Abstract
The plexiform variant of uterine leiomyomata (UL) is named for its ribbons or nests of smooth muscle cells that have a rounded, epithelioid shape caused by their entrapment in abundant extracellular matrix. Plexiform UL are currently classified as epithelioid smooth muscle tumors alongside the less predictable, “true” epithelioid tumors (ie, leiomyoblastomas). Karyotypes of six plexiform UL cases were studied, and their abnormalities were found to differ from those of leiomyoblastomas. Analyses using real-time polymerase chain reaction, immunohistochemistry, and fluorescence in situ hybridization demonstrated elevated mRNA and protein levels of the architectural factor HMGA2 and, in some cases, increased DNA copy number. Four of these plexiform UL were profiled with Affymetrix human U133 plus 2.0 expression arrays. Cluster analysis using genes previously shown to discriminate benign and malignant uterine smooth muscle tissues revealed that the plexiform tumors form an isolated group in the benign branch. This is in contrast to an earlier finding in which another variant, cellular UL characterized by loss of a portion of the short arm of chromosome 1, clustered with malignant leiomyosarcomas. These results provide additional evidence of genetic heterogeneity underlying UL of various histological types. We further suggest that plexiform UL should be classified among tumors with extensive hyalinization rather than with “true” epithelioid smooth muscle neoplasms.
Uterine leiomyomata (UL), frequently referred to as fibroids, are the most common tumors of the female genital tract.1 Although considered benign, UL cause a range of symptoms including urinary incontinence, constipation, abdominal pain, abnormal uterine bleeding, and impaired fertility.2,3 As a result of such high morbidity, UL are the most frequent indication for hysterectomy, cause approximately one in five visits to a gynecologist, and require annual expenditures of greater than 2.1 billion health care dollars in the US (based on data from the year 2000).4,5,6
Approximately 25 to 40% of UL have simple and nonrandom cytogenetic abnormalities.7,8 One of the most common of these aberrations is a t(12;14)(q15;q23-24),9 which leaves the coding sequence for the high mobility group (HMG) protein family member HMGA2 intact but up-regulates its expression in UL.10,11,12 HMGA2, a nonhistone component of chromatin and architectural factor, functions to influence transcription and thereby affect diverse cellular processes such as differentiation and proliferation,13,14,15,16 and has most recently been associated with height in humans.17
UL arise from the uterine myometrium and typically are comprised of fascicles of smooth muscle cells with abundant pink cytoplasm and uniform spindle-shaped nuclei. In contrast, an uncommon variant of UL, the plexiform type, is named for its nonfascicular component of small ribbons, branching strands, or nests of rounded smooth muscle cells. No significant nuclear pleomorphism, mitotic activity, or necrosis is associated with either. In plexiform UL, abundant collagen-rich matrix is present between the cords of cells entrapping them, resulting in loss of the typical spindle shape and gain of an epithelioid (rounded) appearance.18 Consequently, plexiform UL have been classified with smooth muscle tumors composed of cells with “true” epithelioid differentiation that were once referred to as leiomyoblastoma.19 Although leiomyoblastoma may recur or metastasize,20,21 plexiform UL have been widely reported to be clinically benign.19,22
In addition to the problematic classification with potentially malignant leiomyoblastoma, plexiform UL can present some diagnostic challenges because of their gross appearance, which is characterized by a yellowish complex nodular composition that is distinctly different from the whorled grayish pattern of typical UL.23,24 This can raise the level of concern for malignancy sufficiently to cause submission of additional tissue sections for analyses such as cytogenetics before arriving at a pathological diagnosis. Also, the presence of multiple plexiform UL has been reported to create an infiltrative pattern that may be confused with low-grade endometrial stromal sarcoma,25,26 and concern that the single Indian-file arrangement of the smooth muscle cells in plexiform UL may be mistaken for a lobular breast cancer metastasis has previously been raised.24
The majority of evidence indicates that plexiform UL originate from smooth muscle cells,25,27,28 particularly myofibroblasts.29 These neoplasms are often small, historically referred to as plexiform tumorlets, and are usually encountered as an incidental finding in hysterectomy specimens. Even when small enough to be called a tumorlet, they can be recognized as plexiform based in part on the amplification of extracellular matrix, which suggests that the abundant accumulation of connective tissue is an intrinsic and distinctive property of these tumors. Plexiform UL can occur as a solitary tumor or more rarely in a group, usually are found in the presence of typical UL, and have no known predilection for anatomical (ie, subserosal, intramural, or submucosal) location.
This study reports cytogenetic and expression analyses of plexiform UL, confirming their benign nature and demonstrating an increased level of HMGA2 expression. The findings have implications for plexiform UL pathological classification and support the evolving recognition of genetic heterogeneity in the pathobiology of UL.
Materials and Methods
Clinical Material and Histology
Four plexiform UL were identified by a gynecological pathologist through analysis of hematoxylin and eosin (H&E)-stained tissue sections of all samples within a tissue bank of more than 100 consented, premenopausal, 25- to 50-year-old women who underwent myomectomy or hysterectomy at Brigham and Women’s Hospital between 2003 and 2007 (cases 1 to 4). Two additional plexiform samples were obtained from the Division of Women’s and Perinatal Pathology at Brigham and Women’s Hospital, one ascertained through the clinical cytogenetics service (case 5) and the other was an archival specimen (case 6); diagnoses were confirmed by independent review of stained tissue sections. Each of these six tumors was shown to consist of smooth muscle tissue with plexiform features comprising >50% of the total tissue mass unless otherwise noted. All six plexiform UL were disaggregated, cultured, and karyotyped by GTG banding according to established protocols.8 Race was self-reported, and no GnRH agonists were used before surgery.
Fluorescence in Situ Hybridization (FISH)
End-sequenced and FISH-verified bacterial artificial chromosomes (BACs)30 were selected using the University of California Santa Cruz Biotechnology Genome Browser and Database (http://genome.ucsc.edu)31 and obtained from the RP11 library (BACPAC Resource Center at the Children’s Hospital Oakland Research Institute, Oakland, CA) or the CTD library (Invitrogen, Carlsbad, CA). DNA was isolated from bacterial cultures following a standard protocol consisting of alkaline lysis, neutralization, and ethanol precipitation.
Probe sets used include an intragenic HMGA2 BAC (RP11-185D13) combined with a commercial probe for the centromere of chromosome 12 (CEP 12) (Abbott Molecular/Vysis Inc., Des Plaines, IL). Two split-apart probe sets were developed to detect intragenic HMGA2 rearrangements, one from BACs RP11-299L9 (5′ HMGA2) and RP11-427K2 (3′ HMGA2), and the other from cosmids 142H1 (5′ HMGA2) and 27E12 (3′ HMGA2).12 The karyotypic abnormality t(12;14)(q15;q23-24), which can be found in typical spindle cell UL, was detected by fusion signals of probes RP11-185D13 located at 12q15 and CTD-3225F7 at 14q24.
FISH was performed as previously described,32 with the exception that three 50-μm sections from the archival paraffin sample ST03-0414 were deparaffinized to isolate nuclei. This was accomplished using 5-minute washes of xylene, 100% ethanol twice, 80% ethanol, and then 50% ethanol followed by incubation in distilled water overnight at 4°C. The sample was then treated with collagenase XI (Sigma, St. Louis, MO) for 2 hours at 37°C, washed with Hanks’ balanced salt solution, incubated with 0.05% trypsin/ethylenediaminetetraacetic acid (Gibco/Invitrogen, Carlsbad, CA) for 1 hour at 37°C, washed, and resuspended in Hanks’ balanced salt solution before applying onto a glass coverslip and baking overnight at 50°C. Slides prepared from sample ST03-0414 were incubated in the HYBrite denaturation/hybridization system (Abbott Molecular/Vysis Inc.) for 10 minutes at 95°C.
Immunohistochemistry
Detection of HMGA2 protein in formalin-fixed, paraffin-embedded plexiform and myometrial tissue sections involved pressure cooker heat-induced antigen retrieval for 2 minutes in citrate buffer followed by a 20-minute cool down, a 5-minute 0.05 mol/L Tris/Tween 20 wash, a 5-minute peroxidase block (DAKO, Carpinteria, CA), and a 5-minute Tris incubation. A 1:2000 dilution of a primary polyclonal anti-HMGA2 antibody (Biocheck Inc., Foster City, CA) was used for 40 minutes. The Envision Plus detection system (DAKO) was then applied, including a 30-minute incubation with goat anti-rabbit immunoglobulin conjugated to a horseradish peroxidase-labeled polymer followed by a 5-minute exposure to the substrate diaminobenzidine to produce a brown precipitate visible by microscopy. Hematoxylin was used as the counterstain. All steps were performed at room temperature unless otherwise noted. HMGA2 protein expression (brown) versus background (blue) staining was evaluated using a semiautomated image analysis system (ACISII; Chromavision, San Juan Capistrano, CA).33 HMGA2 staining for each plexiform sample is expressed as a fold change compared to matched myometrium from the same patient.
RNA Isolation and Quantitative Real-Time Polymerase Chain Reaction
A portion of each sample was frozen in liquid nitrogen immediately after surgical removal. RNA was isolated from cases 1 to 4 from both the tumor and the corresponding myometrial samples using the RNeasy fibrous tissue kit (Qiagen, Valencia, CA). Real-time PCR was performed as previously described,10 using the standard curve method and normalizing the level of HMGA2 in each tissue to that of GAPDH. HMGA2 expression for each plexiform RNA sample is shown as a fold change compared to myometrium from the same patient.
Transcriptional Profiling
RNA isolated from the plexiform UL of cases 1 to 4 was hybridized to Affymetrix Human U133 Plus 2.0 GeneChip oligonucleotide expression microarrays (Santa Clara, CA) using standard protocols at the Harvard Medical School–Partners Health Care Center for Genetics and Genomics. These expression profiles were compared with those previously acquired for myometrium, UL with typical histology, UL with loss of the short arm of chromosome 1 and hypercellularity or atypia, and uterine leiomyosarcomas using the Affymetrix HuFL microarray34,35 as follows. Descriptions of the U133 Plus 2.0 and HuFL microarrays (GPL570 and GPL80 files, respectively) were downloaded from the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/). For each probe set in the GPL80 table, value(s) of the numerical Entrez Gene field were used to query the GPL570 table. If more than one probe set was identified in the GPL570 table, the corresponding expression values were averaged and the data for genes represented on both microarrays were deposited into a single database table. Finally, the resulting raw data for the newly and previously analyzed samples were normalized as described.35 These normalized expression data were deposited at the NCBI Gene Expression Omnibus; the series entry number is GSE9511 and the specific accession identifiers for PLEX1 to PLEX4 are listed under cases 1 to 4 in Table 1. Expression profiles of plexiform UL were compared to those previously determined for a collection of normal myometrium and benign and malignant uterine smooth muscle tumors using a subset of 134 genes (ie, those represented on both Affymetrix microarrays used in this study) by hierarchical cluster analysis using the statistical software package SYSTAT, version 10.2 (Systat Software, Inc., Richmond, CA) with the previously described parameters.35
Table 1.
Clinical Features of Uterine Leiomyomata with Plexiform Histology
| Case no. | Accession no. | Histopathology | Tumor size (cm) | Total number of tumors | Race | Age at surgery (yr) | Menstrual cycle* | Gene Expression Omnibus (GEO) identifier† |
|---|---|---|---|---|---|---|---|---|
| 1 | ST04-074F-2 | Plexiform | 4 × 3.5 × 3 | 2 | White | 44 | Menstruation | GSM241169 |
| 2 | ST06-015F | Focal plexiform | 8.5 × 3 × 3 | 11 | Black | 34 | Secretory | GSM241170 |
| 3 | ST06-004F-1 | Plexiform | 19.5 × 14.5 × 10 | 2 | White | 46 | Menstruation | GSM241171 |
| 4 | ST05-001F-2 | Focal plexiform | 9.3 × 7.6 × 7.2 | 1 | Asian | 53 | Perimenopausal | GSM241172 |
| 5 | ST07-001 | Focal plexiform | 11.5 | TNTC | White | 59 | Menopausal | N.D. |
| 6 | ST03-0414 | Plexiform | N.D. | 13 | White | 26 | N.D. | N.D. |
N.D., not determined; TNTC, too numerous to count.
Based on day one of last menstrual period relative to surgery date (days 1 to 5, menstruation; 6 to 14, proliferative; 14 to 28+, secretory).
http://www.ncbi.nlm.nih.gov/geo/.
Results
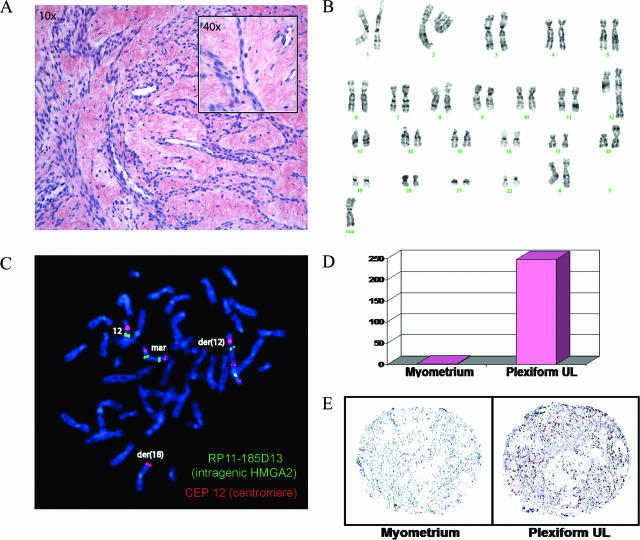
Six UL cases were identified by histopathological analysis of H&E-stained tissue sections as having focal or diffuse plexiform features. These tumors ranged in size from 4 to 19.5 cm and presented in women of varying race, age, and menstrual cycle status (Table 1). Case 1 has the classic plexiform histology and a complex karyotype of 47,XX,add(12)(q24),add(18)(p11.3),+mar[11]/idem,t(8;11)(p23;p13)[3] (Figure 1). Follow-up with metaphase FISH with the intragenic HMGA2 probe RP11-185D13 and the chromosome 12 centromeric probe CEP 12 revealed five copies of HMGA2: one on the apparently normal chromosome 12, two on the der(12), and two on the marker chromosome. Real-time PCR and immunohistochemistry of paraffin sections revealed a 247-fold increase in HMGA2 mRNA and a 3.7-fold elevation of HMGA2 protein in the tumor relative to its matched myometrium. These results as well as those of five additional plexiform UL cases are summarized in Table 2.
Figure 1.
Characterization of plexiform UL case 1 (ST04-074F-2). A: Plexiform histology with characteristic cords of round epithelial-like cells and abundant extracellular matrix (H&E). B: GTG-banded karyotype of 47,XX,add(12)(q24),add(18)(p11.3),+mar[11]/idem,t(8;11)(p23;p13)[3]. C: Metaphase FISH with RP11-185D13 (intragenic HMGA2) (SpectrumGreen) and CEP 12 (centromere of 12) (SpectrumOrange) indicates the presence of five copies of HMGA2, one on the apparently normal chromosome 12, two on the der(12), and two on the marker in 13 of 13 metaphases. D: Real-time PCR demonstrates a 247-fold increase in HMGA2 mRNA compared to myometrium from the same patient. E: Immunohistochemistry on paraffin sections reveals a 3.7-fold elevation of HMGA2 protein in UL relative to the matched myometrium. Data from five additional cases can be found online at http://ajp.amjpathol.org (Supplementary Figures S1 thru S5). Original magnifications: ×10 (A); ×400 (A, inset).
Table 2.
Karyotype, FISH, and HMGA2 mRNA and Protein Expression of Uterine Leiomyomata with Plexiform Histology
| Case no. | Accession no. | GTG-banded karyotype |
|---|---|---|
| 1 | ST04-074F-2 | 47,XX,add(12)(q24),add(18)(p11.3),idem,t(8;11)(p23;p13)[3] |
| 2 | ST06-015F | 46,XX,t(12;14)(q15;q24)[6] |
| 3 | ST06-004F-1 | 46,XX,add(16)(p13.3)[13] |
| 4 | ST05-001F-2 | 47,XX,+mar[2]/46,XX[9] |
| 5 | ST07-001 | 48,XX,add(12)(p1?3),+19,+mar[cp5] |
| 6 | ST03-0414 | 46,XX,der(4)inv(4)(p?12q?21),del(11)(q13),der(14)t(12;14)(q15;q24)[7] |
N.D., not determined.
Table 2A.
Continued
| No. of HMGA2 signals by metaphase FISH | No. of HMGA2 signals by interphase FISH | t(12;14)(q15; q23–24) detected by FISH | Intragenic HMGA2 break detected by FISH | HMGA 2 mRNA (fold-change compared to matched myometrium) | HMGA2 protein (fold-change compared to matched myometrium) |
|---|---|---|---|---|---|
| 1 on normal 12 | 5 | No | No | 247 | 3.6 |
| 2 on add(12q) | |||||
| 2 on marker | |||||
| 1 on normal 12 | 2 | Yes | Yes | 66,467 | 4.1 |
| 1 on der(14) | |||||
| 1 on each normal 12 | 2 | No | No | 2017 | 5 |
| 1 on each normal 12 | 2 | No | N.D. | 568 | 0 |
| 1 on normal 12 | N.D. | N.D. | Yes | N.D. | N.D. |
| 3 on add(12p) | |||||
| 4 on marker | |||||
| N.D. | 3 | Yes | No | N.D. | N.D. |
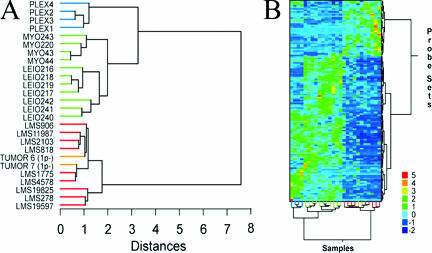
The microarray expression profile of this plexiform UL and others (cases 1 to 4) were included in a hierarchical cluster analysis using 134 probe sets that were previously identified by comparison of myometrium, typical (spindle cell) UL, and leiomyosarcoma (LMS) expression signatures to differentiate benign from malignant uterine smooth muscle tissues.35 Also included in the analysis were the expression profiles of two cases of another type of UL variant, namely cellular UL with partial loss of the short arm of chromosome 1, which were previously shown to cluster within the malignant group.34 In the current analysis, the four plexiform tumors were found to cluster together on a separate node within the benign branch on the sample dendogram (Figure 2A) and matrix plot (Figure 2B). Genes with increased expression in plexiform UL and LMS relative to benign myometrium and typical UL are UBC, RDBP, CYC1, COX5B, CKS1B, DSS1, and TMSB10 whereas those with decreased expression include PIPPIN, ENPP1, GRIA2, CRMP1, TGFB3, NBL1, LAMA3, MORF4L2, KANK, CTNND1, SETDB1, TACR2, GSTM4, APOD, TIMP3, DPP6, DVL3, VCL, TPM4, GTF2I, THRSP, ID2, STAT4, and MMP2. In a separate analysis in which the entire expression profiles of the plexiform UL were directly compared to those of typical UL, the most up-regulated gene in plexiform UL was the α2 chain for type I collagen (COL1A2), with a 361-fold increase.
Figure 2.
Expression analysis of UL with plexiform histology. A: Comparison of the expression profiles of four plexiform UL to a previously defined smooth muscle tumor expression signature using hierarchical clustering. The horizontal length of each arm reflects the relatedness of clusters. Samples are indicated by green terminal segments for benign myometrium (MYO) and UL with normal histology (LEIO), by red terminal segments for malignant leiomyosarcoma (LMS), by orange terminal segments for UL with cellular histology and loss of 1p (TUMOR 6 and TUMOR 7), and by blue terminal segments for UL with plexiform histology (PLEX1 through PLEX4 for cases 1 to 4, respectively). Although cellular UL have previously been shown to cluster with malignant LMS, plexiform UL form their own subcluster within the benign branch. B: Matrix plot displaying clustering of samples (columns) by oligonucleotide sets (rows). The color of each rectangle denotes the corresponding range of normalized (by SD) expression values as shown at the bottom right.
The karyotype for case 2 (see Supplementary Figure S1 at http://ajp.amjpathol.org) is 46,XX,t(12;14)(q15;q24)[6]. This was supported by a metaphase FISH using an intragenic HMGA2 probe RP11-185D13 and chromosome 12 centromere probe CEP 12 that indicated the presence of one copy of HMGA2 on the apparently normal chromosome 12 and translocation of the other copy to the der(14). Surprisingly, metaphase FISH with a split-apart HMGA2 probe set of RP11-299L9 (5′ HMGA2) and RP11-427K2 (3′ HMGA2) showed the presence of one intact copy of HMGA2 and one split copy of HMGA2. To refine the rearrangement, a split-apart HMGA2 probe set of cosmid 142H1 (5′ HMGA2) and cosmid 27E12 (3′ HMGA2) was used and suggested that one copy of HMGA2 is disrupted within a region that includes the 3′ UTR to the beginning portion of intron 3. Real-time PCR demonstrated a 66,467-fold increase in HMGA2 mRNA and paraffin section immunohistochemistry showed a 4.1-fold elevation of HMGA2 protein relative to the matched myometrium.
The karyotype in case 3 (see Supplementary Figure S2 at http://ajp.amjpathol.org) of 46,XX,add(16)(p13.3)[12] as well as the follow-up FISH studies do not suggest a chromosomal alteration of HMGA2. Metaphase FISH with RP11-185D13 (intragenic HMGA2) and CEP 12 (centromere of 12) indicate the presence of two apparently normal chromosomes 12. Metaphase FISH with a split-apart HMGA2 probe set of RP11-299L9 (5′ HMGA2) and RP11-427K2 (3′ HMGA2) shows the presence of two intact copies of HMGA2. Interphase FISH with RP11-185D13 (intragenic HMGA2) and CTD-3225F7 (chromosome 14q24) does not produce a fusion signal, consistent with exclusion of a cryptic t(12;14)(q15;q24) rearrangement. Real-time PCR, however, demonstrated a 2017-fold increase in HMGA2 mRNA compared to the matched myometrium and immunohistochemistry on paraffin sections showed a fivefold elevation of HMGA2 protein in UL relative to the matched myometrium.
Case 4 (see Supplementary Figure S3 at http://ajp.amjpathol.org) had focal areas of plexiform features comprising ∼25% of the tissue examined. Although the remainder of the tumor could not be strictly classified as being plexiform, there was marked, diffuse deposition of extracellular matrix material. The karyotype was 47,XX,+mar[2]/46,XX[9]. Metaphase FISH with RP11-185D13 (intragenic HMGA2) and CEP 12 (centromere of 12) indicates the presence of two apparently normal chromosomes 12 and no expression of HMGA2 on the marker chromosome. Similar to case 3, evidence of HMGA2 involvement was found by real-time PCR, which demonstrated a 568-fold increase in HMGA2 mRNA compared to matched myometrium. Increased HMGA2 protein expression, however, could not be confirmed by immunohistochemistry in stained sections.
The complex karyotype of case 5 (see Supplementary Figure S4 at http://ajp.amjpathol.org), 46,XX,der(?4)?inv(4)(p?12q?21),del(11)(q13),der(14)t(12;14)(q15;q24)[7], involves the 12q15 region where HMGA2 is located. Interphase FISH with RP11-185D13 (intragenic HMGA2) and CEP 12 (centromere of 12) indicates the presence of three copies of HMGA2 and two copies of the chromosome 12 centromere. Interphase FISH with RP11-185D13 (intragenic HMGA2) and CTD-3225F7 (chromosome 14q24) to detect t(12;14)(q15;q24) rearrangements demonstrates the presence of two copies of HMGA2 and a third copy involved in a t(12;14) as shown by a fusion signal. Interphase FISH with a split-apart HMGA2 probe set of RP11-299L9 (5′ HMGA2) and RP11-427K2 (3′ HMGA2) reveals three intact copies of HMGA2. Because of limited material, further investigation was not feasible.
Case 6 (see Supplementary Figure S5 at http://ajp.amjpathol.org) has a karyotype of 48,XX,add(12)(p1?3),+19,+mar[cp5]. Metaphase FISH with the split-apart HMGA2 probe set of RP11-299L9 (5′ HMGA2) and RP11-427K2 (3′ HMGA2) plus CEP 12 (centromere of 12) indicates the presence of eight copies of HMGA2. One intact HMGA2 copy is seen on the presumably normal chromosome 12, two intact copies and one split-apart copy on the der(12), and two intact and two split-apart copies on the marker. Because of limited material, no further analyses were possible.
Discussion
This study presents cytogenetic and expression analyses of six plexiform UL. The results suggest these tumors have been misclassified amid neoplasms with “true” epithelioid differentiation and should instead be grouped among those with extensive hyalinization. This separation would emphasize their prognostic distinction.
The six plexiform UL studied were highly divergent in size and occurred in women of varying race, age, and menstrual cycle status. The unifying finding in each of the six cases was increased expression of HMGA2, a chromatin architectural factor that regulates multiple DNA-dependent activities such as gene transcription and plays a role in cell proliferation and differentiation.15,36 The elevation in HMGA2 mRNA ranged from 284- to more than 66,000-fold and the rise in immunodetectable protein ranged from zero- to fivefold in plexiform UL relative to their adjacent myometria. The lack of detectable protein expression in case 4 is discordant with the 568-fold increase in mRNA, and may represent an artifact because of archival storage. In comparison, chromosomal rearrangement at the HMGA2 locus and aberrant protein expression are found in 10% and 27% of typical (spindle cell) UL, respectively.9,37 The elevation in HMGA2 expression in the plexiform UL appears to result from multiple different mechanisms, including both structural (cases 1, 2, 5, and 6) and submicroscopic (cases 3 and 4). Of note, there is a striking similarity in the karyotypes of cases 1 and 5, both including a der(12) and inverted duplicated marker derived from chromosome 12, which may indicate a similar pathogenetic mechanism for their plexiform phenotype.
Interestingly, the karyotype of the plexiform UL in case 6, which includes two apparently normal chromosomes 12 plus a der(14)t(12;14)(q15;q24) without the reciprocal der(12), closely resembles that of two cases of intravenous leiomyomatosis; IVL is a rare smooth muscle proliferation that invades vascular spaces but is not clinically malignant.38,39 Of note, a plexiform pattern has been observed in some cases of IVL. Although this suggests that significant HMGA2 overexpression is necessary, it is not likely to be sufficient to cause the plexiform phenotype in UL. Further illustration of this observation can be found by inclusion of expression signatures of four of the plexiform UL in a cluster analysis using a previously reported gene set that resolves the spectrum of uterine smooth muscle tissues (myometrium, typical UL, and leiomyosarcoma) into benign versus malignant groups.35 In Figure 2, all plexiform UL clustered into a distinct branch within the benign group. This node did not include a histologically normal UL (Leio240) with a t(12;14)(q15;q23-q24) and proven aberrant HMGA2 expression.35 In fact, nearly 17% of karyotypically abnormal UL have a t(12;14)(q15;q23-q24) associated with significantly increased HMGA2 expression,9,10 and most of these tumors have typical, nonplexiform histology.
A similar cluster analysis using the same gene list was previously performed for two cases of another histological variant of UL, the cellular type with loss of 1p, which differs from plexiform UL by the extent of cellularity and extracellular matrix.34 These cases, TUMOR 6 and TUMOR 7 in Figure 2, grouped with the leiomyosarcomas on the malignant branch. Thus, microarray expression data support the clinical observations that plexiform UL are benign and illustrate that UL are not a single disease process.
The characteristic pseudo-epithelioid/rounded appearance of the plexiform subgroup of UL is secondary to matrix deposition and constrictive pressure, suggesting they are created through their remarkable capacity to synthesize extracellular matrix.18 In fact, the most significantly up-regulated gene in plexiform UL compared to typical UL is one of the type I collagen chain genes, COL1A. Plexiform UL are also generally solitary and benign with an excellent clinical prognosis19; only a single case of a malignant plexiform tumor with increased mitotic activity has been reported.40 This is in contrast to other variants of epithelioid leiomyomata, those with “true” epithelioid differentiation historically called leiomyoblastoma, which have been known to invade locally, metastasize, and recur.20,21 In a series of five of these nonplexiform epithelioid UL, multiple karyotypic abnormalities were reported ranging from normal in one tumor to one tumor with complex changes including t(10;12)(q22;q15) and del(7)(q21.2q31.2).41 The stemline t(10;12) in this latter tumor has breakpoints corresponding to the bands in which MYST4 (MORF) and HMGA2 reside, and the secondary deletion of 7q includes the critical 7q22 region characteristically deleted in UL.32,42 The karyotypes in these nonplexiform epithelioid tumors, however, did not overlap with those of the plexiform UL found in the present study. Despite the malignant potential of leiomyoblastoma, the current classification system combines them with plexiform UL into a single group.19
In conclusion, because of the lower threshold for clinical suspicion of malignancy for “true” epithelioid tumors and based on both clinical observation and the cytogenetic and molecular analyses presented here, plexiform UL may most appropriately be classified among tumors with extensive hyalinization rather than those with “true” epithelioid differentiation. In addition, efforts to dissect the molecular mechanisms of UL should include stratification by histopathology and, ideally, by karyotype and race.
Acknowledgments
We thank Dr. Christopher Fletcher for his facilitation of HMGA2 immunohistochemistry and Christopher Lafargue and Zuned Khalifa in Dr. Mark Rubin’s laboratory for technical assistance in quantitation of the resulting stained slides.
Footnotes
Address reprint requests to Cynthia C. Morton, Ph.D., Brigham and Women’s Hospital, Department of Obstetrics and Gynecology and Reproductive Biology, 77 Avenue Louis Pasteur, New Research Building Room 160, Boston, MA 02115. E-mail: cmorton@partners.org.
Supported by the National Institutes of Health (grants RO1HD046226, RO1CA78895, and T32GM007748 to C.C.M.).
Supplementary material for this article can be found on http://ajp.amjpathol.org.
References
- Cramer SF, Patel A. The frequency of uterine leiomyomas. Am J Clin Pathol. 1990;94:435–438. doi: 10.1093/ajcp/94.4.435. [DOI] [PubMed] [Google Scholar]
- Rein MS, Nowak RA. Biology of uterine myomas and myometrium in vitro. Semin Reprod Endocrinol. 1992;10:310–319. [Google Scholar]
- Coronado GD, Marshall LM, Schwartz SM. Complications in pregnancy, labor, and delivery with uterine leiomyomas: a population-based study. Obstet Gynecol. 2000;95:764–769. doi: 10.1016/s0029-7844(99)00605-5. [DOI] [PubMed] [Google Scholar]
- Hartmann KE, Birnbaum H, Ben-Hamadi R, Wu EQ, Farrell MH, Spalding J, Stang P. Annual costs associated with diagnosis of uterine leiomyomata. Obstet Gynecol. 2006;108:930–937. doi: 10.1097/01.AOG.0000234651.41000.58. [DOI] [PubMed] [Google Scholar]
- Lepine LA, Hillis SD, Marchbanks PA, Koonin LM, Morrow B, Kieke BA, Wilcox LS. Hysterectomy surveillance—United States, 1980–1993. MMWR CDC Surveill Summ. 1997;46:1–15. [PubMed] [Google Scholar]
- Flynn M, Jamison M, Datta S, Myers E. Health care resource use for uterine fibroid tumors in the United States. Am J Obstet Gynecol. 2006;195:955–964. doi: 10.1016/j.ajog.2006.02.020. [DOI] [PubMed] [Google Scholar]
- Nibert M, Heim S. Uterine leiomyoma cytogenetics. Genes Chromosom Cancer. 1990;2:3–13. doi: 10.1002/gcc.2870020103. [DOI] [PubMed] [Google Scholar]
- Rein MS, Friedman AJ, Barbieri RL, Pavelka K, Fletcher JA, Morton CC. Cytogenetic abnormalities in uterine leiomyomata. Obstet Gynecol. 1991;77:923–926. [PubMed] [Google Scholar]
- Meloni AM, Surti U, Contento AM, Davare J, Sandberg AA. Uterine leiomyomas: cytogenetic and histologic profile. Obstet Gynecol. 1992;80:209–217. [PubMed] [Google Scholar]
- Gross KL, Neskey DM, Manchanda N, Weremowicz S, Kleinman MS, Nowak RA, Ligon AH, Rogalla P, Drechsler K, Bullerdiek J, Morton CC. HMGA2 expression in uterine leiomyomata and myometrium: quantitative analysis and tissue culture studies. Genes Chromosom Cancer. 2003;38:68–79. doi: 10.1002/gcc.10240. [DOI] [PubMed] [Google Scholar]
- Schoenberg Fejzo M, Ashar HR, Krauter KS, Powell WL, Rein MS, Weremowicz S, Yoon SJ, Kucherlapati RS, Chada K, Morton CC. Translocation breakpoints upstream of the HMGIC gene in uterine leiomyomata suggest dysregulation of this gene by a mechanism different from that in lipomas. Genes Chromosom Cancer. 1996;17:1–6. doi: 10.1002/(SICI)1098-2264(199609)17:1<1::AID-GCC1>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Schoenmakers EF, Wanschura S, Mols R, Bullerdiek J, Van den Berghe H, Van de Ven WJ. Recurrent rearrangements in the high mobility group protein gene. HMGI-C, in benign mesenchymal tumours. Nat Genet. 1995;10:436–444. doi: 10.1038/ng0895-436. [DOI] [PubMed] [Google Scholar]
- Grosschedl R, Giese K, Pagel J. HMG domain proteins: architectural elements in the assembly of nucleoprotein structures. Trends Genet. 1994;10:94–100. doi: 10.1016/0168-9525(94)90232-1. [DOI] [PubMed] [Google Scholar]
- Wolffe AP. Architectural transcription factors. Science. 1994;264:1100–1101. doi: 10.1126/science.8178167. [DOI] [PubMed] [Google Scholar]
- Reeves R. Molecular biology of HMGA proteins: hubs of nuclear function. Gene. 2001;277:63–81. doi: 10.1016/s0378-1119(01)00689-8. [DOI] [PubMed] [Google Scholar]
- Ligon AH, Moore SD, Parisi MA, Mealiffe ME, Harris DJ, Ferguson HL, Quade BJ, Morton CC. Constitutional rearrangement of the architectural factor HMGA2: a novel human phenotype including overgrowth and lipomas. Am J Hum Genet. 2005;76:340–348. doi: 10.1086/427565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weedon MN, Lettre G, Freathy RM, Lindgren CM, Voight BF, Perry JR, Elliott KS, Hackett R, Guiducci C, Shields B, Zeggini E, Lango H, Lyssenko V, Timpson NJ, Burtt NP, Rayner NW, Saxena R, Ardlie K, Tobias JH, Ness AR, Ring SM, Palmer CN, Morris AD, Peltonen L, Salomaa V, Smith GD, Groop LC, Hattersley AT, McCarthy MI, Hirschhorn JN, Frayling TM. A common variant of HMGA2 is associated with adult and childhood height in the general population. Nat Genet. 2007;39:1245–1250. doi: 10.1038/ng2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nucci MR, Quade BJ. Uterine mesenchymal tumors, chapter 20. Crum CP, Lee KR, editors. Frankfurt: Elsevier Saunders,; Diagnostic Gynecologic and Obstetric Pathology. 2005 [Google Scholar]
- Kurman RJ, Norris HJ. Mesenchymal tumors of the uterus. VI. Epithelioid smooth muscle tumors including leiomyoblastoma and clear-cell leiomyoma: a clinical and pathologic analysis of 26 cases. Cancer. 1976;37:1853–1865. doi: 10.1002/1097-0142(197604)37:4<1853::aid-cncr2820370433>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- Lavin P, Hajdu SI, Foote FW., Jr Gastric and extragastric leiomyoblastomas: clinicopathologic study of 44 cases. Cancer. 1972;29:305–311. doi: 10.1002/1097-0142(197202)29:2<305::aid-cncr2820290206>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- Kyriazis AP, Kyriazis AA. Uterine leiomyoblastoma (epithelioid leiomyoma) neoplasm of low-grade malignancy. A histopathologic study. Arch Pathol Lab Med. 1992;116:1189–1191. [PubMed] [Google Scholar]
- Evans HL, Chawla SP, Simpson C, Finn KP. Smooth muscle neoplasms of the uterus other than ordinary leiomyoma. A study of 46 cases, with emphasis on diagnostic criteria and prognostic factors. Cancer. 1988;62:2239–2247. doi: 10.1002/1097-0142(19881115)62:10<2239::aid-cncr2820621028>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- Mazur MT, Kraus FT. Histogenesis of morphologic variations in tumors of the uterine wall. Am J Surg Pathol. 1980;4:59–74. [PubMed] [Google Scholar]
- Nagel H, Brinck U, Luthje D, Fuzesi L. Plexiform leiomyoma of the uterus in a patient with breast carcinoma: case report and review of the literature. Pathology. 1999;31:292–294. doi: 10.1080/003130299105197. [DOI] [PubMed] [Google Scholar]
- Kaminski PF, Tavassoli FA. Plexiform tumorlet: a clinical and pathologic study of 15 cases with ultrastructural observations. Int J Gynecol Pathol. 1984;3:124–134. [PubMed] [Google Scholar]
- Seidman JD, Thomas RM. Multiple plexiform tumorlets of the uterus. Arch Pathol Lab Med. 1993;117:1255–1256. [PubMed] [Google Scholar]
- Goodhue WW, Susin M, Kramer EE. Smooth muscle origin of uterine plexiform tumors. Arch Pathol. 1974;97:263–268. [PubMed] [Google Scholar]
- Nunez-Alonso C, Battifora HA. Plexiform tumors of the uterus: ultrastructural study. Cancer. 1979;44:1707–1714. doi: 10.1002/1097-0142(197911)44:5<1707::aid-cncr2820440526>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- Fisher ER, Paulson JD, Gregorio RM. The myofibroblastic nature of the uterine plexiform tumor. Arch Pathol Lab Med. 1978;102:477–480. [PubMed] [Google Scholar]
- Cheung VG, Nowak N, Jang W, Kirsch IR, Zhao S, Chen XN, Furey TS, Kim UJ, Kuo WL, Olivier M, Conroy J, Kasprzyk A, Massa H, Yonescu R, Sait S, Thoreen C, Snijders A, Lemyre E, Bailey JA, Bruzel A, Burrill WD, Clegg SM, Collins S, Dhami P, Friedman C, Han CS, Herrick S, Lee J, Ligon AH, Lowry S, Morley M, Narasimhan S, Osoegawa K, Peng Z, Plajzer-Frick I, Quade BJ, Scott D, Sirotkin K, Thorpe AA, Gray JW, Hudson J, Pinkel D, Ried T, Rowen L, Shen-Ong GL, Strausberg RL, Birney E, Callen DF, Cheng JF, Cox DR, Doggett NA, Carter NP, Eichler EE, Haussler D, Korenberg JR, Morton CC, Albertson D, Schuler G, de Jong PJ, Trask BJ. Integration of cytogenetic landmarks into the draft sequence of the human genome. Nature. 2001;409:953–958. doi: 10.1038/35057192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D, Baertsch R, Diekhans M, Furey TS, Hinrichs A, Lu YT, Roskin KM, Schwartz M, Sugnet CW, Thomas DJ, Weber RJ, Haussler D, Kent WJ. The UCSC Genome Browser Database. Nucleic Acids Res. 2003;31:51–54. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore SD, Herrick SR, Ince TA, Kleinman MS, Cin PD, Morton CC, Quade BJ. Uterine leiomyomata with t(10;17) disrupt the histone acetyltransferase MORF. Cancer Res. 2004;64:5570–5577. doi: 10.1158/0008-5472.CAN-04-0050. [DOI] [PubMed] [Google Scholar]
- Bismar TA, Demichelis F, Riva A, Kim R, Varambally S, He L, Kutok J, Aster JC, Tang J, Kuefer R, Hofer MD, Febbo PG, Chinnaiyan AM, Rubin MA. Defining aggressive prostate cancer using a 12-gene model. Neoplasia. 2006;8:59–68. doi: 10.1593/neo.05664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christacos NC, Quade BJ, Dal Cin P, Morton CC. Uterine leiomyomata with deletions of Ip represent a distinct cytogenetic subgroup associated with unusual histologic features. Genes Chromosom Cancer. 2006;45:304–312. doi: 10.1002/gcc.20291. [DOI] [PubMed] [Google Scholar]
- Quade BJ, Wang TY, Sornberger K, Dal Cin P, Mutter GL, Morton CC. Molecular pathogenesis of uterine smooth muscle tumors from transcriptional profiling. Genes Chromosom Cancer. 2004;40:97–108. doi: 10.1002/gcc.20018. [DOI] [PubMed] [Google Scholar]
- Bustin M. Regulation of DNA-dependent activities by the functional motifs of the high-mobility-group chromosomal proteins. Mol Cell Biol. 1999;19:5237–5246. doi: 10.1128/mcb.19.8.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klotzbücher M, Wasserfall A, Fuhrmann U. Misexpression of wild-type and truncated isoforms of the high-mobility group I proteins HMGI-C and HMGI(Y) in uterine leiomyomas. Am J Pathol. 1999;155:1535–1542. doi: 10.1016/S0002-9440(10)65469-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quade BJ, Dal Cin P, Neskey DM, Weremowicz S, Morton CC. Intravenous leiomyomatosis: molecular and cytogenetic analysis of a case. Mod Pathol. 2002;15:351–356. doi: 10.1038/modpathol.3880529. [DOI] [PubMed] [Google Scholar]
- Dal Cin P, Quade BJ, Neskey DM, Kleinman MS, Weremowicz S, Morton CC. Intravenous leiomyomatosis is characterized by a der(14)t(12;14)(q15;q24). Genes Chromosom Cancer. 2003;36:205–206. doi: 10.1002/gcc.10159. [DOI] [PubMed] [Google Scholar]
- Kuroda H, Konishi I, Nanbu K, Mandai M, Komatsu T, Yamamoto S, Yamabe H, Kida A, Mori T. Malignant plexiform tumor of the uterus: an unusual variant of epithelioid leiomyosarcoma. Gynecol Oncol. 1996;63:270–275. doi: 10.1006/gyno.1996.0319. [DOI] [PubMed] [Google Scholar]
- Karaiskos C, Pandis N, Bardi G, Sfikas K, Tserkezoglou A, Fotiou S, Heim S. Cytogenetic findings in uterine epithelioid leiomyomas. Cancer Genet Cytogenet. 1995;80:103–106. doi: 10.1016/0165-4608(94)00167-a. [DOI] [PubMed] [Google Scholar]
- Sargent MS, Weremowicz S, Rein MS, Morton CC. Translocations in 7q22 define a critical region in uterine leiomyomata. Cancer Genet Cytogenet. 1994;77:65–68. doi: 10.1016/0165-4608(94)90151-1. [DOI] [PubMed] [Google Scholar]