (H; 20 ng/ml). A total of 1000 to 2000 cells were counted per dish after PI staining under a fluorescence microscope in a blinded manner. Black bars, Metflx/flx cells. White bars, Met−/− cells. Data are mean ± SEM of three experiments. ***P < 0.001 (versus T, Metflx/flx) ++P < 0.01 (versus T, Metflx/flx). B: Representative images of oval cells treated for 48 hours with TGF-β or TGF-β plus HGF were taken under a fluorescence microscope after PI staining. Arrows point to apoptotic nuclei. C: Caspase-3 activity in Metflx/flx oval cells treated for 24 hours with TGF-β or TGF-β plus HGF. A representative experiment of two is shown.
An HGF/Met Autocrine Loop Promotes Survival of Oval Cells
In light of our observation concerning a differential response to pro-apoptotic stimuli between Metflx/flx and Met−/− in serum-free conditions, we hypothesized that oval cells may produce survival factors, including HGF, which may account for the differences in cell susceptibility to apoptosis. To test this hypothesis, we analyzed the endogenous expression of hgf in oval cells by RT-PCR. HGF mRNA was detected both in Metflx/flx and Met−/− oval cells (Figure 7A). In addition, phosphorylation of Met was readily detected and occurred in a time-dependent fashion when Metflx/flx cells were grown in serum-free medium (Figure 7B). Furthermore, substitution of fresh serum-free medium with medium conditioned by oval cells for 48 hours, caused an earlier Met phosphorylation in Metflx/flx cells that was observed at 12 hours, as compared with 24 hours in cells cultured in fresh serum-free medium (Figure 7B).
Figure 7.
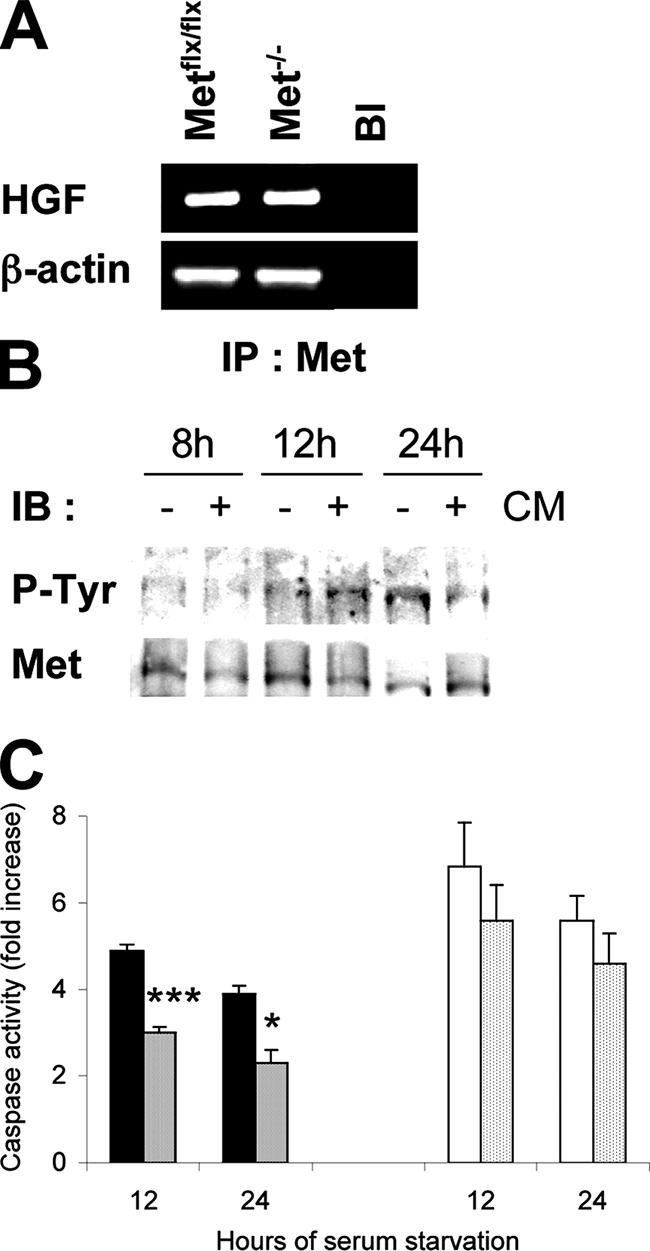
HGF/Met autocrine loop is active in Metflx/flx oval cells. A: Basal expression of the hgf mRNA as detected by RT-PCR analysis in Metflx/flx and Met−/− oval cells. Bl, Blank, no reverse transcription. β-actin was used for normalization. B: Kinetics of Met activation in Metflx/flx cells cultured in the serum-free medium or conditioned serum-free medium (CM). Whole protein extracts were used for immunoprecipitation of Met protein. Phosphorylation was detected by immunoblotting with anti-P-tyrosine antibody using Met as a loading control. C: Effect of conditioned medium on caspase-3 activity in oval cells cultured under serum deprivation for 12 hours and 24 hours. Black bars, Metflx/flx cells, serum-free medium. Dense dotted-bars, Metflx/flx cells, conditioned medium. White bars, Met−/− cells, serum-free medium. Light dotted bars, Met−/− cells, conditioned medium. Data are mean ± SEM of three independent experiments. ***P < 0.001. *P < 0.05.
Both Metflx/flx and Met−/− oval cell lines were negative for glial fibrillary acidic protein, a marker of stellate cells, as judged by PCR and immunohistochemistry (see Supplemental Figure S2A and S2B at http://ajp.amjpathol.org), thus excluding the possibility that a contaminating population of stellate cells could be responsible for hgf expression. Also, we did not find any evidence that spontaneous epithelial mesenchymal transition in culture could have contributed to HGF expression. Although oval cells expressed the mesenchymal marker vimentin, they were positive for the epithelial marker E-cadherin and did not express the characteristic marker of epithelial mesenchymal transition, Snail (see Supplemental Figure S3A and S3B at http://ajp.amjpathol.org).
We next tested the anti-apoptotic potency of serum-free medium conditioned by oval cells as judged by caspase activation (Figure 7C). In Metflx/flx cells, the conditioned medium decreased the apoptotic response to serum deprivation by about 40% at 12 hours. In contrast, in Met−/− cells, the presence of the conditioned medium had minimal if any effect on the caspase activity (Figure 7C). Together, these results suggest that HGF/Met autocrine activation contributes to survival of oval cells.
Discussion
Previously, it has been established that liver progenitor cells express Met and respond to HGF by a plethora of mitogenic, morphogenic, and survival activities.22,23,24,25 In this work, we have for the first time taken a direct genetic approach to address the relevance of the HGF/Met signaling pathway to oval cell biology. For this purpose, we have isolated and characterized oval cell lines harboring a genetically inactivated Met tyrosine kinase. Our results demonstrate that loss of Met did not affect the proliferative capacity of oval cells, but instead increased the sensitivity to apoptosis caused by serum deprivation or treatment with TGF-β. Furthermore, Met mutant oval cells had a significantly reduced capacity to adapt to unfavorable conditions in vitro through loss of the Met-dependent autocrine stimulation.
These findings have important implications in the context of oval cell-mediated liver regeneration. HGF and TGF-β are key molecules in liver physiology that exert the opposite biological effects. Thus, while HGF works as a pro-regenerative stimulus, promoting cell growth and protecting against apoptosis, among other hepatotrophic activities, TGF-β is involved in termination of liver regeneration,38 acting as a growth inhibitor39 and pro-apoptotic31,37 factor. It has been shown that Met levels increase in rat oval cells induced by the N-acetyl-2-amino-fluorene/partial hepatectomy protocol.20 This, together with the increase in oval cell number observed after in vivo HGF infusion,21 suggests that HGF is involved in the expansion of oval cells following liver injury. TGF-β is synthesized by oval cells during their early differentiation in vivo,40 and its overexpression leads to impairment of oval cell expansion induced by 3,5-diethoxycarbonyl-1,4-dihydro-collidine.27 Moreover, oval cells are growth-inhibited by TGF-β in vitro.41 Thus, TGF-β appears to function as an inhibitory cytokine during oval cell growth. In this report, we demonstrate that TGF-β, in addition to inhibition of the oval cell growth, induces apoptosis in oval cells, and more importantly, that the loss of Met tyrosine kinase activity increases sensitivity to the TGF-β-induced apoptosis (Figure 5). Together, our results suggest that the Met-driven anti-apoptotic activity might be critical to support the expansion of oval cells following liver injury by permitting to overcome the local tissue insults and inhibitory signals. The powerful anti-apoptotic activity of HGF/Met signaling was first demonstrated in adult hepatocytes,12 and was proved to be important for the proregenerative action of HGF/Met.18 Very recently, it has been also shown that Met controls hepatocytes survival during development.13 Based on these data, we propose that the HGF/Met signaling is a major survival pathway in liver that operates during liver development, homeostasis, and hepatocyte or oval cell-mediated regeneration.
We also provide evidence that HGF/Met autocrine loop is operational in liver progenitor cell lines (Figure 7). It is generally thought that HGF is produced by mesenchymal cells of various tissues, including liver, which acts on Met-expressing epithelial cells in an endocrine and/or paracrine fashion.42 Indeed, in experimental models of oval cell activation in vivo, HGF mRNA was identified in the desmin-positive stellate cells localized in the immediate proximity to proliferating oval cells, whereas Met was strongly expressed by oval cells,20,43 suggesting a paracrine regulatory mechanism. We verified that the oval cell lines generated in this study were not contaminated with stellate cells responsible for hgf expression (see Supplemental Figure S2 at http://ajp.amjpathol.org). The oval cell lines express vimentin transcript, a typical mesenchymal marker (see Supplemental Figure S3 at http://ajp.amjpathol.org). This finding is not unique for our oval cells. The co-expression of epithelial and mesenchymal markers has been previously described in rat oval cells induced by AAF-PH protocol.44,45 Apart from vimentin expression, our oval cell lines did not show any signs of epithelial mesenchymal transition, as judged by morphology, lack of expression of Snail, a transcription factor that plays a critical role during the epithelial mesenchymal transition, and the presence of high levels of E-cadherin, a Snail-repressed gene involved in cell-cell adhesion.46 Our results suggest that in addition to paracrine mechanism, HGF/Met might also operate through autocrine mechanism during oval cell activation. The co-expression of HGF and Met is not exclusive to oval cells. Evidence for an autocrine mechanism of action for HGF/Met can be found in other cell systems. Thus, HGF is involved in neuron development by enhancing the survival and differentiation of sympathetic neuroblasts, partly acting via an autocrine mechanism.47 A paracrine regulatory mechanism for HGF/Met in oval cells was suggested based on the data obtained in rat models of oval cell activation. Therefore, it seems plausible that mechanisms regulating oval cell expansion may differ across species or depend on the protocol of oval cell activation. In support of this, a recent work has delineated the profound phenotypic heterogeneity displayed by oval cells in different rat and mouse models of oval cell-mediated liver regeneration.48 It seems reasonable that the same diversity may apply to the regulatory mechanisms operating in these cells. Thus, mouse oval cells isolated from CDE-treated mouse livers express cytokines such as lymphotoxin-β, interferon-γ, interleukin-6,49,50 suggesting that they may respond both to autocrine and paracrine cytokine stimulation. Another possibility is that oval cells may acquire the capacity to produce growth factors when they are isolated from their normal environment, and no other source of growth factors is available. Additional experiments will help to clarify these issues.
The HGF/Met autocrine loop has been identified in a subset of spontaneous transformed WB-F344 rat liver stem-like cells, and it seems to be partially responsible for driving autonomous proliferation of these tumor cell lines.51 This finding, together with the putative oncogenic role for Met during development and invasion of hepatocellular carcinomas,52,53 would argue in favor of an active role for Met in promoting the malignant conversion of oval cells. Our results indicate that oval cell lines can adapt to serum-free conditions (Figure 4), a capacity that is significantly reduced in Met−/− cells. However, neither Metflx/flx nor Met−/− oval cells showed signs of transformation. They undergo contact inhibition, do not have the ability to grow in soft agar, and do not form tumors when transplanted subcutaneously into nude mice (data not shown). These findings suggest that the establishment of a functional HGF/Met autocrine loop cannot be considered per se as an indication of malignancy, but rather seems to function as a normal regulatory mechanism to support the survival of oval cells in a hostile environment. Nevertheless, the potential of oval cells to become transformed under certain conditions is known,54,55 and further work is necessary to test the relevance of Met to such phenomenon.
In conclusion, this is the first study to report that HGF/Met autocrine regulation plays a critical role in hepatic progenitor cell survival. Oval cell-derived cell lines expressing a kinase-inactive Met receptor represent a novel in vitro model for dissecting molecular mechanisms of HGF/Met signaling in hepatic progenitor cells.
Acknowledgments
A. Sánchez is recipient of a research contract (Ramón y Cajal Program) from the Ministerio de Educación y Ciencia/Universidad Complutense de Madrid, Spain. G. del Castillo is recipient of a student contract from the Ministerio de Educación y Ciencia/Universidad Complutense de Madrid, Spain. CNIC is supported by the Spanish Ministry of Health and Consumer Affairs and the Pro-CNIC Foundation.
We express appreciation to Dr. T. Nakamura for his generous gift of human recombinant HGF. We also acknowledge E. Arza, P. Torralbo, and R. Nieto for technical assistance in flow cytometry and confocal microscopy; P. González, I. del Valle, and A. Silva for generously providing positive control samples for glial fibrillary acidic protein; and C. Roncero and A. Porras for helpful discussions. Special thanks to M. Murillo for his assistance in preparing the digital images.
Footnotes
Address reprint requests to Dr. A. Sánchez. Dep. Bioquímica y Biología Molecular II, Facultad de Farmacia, Universidad Complutense de Madrid, Plaza de Ramón y Cajal S/N, 28040-Madrid, Spain. E-mail: munozas@farm.ucm.es.
This work was supported by grants from Comunidad Autónoma de Madrid (GR/SAL/0578/2004) and from the Ministerio de Educación y Ciencia (SAF2006-12025).
Supplemental material for this article can be found on http://ajp.amjpathol.org.
References
- Knight B, Matthews VB, Olynyk JK, Yeoh GC. Jekyll and Hyde: evolving perspectives on the function and potential of the adult liver progenitor (oval) cell. BioEssays. 2005;27:1192–1202. doi: 10.1002/bies.20311. [DOI] [PubMed] [Google Scholar]
- Fausto N, Campbell JS. The role of hepatocytes and oval cells in liver regeneration and repopulation. Mech Dev. 2003;120:117–130. doi: 10.1016/s0925-4773(02)00338-6. [DOI] [PubMed] [Google Scholar]
- Alison MR. Liver stem cells: implications for hepatocarcinogenesis. Stem Cell Rev. 2005;1:253–260. doi: 10.1385/SCR:1:3:253. [DOI] [PubMed] [Google Scholar]
- Roskams T. Liver stem cells and their implications in hepatocellular and cholangiocarcinoma. Oncogene. 2006;25:3818–3822. doi: 10.1038/sj.onc.1209558. [DOI] [PubMed] [Google Scholar]
- Santoni-Rugiu E, Jelnes P, Thorgeirsson SS, Bisgaard HC. Progenitor cells in liver regeneration: molecular responses controlling their activation and expansion. APMIS. 2005;113:876–902. doi: 10.1111/j.1600-0463.2005.apm_386.x. [DOI] [PubMed] [Google Scholar]
- Zarnegar R, Michalopouls GK. The many faces of hepatocyte growth factor: from hepatopoiesis to hematopoiesis. J Cell Biol. 1995;129:1177–1180. doi: 10.1083/jcb.129.5.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto K, Nakamura T. Emerging multipotent aspects of hepatocyte growth factor. J Biochem (Tokyo) 1996;119:591–600. doi: 10.1093/oxfordjournals.jbchem.a021283. [DOI] [PubMed] [Google Scholar]
- Furge KA, Zhang YW, Vande Woude GF. Met receptor tyrosine kinase: enhanced signaling through adapter proteins. Oncogene. 2000;19:5582–5589. doi: 10.1038/sj.onc.1203859. [DOI] [PubMed] [Google Scholar]
- Fausto N, Laird AD, Webber EM. Liver regeneration. 2. Role of growth factors and cytokines in hepatic regeneration. FASEB J. 1995;9:1527–1536. doi: 10.1096/fasebj.9.15.8529831. [DOI] [PubMed] [Google Scholar]
- Stolz DB, Mars WM, Petersen BE, Kim TH, Michalopoulos GK. Growth factor signal transduction immediately after two-thirds partial hepatectomy in the rat. Cancer Res. 1999;59:3954–3960. [PubMed] [Google Scholar]
- Fabregat I, de Juan C, Nakamura T, Benito M. Growth stimulation of rat fetal hepatocytes in response to hepatocyte growth factor: modulation of c-myc and c-fos expression. Biochem Biophys Res Commun. 1992;189:684–690. doi: 10.1016/0006-291x(92)92255-v. [DOI] [PubMed] [Google Scholar]
- Kosai K, Matsumoto K, Nagata S, Tsujimoto Y, Nakamura T. Abrogation of Fas-induced fulminant hepatic failure in mice by hepatocyte growth factor. Biochem Biophys Res Commun. 1998;244:683–690. doi: 10.1006/bbrc.1998.8293. [DOI] [PubMed] [Google Scholar]
- Moumen A, Patane S, Porras A, Dono R, Maina F. Met acts on Mdm2 via mTOR to signal cell survival during development. Development. 2007;134:1443–1451. doi: 10.1242/dev.02820. [DOI] [PubMed] [Google Scholar]
- Schmidt C, Bladt F, Goedecke S, Brinkmann V, Zschiesche W, Sharpe M, Gherardi E, Birchmeier C. Scatter factor/hepatocyte growth factor is essential for liver development. Nature. 1995;373:699–702. doi: 10.1038/373699a0. [DOI] [PubMed] [Google Scholar]
- Uehara Y, Minowa O, Mori C, Shiota K, Kuno J, Noda T, Kitamura N. Placental defect and embryonic lethality in mice lacking hepatocyte growth factor/scatter factor. Nature. 1995;373:702–705. doi: 10.1038/373702a0. [DOI] [PubMed] [Google Scholar]
- Bladt F, Riethmacher D, Isenmann S, Aguzzi A, Birchmeier C. Essential role for the c-met receptor in the migration of myogenic precursor cells into the limb bud. Nature. 1995;376:768–771. doi: 10.1038/376768a0. [DOI] [PubMed] [Google Scholar]
- Borowiak M, Garratt AN, Wüstefeld T, Strehle M, Trautwein C, Birchmeier C. Met provides essential signals for liver regeneration. Proc Natl Acad Sci USA. 2004;101:10608–10613. doi: 10.1073/pnas.0403412101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh C-G, Factor VM, Sánchez A, Uchida K, Conner EA, Thorgeirsson SS. Hepatocyte growth factor/c-met signaling pathway is required for efficient liver regeneration and repair. Proc Natl Acad Sci USA. 2004;101:4477–4482. doi: 10.1073/pnas.0306068101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phaneuf D, Moscioni AD, LeClair C, Raper SE, Wilson JM. Generation of a mouse expressing a conditional knockout of the hepatocyte growth factor gene: demonstration of impaired liver regeneration. DNA Cell Biol. 2004;23:592–603. doi: 10.1089/dna.2004.23.592. [DOI] [PubMed] [Google Scholar]
- Hu Z, Evarts RP, Fujio K, Marsden ER, Thorgeirsson SS. Expression of hepatocyte growth factor and c-met genes during hepatic differentiation and liver development in the rat. Am J Pathol. 1993;142:1823–1830. [PMC free article] [PubMed] [Google Scholar]
- Nagy P, Bisgaard HC, Santoni-Rugiu E, Thorgeirsson SS. In vivo infusion of growth factors enhances the mitogenic response of rat hepatic ductal (oval) cells after administration of 2-acetylaminofluorene. Hepatology. 1996;23:71–79. doi: 10.1002/hep.510230111. [DOI] [PubMed] [Google Scholar]
- Johnson M, Koukoulis G, Matsumoto K, Nakamura T, Iyer A. Hepatocyte growth factor induces proliferation and morphogenesis in nonparenchymal epithelial liver cells. Hepatology. 1993;17:1052–1061. [PubMed] [Google Scholar]
- Jeffers M, Sambasiva Rao M, Rulong S, Reddy JK, Subbarao V, Hudson E, Vande Woude GF, Resau JH. Hepatocyte growth factor/scatter factor-met signaling induces proliferation, migration, and morphogenesis of pancreatic oval cells. Cell Growth & Differ. 1996;7:1805–1813. [PubMed] [Google Scholar]
- Okano J, Shiota G, Matsumoto K, Yasui S, Kurimasa A, Hisatome I, Steinberg P, Murawaki Yoshikazu. Hepatocyte growth factor exerts a proliferative effect on oval cells through the PI3K/AKT signaling pathway. Biochem Biophys Res Commun. 2003;309:298–304. doi: 10.1016/j.bbrc.2003.04.002. [DOI] [PubMed] [Google Scholar]
- Yao P, Zhan Y, Xu W, Li C, Yue P, Xu C, Hu D, Qu CK, Yang X. Hepatocyte growth factor-induced proliferation of hepatic stem-like cells depends on activation of NF-kappaB. J Hepatol. 2004;40:391–398. doi: 10.1016/j.jhep.2003.11.001. [DOI] [PubMed] [Google Scholar]
- Kaposi-Novak P, Lee JS, Gomez-Quiroz L, Coulouarn C, Factor VM, Thorgeirsson SS. Met-regulated expression signature defines a subset of human hepatocellular carcinomas with poor prognosis and aggressive phenotype. J Clin Invest. 2006;116:1582–1595. doi: 10.1172/JCI27236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preisegger KH, Factor VM, Fuchsbichler A, Stumptner C, Kenk H, Thorgeirsson SS. Atypical ductular proliferation and its inhibition by transforming growth factor beta 1 in the 3,5-diethoxycarbonyl-1,4-dihydrocollidine mouse model for chronic alcoholic liver disease. Lab Invest. 1999;79:103–109. [PubMed] [Google Scholar]
- Bisgaard HC, Santoni-Rugiu E, Nagy P, Thorgeirsson SS. Modulation of the plasminogen activator/plasmin system in rat liver regenerating by recruitment of oval cells. Lab Invest. 1998;78:237–246. [PubMed] [Google Scholar]
- Engelhardt NV, Factor VM, Yasova AK, Poltoranina VS, Baranov VN, Lasareva MN. Common antigens of mouse oval and biliary epithelial cells. Expression on newly formed hepatocytes. Differentiation. 1990;45:29–37. doi: 10.1111/j.1432-0436.1990.tb00453.x. [DOI] [PubMed] [Google Scholar]
- de Juan C, Benito M, Alvarez A, Fabregat I. Differential proliferative response of cultured fetal and regenerating hepatocytes to growth factors and hormones. Exp Cell Res. 1992;202:495–500. doi: 10.1016/0014-4827(92)90104-g. [DOI] [PubMed] [Google Scholar]
- Sanchez A, Alvarez AM, Benito M, Fabregat I. Apoptosis induced by transforming growth factor-beta in fetal hepatocyte primary cultures: involvement of reactive oxygen intermediates. J Biol Chem. 1996;271:7416–7422. doi: 10.1074/jbc.271.13.7416. [DOI] [PubMed] [Google Scholar]
- Conner EA, Wirth PJ, Kiss A, Santoni-Rugiu E, Thorgeirsson SS. Growth inhibition and induction of apoptosis by HGF in transformed rat liver epithelial cells. Biochem Biophys Res Commun. 1997;236:396–401. doi: 10.1006/bbrc.1997.6938. [DOI] [PubMed] [Google Scholar]
- Murillo MM, del Castillo G, Sanchez A, Fernandez M, Fabregat I. Involvement of EGF receptor and c-Src in the survival signals induced by TGF-beta1 in hepatocytes. Oncogene. 2005;24:4580–4587. doi: 10.1038/sj.onc.1208664. [DOI] [PubMed] [Google Scholar]
- Petersen BE, Goff JP, Greenberger JS, Michalopoulos GK. Hepatic oval cells express the hematopoietic stem cell marker Thy-1 in the rat. Hepatology. 1998;27:433–445. doi: 10.1002/hep.510270218. [DOI] [PubMed] [Google Scholar]
- Omori N, Omori M, Evarts RP, Teramoto T, Miller MJ, Hoang TN, Thorgeirsson SS. Partial cloning of rat CD34 cDNA and expression during stem cell-dependent liver regeneration in the adult rat. Hepatology. 1997;26:720–727. doi: 10.1002/hep.510260325. [DOI] [PubMed] [Google Scholar]
- Thoresen GH, Guren TK, Sandnes D, Peak M, Agius L, Christoffersen T. Response to transforming growth factor alpha (TGFalpha) and epidermal growth factor (EGF) in hepatocytes: lower EGF receptor affinity of TGF-alpha is associated with more sustained activation of p42/p44 mitogen-activated protein kinase and greater efficacy in stimulation of DNA synthesis. J Cell Physiol. 1998;175:10–18. doi: 10.1002/(SICI)1097-4652(199804)175:1<10::AID-JCP2>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Oberhammer F, Bursch W, Parzefall W, Breit P, Erber E, Stadler M, Schulte-Hermann R. Effect of transforming growth factor beta on cell death of cultured rat hepatocytes. Cancer Res. 1991;51:2478–2485. [PubMed] [Google Scholar]
- Braun L, Mead JE, Panzica M, Mikumo R, Bell GI, Fausto N. Transforming growth factor beta mRNA increases during liver regeneration: a possible paracrine mechanism of growth regulation. Proc Natl Acad Sci USA. 1988;85:1539–1543. doi: 10.1073/pnas.85.5.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Tomita Y, Hirai R, Yamaoka K, Kaji K, Ichihara A. Inhibitory effect of transforming growth factor-beta on DNA synthesis of adult rat hepatocytes in primary culture. Biochem Biophys Res Commun. 1985;133:1042–1050. doi: 10.1016/0006-291x(85)91241-0. [DOI] [PubMed] [Google Scholar]
- Nagy P, Evarts RP, McMahon JB, Thorgeirsson SS. Role of TGF-beta in normal differentiation and oncogenesis in rat liver. Mol Carcinog. 1989;2:345–354. doi: 10.1002/mc.2940020609. [DOI] [PubMed] [Google Scholar]
- Nguyen LN, Furuya MH, Wolfraim LA, Nguyen AP, Holdren MS, Campbell JS, Knight B, Yeoh GC, Fausto N, Parks WT. Transforming growth factor-beta differentially regulates oval cell and hepatocyte proliferation. Hepatology. 2007;45:31–41. doi: 10.1002/hep.21466. [DOI] [PubMed] [Google Scholar]
- Sonnenberg E, Meyer D, Weidner KM, Birchmeier C. Scatter factor/hepatocyte growth factor and its receptor, the c-met tyrosine kinase, can mediate a signal exchange between mesenchyme and epithelia during mouse development. J Cell Biol. 1993;123:223–235. doi: 10.1083/jcb.123.1.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alison MR, Poulsom R, Jeffery R, Anilkumar TV, Jagoe R, Sarraf CE. Expression of hepatocyte growth factor mRNA during oval cell activation in the rat liver. J Pathol. 1993;171:291–299. doi: 10.1002/path.1711710410. [DOI] [PubMed] [Google Scholar]
- Sarraf C, Lalani EN, Golding M, Anilkumar TV, Poulsom R, Alison M. Cell behaviour in the acetylaminofluorene-treated regenerating rat liver. Light and electron microscopic observations. Am J Pathol. 1994;145:1114–1126. [PMC free article] [PubMed] [Google Scholar]
- Yovchev MI, Grozdanov PN, Joseph B, Gupta S, Dabeva MD. Novel hepatic progenitor cell surface markers in the adult rat liver. Hepatology. 2007;45:139–149. doi: 10.1002/hep.21448. [DOI] [PubMed] [Google Scholar]
- Cano A, Pérez-Moreno MA, Rodrigo I, Locascio A, Blanco MJ, del Barrio MG, Portillo F, Nieto MA. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol. 2000;2:76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- Maina F, Hilton MC, Andres R, Wyatt S, Klein R, Davies AM. Multiple roles for hepatocyte growth factor in sympathetic neuron development. Neuron. 1998;20:835–846. doi: 10.1016/s0896-6273(00)80466-3. [DOI] [PubMed] [Google Scholar]
- Jelnes P, Santoni-Rugiu E, Rasmussen M, Friis SL, Nielsen JH, Tygstrup N, Bisgaard HC. Remarkable heterogeneity displayed by oval cells in rat and mouse models of stem cell-mediated liver regeneration. Hepatology. 2007;45:1462–1470. doi: 10.1002/hep.21569. [DOI] [PubMed] [Google Scholar]
- Akhurst B, Matthews V, Husk K, Smyth MJ, Abraham LJ, Yeoh GC. Differential lymphotoxin-beta and interferon gamma signaling during mouse liver regeneration induced by chronic and acute injury. Hepatology. 2005;41:327–335. doi: 10.1002/hep.20520. [DOI] [PubMed] [Google Scholar]
- Matthews VB, Klinken E, Yeoh GC. Direct effects of interleukin-6 on liver progenitor oval cells in culture. Wound Repair Regen. 2004;12:650–656. doi: 10.1111/j.1067-1927.2004.12605.x. [DOI] [PubMed] [Google Scholar]
- Presnell SC, Hooth MJ, Borchert KM, Coleman WB, Grisham JW, Smith GJ. Establishment of a functional HGF/c-Met autocrine loop in spontaneous transformants of WB-F344 rat liver stem-like cells. Hepatology. 1998;28:1253–1259. doi: 10.1002/hep.510280513. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Xu W, Lu J, He F, Yang X. Invasiveness of hepatocellular carcinoma cell lines: contribution of hepatocyte growth factor, c-met, and transcription factor Ets-1. Biochem Biophys Res Commun. 2001;286:1123–1130. doi: 10.1006/bbrc.2001.5521. [DOI] [PubMed] [Google Scholar]
- Daveau M, Scotte M, Francois A, Coulouarn C, Ros G, Tallet Y, Hiron M, Hellot MF, Salier JP. Hepatocyte growth factor, transforming growth factor alpha, and their receptors as combined markers of prognosis in hepatocellular carcinoma. Mol Carcinog. 2003;36:130–141. doi: 10.1002/mc.10103. [DOI] [PubMed] [Google Scholar]
- Pack R, Heck R, Dienes HP, Oesch F, Steinberg P. Isolation, biochemical characterization, long term culture, and phenotype modulation of oval cells from carcinogen-fed rats. Exp Cell Res. 1993;20:198–209. doi: 10.1006/excr.1993.1025. [DOI] [PubMed] [Google Scholar]
- Hooth MJ, Coleman WB, Presnell SC, Borchert KM, Grisham JW, Smith GJ. Spontaneous neoplastic transformation of WB-F344 rat liver epithelial cells. Am J Pathol. 1998;153:1913–1921. doi: 10.1016/S0002-9440(10)65705-7. [DOI] [PMC free article] [PubMed] [Google Scholar]