Abstract
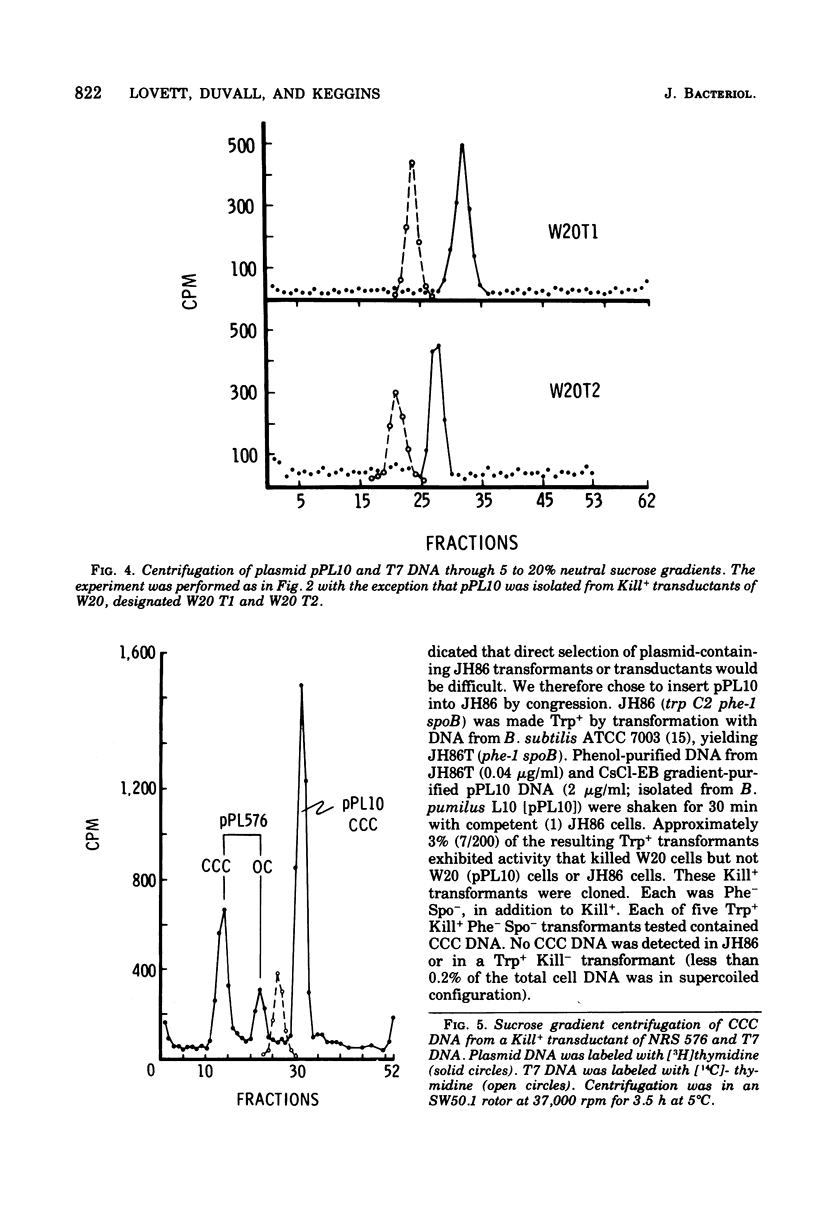
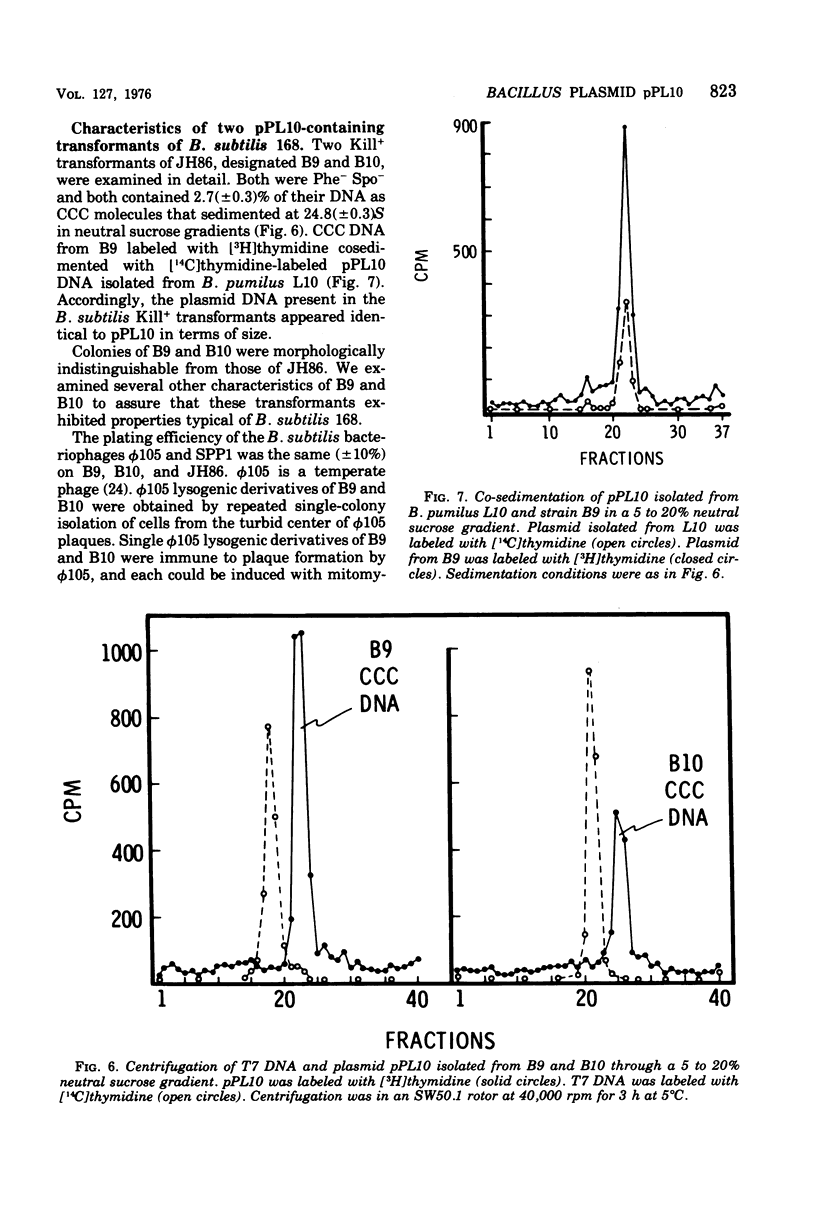
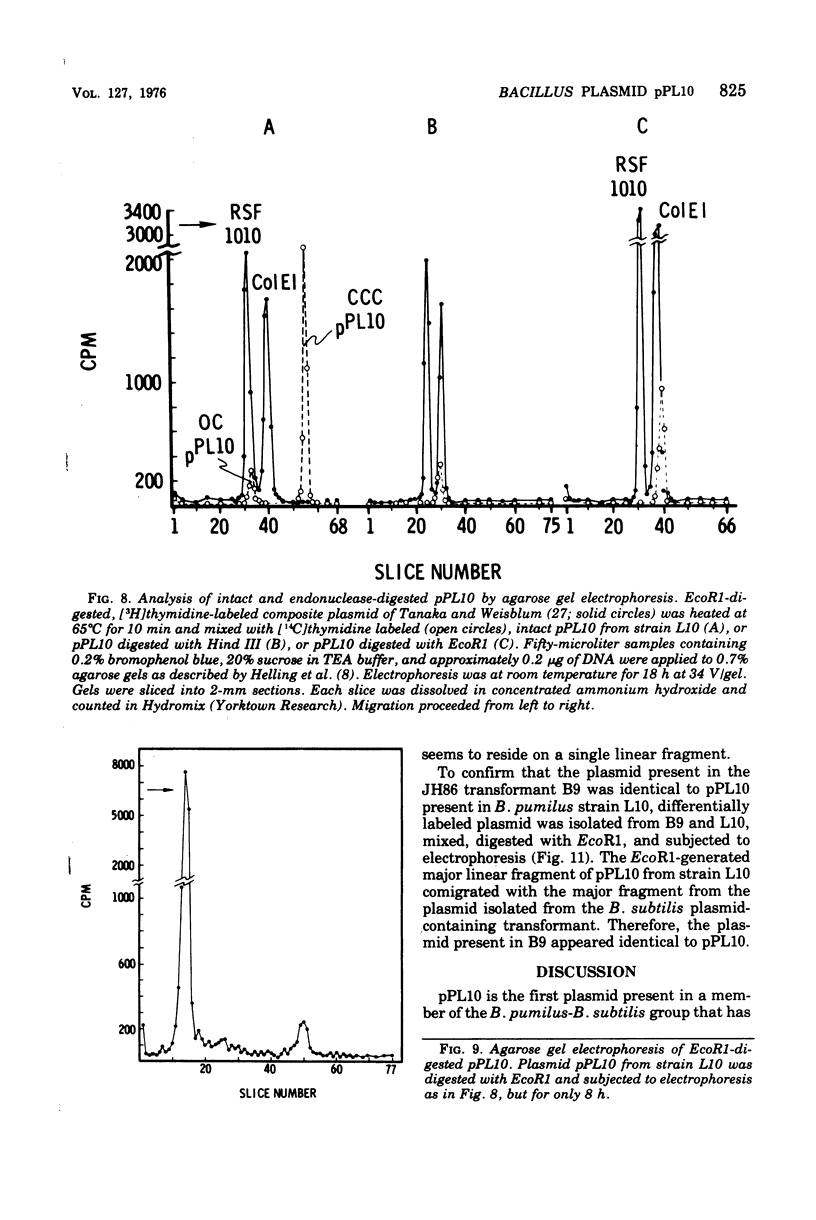
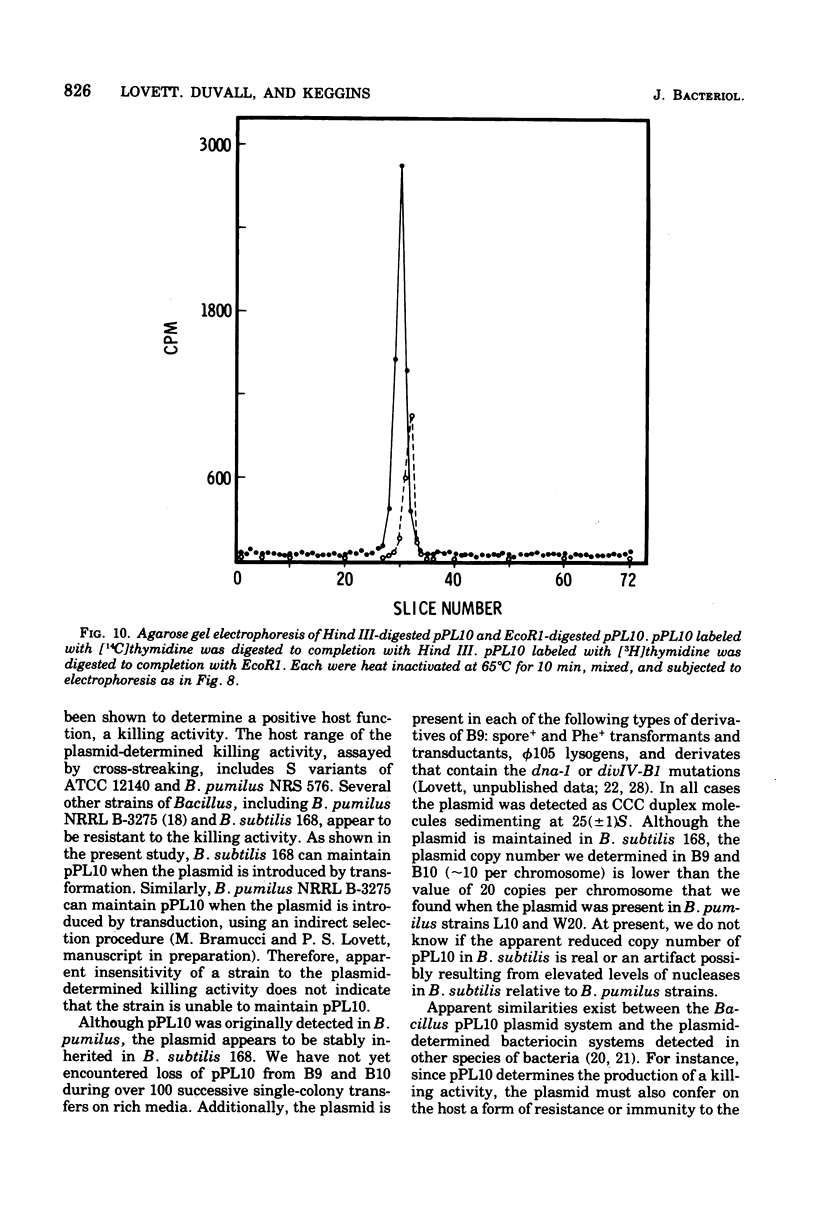
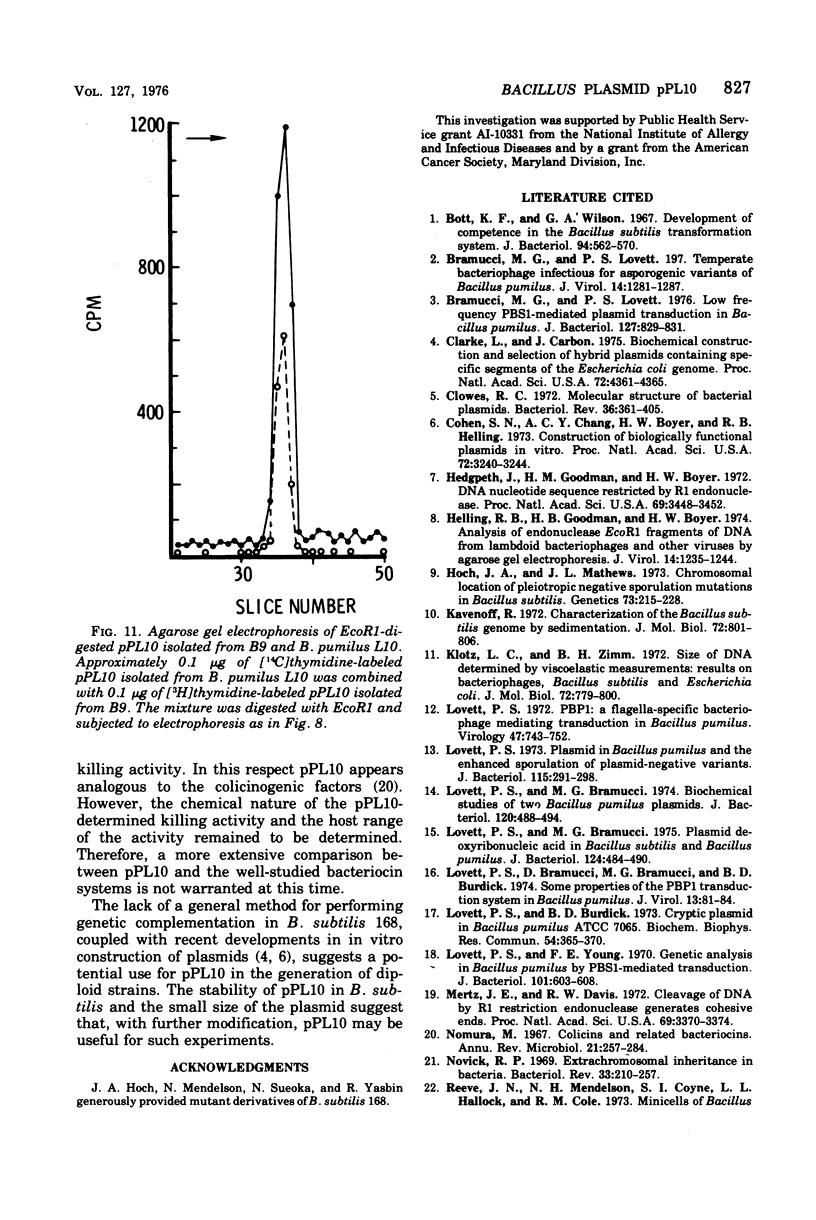
Bacillus pumilus ATCC 12140 harbors 10 or more copies per chromosome of each two small plasmids. Variants of this strain were isolated which were sensitive to a killing activity produced by the plasmid-containing parent. Each of 24 such sensitive (S) variants tested lacked detectable levels of supercoiled deoxyribonucleic acid. Transduction of S variants to the Kill+ phenotype was performed using phage PBP1 propagated on a mutant of ATCC 12140, designated strain L10, that remained Kill+ but retained only a single plasmid species (plasmid pPL10; molecular weight, approximately 4.4 X 10(6), approximately 20 copies per chromosome; RHO = 1.698). Resulting Kill+ transductants of S variants contained a single plasmid species having a size and copy number comparable to that of pPL10. Transfer of pPL10 from strain L10 TO B. pumilus strain NRS 576 was accomplished by transduction with selection for the Kill+ phenotype. Strain NRS 576 naturally harbors about two copies per chromosome of a 28-million-dalton plasmid, pPL576. In Kill + transductants of NRS 576, plasmids pPL10 and pPL576 stably coexisted at a ratio of about 11 molecules of pPL10 to 1 molecule of pPL576. Therefore, pPL576 and pPL10 are compatible plasmids. B. subtilis 168 is naturally resistant to pPl10- determined killing activity. Plasmid pPl10 was therefore inserted into B-subtilis 168 by transformation, using an indirect selection procedure and a spoB mutant as recipient. The plasmid is stably maintained at an estimated 10 copies per chromosome in the spore- recipient and in spore+ transformants. pPL10 is sensitive to cleavage by the endonucleases Hind III and EcoR1.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bott K. F., Wilson G. A. Development of competence in the Bacillus subtilis transformation system. J Bacteriol. 1967 Sep;94(3):562–570. doi: 10.1128/jb.94.3.562-570.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramucci M. G., Lovett P. S. Low-frequency, pbsi-mediated plasmid transduction in Bacillus pumilus. J Bacteriol. 1976 Aug;127(2):829–831. doi: 10.1128/jb.127.2.829-831.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke L., Carbon J. Biochemical construction and selection of hybrid plasmids containing specific segments of the Escherichia coli genome. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4361–4365. doi: 10.1073/pnas.72.11.4361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clowes R. C. Molecular structure of bacterial plasmids. Bacteriol Rev. 1972 Sep;36(3):361–405. doi: 10.1128/br.36.3.361-405.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C., Boyer H. W., Helling R. B. Construction of biologically functional bacterial plasmids in vitro. Proc Natl Acad Sci U S A. 1973 Nov;70(11):3240–3244. doi: 10.1073/pnas.70.11.3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedgpeth J., Goodman H. M., Boyer H. W. DNA nucleotide sequence restricted by the RI endonuclease. Proc Natl Acad Sci U S A. 1972 Nov;69(11):3448–3452. doi: 10.1073/pnas.69.11.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helling R. B., Goodman H. M., Boyer H. W. Analysis of endonuclease R-EcoRI fragments of DNA from lambdoid bacteriophages and other viruses by agarose-gel electrophoresis. J Virol. 1974 Nov;14(5):1235–1244. doi: 10.1128/jvi.14.5.1235-1244.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch J. A., Mathews J. L. Chromosomal location of pleiotropic negative sporulation mutations in Bacillus subtilis. Genetics. 1973 Feb;73(2):215–228. doi: 10.1093/genetics/73.2.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavenoff R. Characterization of the Bacillus subtilis W23 genome by sedimentation. J Mol Biol. 1972 Dec 30;72(3):801–806. doi: 10.1016/0022-2836(72)90192-1. [DOI] [PubMed] [Google Scholar]
- Klotz L. C., Zimm B. H. Size of DNA determined by viscoelastic measurements: results on bacteriophages, Bacillus subtilis and Escherichia coli. J Mol Biol. 1972 Dec 30;72(3):779–800. doi: 10.1016/0022-2836(72)90191-x. [DOI] [PubMed] [Google Scholar]
- Lovett P. S., Bramucci D., Bramucci M. G., Burdick B. D. Some properties of the PBP1 transduction system in Bacillus pumilus. J Virol. 1974 Jan;13(1):81–84. doi: 10.1128/jvi.13.1.81-84.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovett P. S., Bramucci M. G. Biochemical studies of two Bacillus pumilus plasmids. J Bacteriol. 1974 Oct;120(1):488–494. doi: 10.1128/jb.120.1.488-494.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovett P. S., Bramucci M. G. Plasmid deoxyribonucleic acid in Bacillus subtilis and Bacillus pumilus. J Bacteriol. 1975 Oct;124(1):484–490. doi: 10.1128/jb.124.1.484-490.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovett P. S., Burdick B. D. Cryptic plasmid in Bacillus pumilus ATCC 7065. Biochem Biophys Res Commun. 1973 Sep 5;54(1):365–370. doi: 10.1016/0006-291x(73)90931-5. [DOI] [PubMed] [Google Scholar]
- Lovett P. S. PBPI: a flagella specific bacteriophage mediating transduction in Bacillus pumilus. Virology. 1972 Mar;47(3):743–752. doi: 10.1016/0042-6822(72)90564-8. [DOI] [PubMed] [Google Scholar]
- Lovett P. S. Plasmid in Bacillus pumilus and the enhanced sporulation of plasmid-negative variants. J Bacteriol. 1973 Jul;115(1):291–298. doi: 10.1128/jb.115.1.291-298.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovett P. S., Young F. E. Genetic analysis in Bacillus pumilus by PBSI-mediated transduction. J Bacteriol. 1970 Feb;101(2):603–608. doi: 10.1128/jb.101.2.603-608.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertz J. E., Davis R. W. Cleavage of DNA by R 1 restriction endonuclease generates cohesive ends. Proc Natl Acad Sci U S A. 1972 Nov;69(11):3370–3374. doi: 10.1073/pnas.69.11.3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura M. Colicins and related bacteriocins. Annu Rev Microbiol. 1967;21:257–284. doi: 10.1146/annurev.mi.21.100167.001353. [DOI] [PubMed] [Google Scholar]
- Novick R. P. Extrachromosomal inheritance in bacteria. Bacteriol Rev. 1969 Jun;33(2):210–263. doi: 10.1128/br.33.2.210-263.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeve J. N., Mendelson N. H., Coyne S. I., Hallock L. L., Cole R. M. Minicells of Bacillus subtilis. J Bacteriol. 1973 May;114(2):860–873. doi: 10.1128/jb.114.2.860-873.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy P. H., Smith H. O. DNA methylases of Hemophilus influenzae Rd. II. Partial recognition site base sequences. J Mol Biol. 1973 Dec 25;81(4):445–459. doi: 10.1016/0022-2836(73)90516-0. [DOI] [PubMed] [Google Scholar]
- Rutberg L. Mapping of a temperate bacteriophage active on Bacillus subtilis. J Virol. 1969 Jan;3(1):38–44. doi: 10.1128/jvi.3.1.38-44.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHILDKRAUT C. L., MARMUR J., DOTY P. Determination of the base composition of deoxyribonucleic acid from its buoyant density in CsCl. J Mol Biol. 1962 Jun;4:430–443. doi: 10.1016/s0022-2836(62)80100-4. [DOI] [PubMed] [Google Scholar]
- STUDIER F. W. SEDIMENTATION STUDIES OF THE SIZE AND SHAPE OF DNA. J Mol Biol. 1965 Feb;11:373–390. doi: 10.1016/s0022-2836(65)80064-x. [DOI] [PubMed] [Google Scholar]
- Tanaka T., Weisblum B. Construction of a colicin E1-R factor composite plasmid in vitro: means for amplification of deoxyribonucleic acid. J Bacteriol. 1975 Jan;121(1):354–362. doi: 10.1128/jb.121.1.354-362.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White K., Sueoka N. Temperature-sensitive DNA synthesis mutants of Bacillus subtilis--appendix: theory of density transfer for symmetric chromosome replication. Genetics. 1973 Feb;73(2):185–214. doi: 10.1093/genetics/73.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]