Abstract
Synthesis of mouse metallothionein (MT)-I and MT-II is transcriptionally induced by the synthetic glucocorticoid, dexamethasone (DEX) or both in vivo as well as in numerous cell lines. However, the location(s) of a glucocorticoid response element (GRE) has not been described. The observation that a marked MT-I gene, as well as heterologous genes, when placed in the context of 17 kb of flanking sequence from the MT locus, are inducible by DEX and lipopolysaccharide in transgenic mice renewed the search for the GRE. Analysis of a series of deletion constructs from this 17-kb region in cultured cells identified a single 455-bp region that conferred DEX induction on a reporter gene. This 455-bp region contains two GREs that bind to the glucocorticoid receptor as assessed by gel mobility shift. Deletion of this fragment from the 17-kb flanking region eliminates the DEX responsiveness of reporter genes. The two GREs, which are located ≈1 kb upstream of the MT-II gene and ≈7 kb upstream of the MT-I gene, are necessary for induction of both genes and can function independently of elements within the proximal promoter region of either gene.
Metallothioneins (MTs) were discovered by virtue of their ability to bind heavy metals such as zinc, copper, and cadmium (reviewed in ref. 1). Subsequently, it was shown that MT mRNA is induced by these and many other heavy metals and that this induction is transcriptional (2), being mediated by a transcription factor, MTF-1, that binds to multiple metal response elements (MREs) located in the proximal promoters of the MT genes (3).
At about the same time that induction of MT by metals was being explored, Karin and Herschman (4) showed that MT expression was also inducible by glucocorticoids. It subsequently was shown that this induction was also transcriptional both in vivo and in cultured cells (5, 6). We were eager to identify the glucocorticoid response element (GRE) that controls mouse MT-I gene induction, but all attempts to demonstrate induction after gene transfer of various MT-I gene constructs with up to 1.8 kb of flanking sequences failed. These experiments were performed in cell culture (7) and in transgenic mice (8); in each case induction by metals was observed but there was no induction by dexamethasone (DEX), a synthetic glucocorticoid that would induce endogenous MT genes in the same experiments (reviewed in ref. 9). These results were particularly baffling because Karin et al. (10) identified a GRE in the promoter of the human MT-IIA gene that was located ≈250 bp 5′ of the transcription start site, which was one of the first GREs to be identified (11). Thus, the location of elements involved in the regulation of the mouse MT-I gene by glucocorticoids appeared to be different than in the human MT-IIA gene. During the last 15 years sporadic studies have been directed toward the localization of GREs that regulate this (12) and other MT genes from various species (13), but no bona fide GREs have been identified for any other MT gene except the human MT-IIA gene.
The MT gene family has grown in the last few years. In the mouse, there are now four MT genes (MT-IV, MT-III, MT-II, and MT-I) with the same transcriptional orientation clustered within a 50-kb region on chromosome 8 (14). Expression of MT-IV and MT-III is restricted to a few cell types, and their expression is relatively unaffected by metals or hormones that induce MT-II and MT-I (15, 16). MT-I and MT-II, which are 6 kb apart, are regulated coordinately by metals, glucocorticoid hormones, and lipopolysaccharide (LPS)-induced cytokines such as interleukin (IL)-1 and IL-6 (17). The MT gene family in humans is more complicated. At least 16 human MT genes are clustered on chromosome 16. Human MT-IV and MT-III are linked (15) and map close to a cluster of 14 MT genes in which the MT-IIA gene precedes 13 MT-I variants (18).
Expression of MT-driven genes in transgenic mice generally has been successful but rather unpredictable in terms of level of expression, inducibility, and tissue distribution (19). In an attempt to improve the expression of MT genes in transgenic mice we assembled a marked MT-I gene (MT-I*) flanked by 10-kb piece of DNA that lies 5′ of the MT-II gene and 7-kb piece that lies 3′ of the MT-I gene (20). These flanking DNAs were chosen because they contain DNaseI hypersensitive sites that might mark the location of important regulatory elements (21). Indeed, constructs with these flanking regions were expressed at higher levels, copy-number-dependent expression was improved, and tissue-specific expression patterns were similar to that of the endogenous MT-I gene (20, 22). Remarkably, these flanking sequences conferred DEX inducibility to the marked MT-I gene (20). This study was designed to find the GREs in this construct.
MATERIALS AND METHODS
Gene Constructs.
The 10-kb EcoRI fragment that lies upstream of the MT-II gene and the 7-kb EcoRI–XbaI fragment that lies downstream of the MT-I gene were combined in a Bluescript (Stratagene) vector with a unique cloning site between them. Either a marked MT-I gene (MT-I*), which has a 2-bp insertion in the 5′ untranslated region that converts a BglII site to an EcoRV site (20), or a MRE-βGeo reporter gene (23) was inserted between the two flanking regions. The 5′3′-human growth hormone (hGH) construct has a hGH gene with only 83 bp of promoter sequence inserted between the two MT-flanking regions. The 5′3′-albumin/tumor growth factor type α (TGFα) construct contains the mouse albumin promoter (0.3 kb) and enhancer (2.0-kb NheI–BamHI), the TGFα cDNA, and the hGH gene (24, 25). The 5′3′ΔGRE construct was prepared by deleting the 455-bp NcoI–EcoRI fragment from the 3′ end of the 10-kb EcoRI fragment. A set of five progressive 5′ deletions was prepared from the 10-kb fragment using convenient restriction sites, the shortest of which was the 2-kb SmaI–EcoRI fragment designated 5′a. This deletion series then was inserted upstream of the MRE-βGeo gene. The SmaI–EcoRI fragment then was subdivided as depicted (5′b, 5′c 5′d, and 5′e), and these smaller regions were inserted upstream of MRE-βGeo. Deletions of the two GREs within the 455-bp fragment were constructed by PCR using primers at the ends of the fragment and adjacent to either GRE1 or GRE2. The PCR products were cut with restriction enzymes whose sites were included in the primers and cloned into Bluescript. DNA fragments containing either wild-type sequences or deleted GREs were excised with convenient restriction sites and reassembled to generate 455-bp fragments with deletions of GRE1, GRE2, or both. These fragments then were moved into the MRE-βGeo reporter construct for functional testing. The plasmid pKH has been described (26); the 5′ KpnI–ApaI region was deleted to create pKHΔ; pKHΔ+GRE was created by inserting a pair of 21-bp oligos containing a consensus GRE, CGGTACAAAATGTTCTGGGCC and CAGAA- CATTTTGTACCGGTAC where the underlined nucleotides represent the consensus GRE.
Assays of Gene Expression.
Transgenic mice were generated by standard techniques (27). Adult transgenic mice of selected lines were injected with either 1 mg/kg LPS or 5 mg/kg DEX dissolved in PBS by intraperitoneal injection. Control littermates received an equal volume of PBS. Six hours later they were killed by CO2 asphyxiation, and their livers were removed, placed on dry ice, and stored at −80°C until total nucleic acid was prepared by homogenization in proteinase K/SDS followed by phenol/chloroform extraction and ethanol precipitation. The abundance of specific mRNAs was determined by solution hybridization as described (20). For cell culture experiments, plasmids (20 μg) were introduced into baby hamster kidney (BHK) cells by the calcium phosphate method (28). Before these experiments, the BHK cells were transfected with a plasmid containing the Rous sarcoma virus promoter driving expression of the rat glucocorticoid receptor (GR) (provided by Keith Yamamoto, University of California, San Francisco) and a hygromycin resistance gene. A clone (2990-5) with high expression of GR mRNA was identified and used for these studies. When constructs contained MRE-βGeo as reporter gene, stable populations of cells were selected by growth in 800 μg/ml of G418 and 100 μM ZnSO4. When MT-I* was the reporter gene, the cells were selected in 20 μM CdSO4. In all cases, pools of at least 50 clones were analyzed to minimize any positional insertion effects. To measure induction, the cells were split into several plates and removed from selection for several days to allow expression to return to basal levels. Then either 100 μM ZnSO4, 100 nM DEX, or both were added. Cells were harvested 18–20 hr later for assay of β-galactosidase activity using o-nitrophenyl β-d-galactopyranoside as substrate (23), or they were harvested 6–8 hr later to measure MT mRNA by solution hybridization (20). All measurements were made in at least triplicate, and the results are expressed as fold induction relative to untreated cells.
Gel Mobility Shift Analysis.
The ability of the identified GREs to interact with the DNA binding domain of the human GR (29) was assayed by mobility shift assay (30). As a positive control, a 375-bp EagI–ScaI fragment of the human MT-IIa gene was isolated while a 335-bp RsaI piece of the mouse MT-I cDNA was used as a negative control. The positive control contains a single GRE (10), whereas the negative control contains no consensus GRE sequences and does not respond to DEX in vitro (7) or in vivo (8). Four different SstII–XhoI fragments were tested that contained either both putative GREs, a single GRE, or neither GRE (wild type, D3, D4, and D5, respectively). The various probes were gel-purified and then radiolabeled by filling in the ends with either [α32P]dATP or dCTP using DNA polymerase. The binding reaction contained 20,000 cpm of probe (≈0.5 ng DNA) in a binding buffer composed of 10 mM Hepes (pH 7.9), 4 mM Tris⋅HCl (pH 7.9), 30 mM KCl, 1 mM EDTA, 1 mM DTT, and 10% glycerol in a final volume of 20 μl. In addition, each reaction contained 0.5 μg of (poly)dI-dC, and the specificity of the interaction was tested by inclusion of 100 ng of unlabeled probe. The binding reaction was conducted by combining all the above reagents and incubating for 10 min on ice after which 250 ng of purified GR was added and kept on ice for an additional 30 min. The binding reaction mixture then was electrophoresed on a 0.5× TBE (45 mM Tris/45 mM boric acid/1 mM EDTA, pH 8.3)-buffered 4% polyacrylamide gel and processed for autoradiography.
RESULTS
MT-Flanking Regions Confer Induction by DEX and LPS in Transgenic Mice.
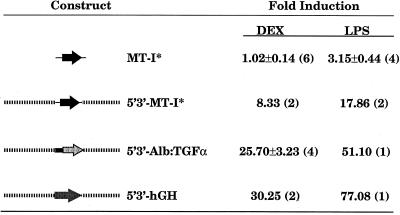
A 3.8-kb marked MT-I gene was used to generate several lines of transgenic mice (20). Adult mice were injected with DEX, LPS, or PBS as a control, and liver MT-I* mRNA was measured 6 hr later by solution hybridization using an allele-specific oligonucleotide. Fig. 1 shows that there was no induction of MT-I* by DEX, whereas LPS induced expression 3- to 4-fold. Previous studies suggested that an LPS response element lies between −180 and −350 of the MT-I transcription start site (31); hence, the induction of MT-I* by LPS was expected. Similar results were obtained with three independent lines of mice. The lack of induction of MT-I* by DEX is consistent with previous results obtained after transfection of the same EcoRI fragment into HeLa cells (7). DEX and LPS typically induce endogenous MT-I mRNA by 5- to 30-fold; most of the variability arises from the basal level of expression, which is difficult to control. The same constructs showed good induction by zinc and cadmium (data not shown), consistent with the location of several MREs in the 150 bp upstream of the TATA box (20).
Figure 1.
Analysis of DEX and LPS response in mice expressing transgenes flanked by DNA from the mouse MT locus. Adult transgenic mice were treated with DEX or LPS for 6 hr, total nucleic acid was harvested from the liver, and transgene expression was measured by solution hybridization. The values represent mean ± SEM relative to littermate controls injected with PBS; (n), number of animals per group. The flanking mouse MT locus sequences are indicated by the dashed lines; for a better depiction of these regions, see Fig. 6A.
When the MT-I* gene was flanked by the 10-kb EcoRI fragment located 5′ of the MT-II gene and a 7-kb EcoRI–XbaI fragment located 3′ of the MT-I gene and introduced into mice, the results were striking. The MT-flanking regions conferred induction by both LPS and DEX (Fig. 1; 5′3′-MT-I*). DEX induction was observed not only in liver but also in kidney, spleen, pancreas, and brain (data not shown). In this construct, GREs in the flanking regions could be acting in concert with elements in the MT-I promoter. To ascertain whether they could act independently of the MT-I promoter, the MT-flanking regions were tested with either an albumin promoter/enhancer (5′3′-albumin/tumor growth factor α) or with the hGH gene and its own minimal (extending to −83) promoter (5′3′-hGH). Fig. 1 shows that expression from both of these constructs was stimulated at least 25-fold by either DEX or LPS. Thus, we conclude that response elements for both DEX and LPS exist within the 10- and 7-kb MT-flanking regions, and these elements can act independently of the MT-I promoter.
Localization of a GRE Within the 5′ Flanking Region of the MT-II Gene.
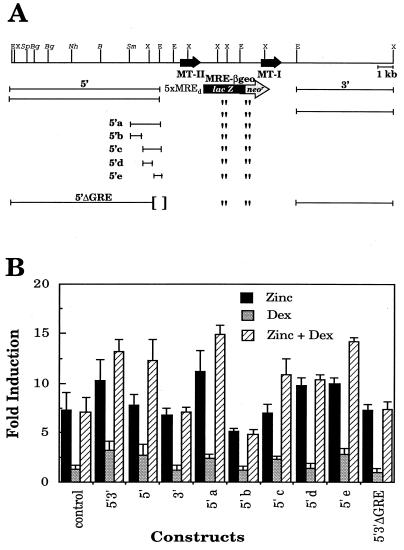
Having obtained DEX-responsive gene expression in vivo, we established a simpler transfection assay with BHK cells to test a series of deletions as a means of locating the GRE(s). To optimize responsiveness to DEX, the BHK cells first were transfected with an Rous sarcoma virus–GR construct, and a clone with maximal expression of GR mRNA was isolated. Then, the 5′ and 3′ flanking regions that gave DEX responsiveness in vivo were added to a MRE-βGeo reporter gene (Fig. 2A). This reporter gene has five MRE-d sequences upstream of a TATA box that drives expression of a gene encoding a β-galactosidase-neomycin phosphotransferase fusion protein (23). This construct can be used as both a reporter and a selectable gene. Various deletions were made in the MT-flanking regions and transfected into the modified BHK cells, and pools of cells resistant to G418 were selected in the presence of 100 μM zinc. Under these selection conditions, MRE-βGeo would be optimally induced. Then, several replica plates of cells were grown for several days without zinc to allow expression to return to basal levels, before being tested with DEX, zinc, or both. Using this approach, selection does not depend upon DEX responsiveness.
Figure 2.
The mouse MT-I/II locus, deletion constructs and their activity in cultured cells as assessed by β-galactosidase activity. (A) The boundary of the 10-kb 5′ flank is indicated by the EcoRI sites (E) whereas the 3′ flank is bounded by a EcoRI and XbaI (X) site. The various deletions of the 5′ region were obtained by restriction digests of the sites indicated (S, SpeI; Bg, BglII; Nh, NheI; B, BamHI; Sm, SmaI). All were tested, but only the results for the largest deletion (5′a) are shown. The construct 5′3′ΔGRE is the same as 5′3′ except that the region defined in 5′e is deleted. (B) Transcriptional response of the MRE–βGeo fusion constructs in BHK cells in response to induction with 100 μM ZnSO4, 100 nM DEX, or both in comparison to untreated cells expressing the tested construct. All results are derived from at least three separate experiments and are expressed as the mean ± SEM.
The presence of both 5′ and 3′ flanking sequences conferred DEX responsiveness to MRE-βGeo, and the response to a combination of DEX plus zinc was additive (Fig. 2B, construct 5′3′). When the two flanking sequences were tested separately, only the 5′ sequence conferred DEX responsiveness (Fig. 2B, compare 5′ and 3′). A series of deletions was prepared from the distal end of the 5′ region using convenient restriction sites. All the deletions were equally responsive to DEX down to the smallest 2-kb SmaI–EcoRI fragment tested in this series (Fig. 2B, 5′a). This region was further subdivided into four fragments that were tested individually. The smallest region tested that conferred DEX responsiveness was a 455-bp NcoI–EcoRI fragment, designated 5′e in Fig. 2A. This region had the same activity as the 10-kb fragment from which it was derived (Fig. 2B), and when it was deleted from the 5′3′ construct (5′3′ΔGRE), DEX responsiveness was lost.
Two Functional GREs Are Located Within the 455-bp Fragment.
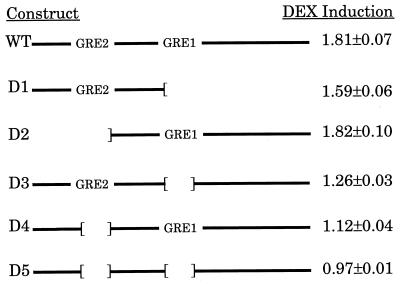
The sequence of the 455-bp NcoI–EcoRI fragment is shown in Fig. 3. Two potential GREs were identified based on similarity to the consensus GRE sequence (GGTACAnnnTGTTCT). The more promoter-proximal one, GRE1, differs at two positions from the consensus, whereas the more distal one, GRE2, differs in three positions. Initially, we thought that only GRE1 was likely to be functional; however, a mutation of two critical bases in this GRE (32) did not eliminate DEX responsiveness of reporter constructs carrying this 455-bp fragment (data not shown). Furthermore, subclones from the 455-bp fragment that included only GRE1 or GRE2 were functional (Fig. 4, constructs D1 and D2), whereas subclones that lacked both had no activity (data not shown). Also, deletion of two small regions including GRE1 and GRE2 (underlined in Fig. 3) from the 455-bp fragment abolished DEX responsiveness (Fig. 4, construct D5). As in previous experiments, when responsiveness to DEX alone was observed, DEX also enhanced the response observed with zinc (data not shown). Surprisingly, subfragments that contained a single GRE (Fig. 4, constructs D1 and D2) produced better DEX responsiveness than larger fragments with small deletions of either GRE (Fig. 4, constructs D3 and D4). These experiments were repeated five times with the same result. The results suggest that sequences upstream of GRE2 and downstream of GRE1 have an inhibitory effect on DEX responsiveness.
Figure 3.
Sequence of the region that confers a DEX response. The locations of the GREs are indicated by the shaded boxes, and the regions deleted in Fig. 4 are underlined. The sequence begins with the NcoI site and ends with the EcoRI site shown in bold. The consensus GRE sequence is GGTACAnnnTGTTCT.
Figure 4.
Deletion analysis within the DEX-responsive fragment. The intact 455-bp NcoI/EcoRI fragment is denoted as wild type whereas the 5′ fragment lacking GRE1 is D1 and the 3′ fragment lacking GRE2 is D2. Constructs D3–5 represent internal deletions of GRE1, GRE2, or both GRE1 and GRE2, respectively. The values shown to the right of each construct represent the mean ± SEM induction by DEX (fold) of MRE–βGeo from five separate experiments of samples assayed in triplicate.
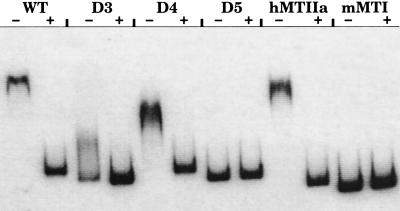
The ability of the DNA fragments used in the functional assays to bind to the GR also was examined in a mobility shift assay. A DNA fragment from the human MT-IIA promoter that has a functional GRE (11) was used as a positive control and the mouse MT-I cDNA was used as a negative control (Fig. 5). The wild-type 455-bp fragment with both GREs was completely retarded by the GR, as was the deletion mutant containing only GRE1, whereas the deletion mutant containing only GRE2 was only partially retarded under these assay conditions. Significantly, there was no retardation of the construct (D5) with both GREs deleted (Fig. 5).
Figure 5.
Gel mobility shift analysis of the DNA fragments tested in Fig. 4. The ability of the DNA-binding domain of the GR to retard the mobility of wild type (WT) and mutants lacking either one (D3 or D4) or both (D5) putative GREs was tested and compared with a sequence known to contain a functional GRE (hMTIIa). The specificity of the interaction was verified by the lack of interaction between GR and a fragment containing the mMTI cDNA and also by the ability to block the shift with excess unlabeled probe as indicated by + above the lanes.
Induction of MT-I* by DEX in Transfected Cells.
The 5′3′ fragments also were tested in BHK cells with the MT-I* reporter gene that had revealed DEX responsiveness in vivo. For these experiments the constructs were transfected into BHK cells modified to express GR, and populations of cells resistant to 20 μM cadmium were selected. BHK cells do not express their endogenous MT genes; hence after mock transfections, all the cells are killed in 20 μM cadmium. As in the previous experiments, cells were plated without cadmium and grown for several days before being tested for induction of MT-I mRNA by treatment with DEX, zinc, or both. MT-I* mRNA was measured by solution hybridization with an oligonucleotide complementary to the 3′ untranslated region of MT-I mRNA. When the MT-I* gene with both 5′ and 3′ flanking regions (Fig. 6A, line 1) was tested in BHK cells, DEX induced MT-I mRNA as well as zinc (≈6-fold), and DEX plus zinc were additive (Fig. 6B; 5′3′-MT-I*).
Figure 6.
Analysis of MT induction by DEX and zinc in cells transfected with various MT-containing constructs. The intact MT locus is shown on the fourth line. The various regions of the MT locus that were tested are indicated in A. The MT-I* constructs depicted in the top three lines contain the 3.8-kb EcoRI fragment of the MT-I gene with a mutation that changes a BglII site in the 5′ untranslated region to a EcoRV site (indicated by a ▾). 5′3′-MT-I* contains the entire flanking region that was tested in transgenic mice (see Fig. 1) and 5′3′ΔGRE-MT-I* has all of the flanking sequence except for a NcoI/EcoRI deletion of the DEX-response region whereas GRE-MT-I* contains only the NcoI/EcoRI fragment with the DEX response region (∗) upstream of MT-I*. The pKH construct contains MT-I and MT-II and includes the region that conferred a DEX response (∗); pKHΔ has the DEX region deleted via a KpnI/ApaI restriction digest; pKHΔ+GRE has a single consensus GRE (see Materials and Methods) in replace of the KpnI/ApaI region. The response of the cells to 100 μM ZnSO4, 100 nM DEX, or both was measured by solution hybridization using oligonucleotides specific for either MT-I (B and C) or MT-II (D). The values (mean ± SEM) for MT-I are derived from four separate samples; the values for MT-II represent means from two populations of cells, each assayed in triplicate.
Induction of Both MT-II and MT-I by the DEX-Responsive Fragment.
Normally, the MT-II gene lies between the GRE element located in the 455-bp NcoI–EcoRI fragment and the MT-I gene. In the previous experiment the MT-II gene and ≈4 kb of downstream flanking regions were removed. To test whether additional GREs are in those regions, we examined the activity of construct pKH, which includes the DNA between the GREs and the MT-I gene (Fig. 6A). This construct produced ≈2-fold induction of both MT-II and MT-I mRNA in response to DEX and slightly more induction of both mRNAs with zinc plus DEX (Fig. 6 C and D). However, when the region containing GRE1 and GRE2 was deleted (Fig. 6A, construct pKHΔ) induction of both mRNAs was lost (Fig. 6 C and D). Replacement of the region deleted with a single consensus GRE (Fig. 6A, construct pKHΔ+GRE) restored induction of both MT-I and MT-II by DEX (Fig. 6 C and D).
DISCUSSION
The only GRE shown previously to regulate any MT gene is the one located ≈250 bp upstream of the human MT-IIA gene. The identification of the two GREs upstream of the mouse MT-II gene relied on analysis of pools of many individual clones. The results were reproducible, and in all cases where DEX induced expression, the combination of DEX plus zinc gave an even larger induction than that achieved by zinc alone. DEX gave the best induction (≈6-fold) with construct 5′3′-MT-I* (which has intact 5′ and 3′ regions flanking MT-I*), and it was comparable to that achieved with zinc. Excellent induction by either metals or DEX also was observed in transgenic mice bearing this construct (20). However, when MT-I* was substituted by MRE-βGeo, induction by DEX was less than that achieved with zinc. Likewise, removal of all the flanking regions, except the 455-bp fragment containing the GREs (construct GRE-MT-I*), resulted in less induction by DEX than by zinc. These results suggest that there may be synergism between GRs bound to the GREs and other transcription factors bound to elements in both the flanking regions and the proximal MT promoter. However, these GREs do not require any MT-specific promoter elements because they function well with either a minimal hGH promoter or with the albumin promoter/enhancer (Fig. 1). These results argue against the possibility that the GREs upstream of the MT-II gene act in concert with cryptic GREs in the mouse MT-I promoter region that have been shown to function only when taken out of context (12).
The results reported here contradict some of our earlier results. In particular, we tested construct pKH previously in HeLa cells and reported that the small increase (1.2- to 2.3-fold) in MT-I and MT-II mRNA accumulation observed after administration of DEX was much lower than the response of the endogenous MT genes (26). The induction of MT-I and MT-II that we observe in BHK cells fortified with GR after transfection of pKH is only 2- to 3-fold; thus, the small effect of the GREs in this context precluded its identification 13 years ago. Another result that seems at odds with our current understanding is that when the MT locus was amplified in S180 cells after selection for cadmium resistance, the amplified genes selectively lost their responsiveness to DEX but not to metals (33). We knew from Southern blot experiments that a large region of DNA contiguous with the MT-I gene was amplified and would have included the GREs that we have identified here. Thus, we now would argue that the loss of DEX responsiveness in those cells probably was due to changes in the amount or activity of the GR or some other factor that confers responsiveness to DEX. In fact, another group (34) was able to preserve hormone responsiveness of MT genes in a different cell line after amplification of the locus.
The human MT-IIA promoter is responsive to DEX after gene transfer into cells or mice (35–37). The sequences flanking the GRE in the hMT-IIA gene do not resemble those flanking either GRE1 or GRE2; thus, it is difficult to know whether the GREs of mouse and human are homologous. In contrast, the sequences upstream of rat and mouse MT-II genes are clearly homologous, and the two GRE sequences are in comparable locations relative to the transcription start sites; the GRE2 sequence is identical in the two species, whereas there are two base differences in GRE1. Thus, it is not clear whether the rat has one or two functional GREs. Although two functional GREs are in the mouse, we have shown that they can be substituted with a single GRE and that it can regulate both MT-I and MT-II genes (Fig. 6 C and D). Considering that none of the MT-I genes from any species have been shown convincingly to respond to glucocorticoids when taken out of the normal chromosomal location, it is possible that in all vertebrate species the MT-I genes respond to GRs interacting with GREs upstream of the MT-II genes.
It is unusual for one GRE (or a pair of GREs in the mouse) to regulate two (or more in human) genes. There are precedents for centrally located enhancers regulating two divergently transcribed genes (38, 39) and for locus control regions to control expression of several tandemly linked genes as in the case of the β-globin locus (40), but we could not find an example of the arrangement we describe here. There does not appear to be any preference for activation of the closer MT-II gene either in vivo (26, 41) or after gene transfer (Fig. 6 C and D). Several possible mechanisms could allow one GRE (or a closely spaced pair) to control two genes. The GR bound to the GRE could interact transiently with basal transcription factors bound to either MT-II or MT-I gene promoters to facilitate transcription. Alternatively, stable interactions could be established, with the probability of forming such a complex with factors bound to either MT-II or MT-I gene promoters being similar. A third possibility is that the promoters for MT-I and MT-II genes are, in fact, drawn together by factors bound to common promoter elements, and the GR may join the complex to activate both promoters simultaneously.
Although not the thrust of this paper, it is clear from the transgenic analysis, that the combined 5′ and 3′ flanking regions also contain elements that can respond to transcription factors that are activated in response to LPS, which induces the expression of interleukins by macrophages. IL-1 and IL-6 have been shown to induce MT-I and MT-II in various organs in vivo (42). Activation of the IL-6 receptor results in the activation of JAK family tyrosine kinases, which then activate STAT transcription factors (reviewed in ref. 43). STAT binding sites have been located in the promoters of several acute phase proteins that respond to IL-6 (44). The present results suggest that similar sequences may reside in the MT flanking regions. Previous transgenic studies (32) indicate that there is also an LPS-responsive element in the proximal MT-I gene promoter, located between −185 and −350, but there are no consensus IL-6 response elements in that region.
The significance of glucocorticoid induction of MT-I and MT-II is uncertain, primarily because the function(s) of MTs themselves are not established, despite the availability of mice that either overexpress or fail to express these ubiquitous proteins (22, 45). The induction of MTs by glucocorticoids results in the synthesis of apo-MT, which then binds available metals, predominantly zinc. This can result in the accumulation of zinc within tissues and a corresponding reduction in serum levels of zinc. MT-bound zinc accumulates in fetal liver during the latter stages of development in response to the rise in maternal corticosterone at that stage of gestation (46). We have shown that mice lacking functional MT genes are born with less than half the normal amount of hepatic zinc, and the mice manifest developmental abnormalities if they are subsequently reared on zinc-deficient diets (47). Thus, glucocorticoid induction of MTs provides a means of sequestering zinc. This function also might be important in response to inflammation or starvation, conditions that lead to elevated glucocorticoid levels.
Acknowledgments
We thank David Koeller for his help in the initial phases of this work, Keith Yamamoto for providing purified GR protein and the Rous sarcoma virus–GR plasmid, and Mary Avarbock for her technical assistance. This work was supported in part by National Institutes of Health Grant CA-61268 (to R.D.P.) and HD-23657 (to R.L.B.). E.J.K. was supported by Public Health Service National Research Service Award T32 GM07270.
ABBREVIATIONS
- MT
metallothionein
- DEX
dexamethasone
- GRE
glucocorticoid response element
- MRE
metal response element
- LPS
lipopolysaccharide
- hGH
human growth hormone
- BHK
baby hamster kidney
- GR
glucocorticoid receptor
- IL
interleukin
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF-012925).
References
- 1.Vallee B L. Neurochem Int. 1995;27:23–33. doi: 10.1016/0197-0186(94)00165-q. [DOI] [PubMed] [Google Scholar]
- 2.Durnam D M, Palmiter R D. J Biol Chem. 1981;256:5712–5716. [PubMed] [Google Scholar]
- 3.Radtke F, Heuchel R, Georgiev O, Hergersberg M, Gariglio M, Dembic Z, Schaffner W. EMBO J. 1993;12:1355–1362. doi: 10.1002/j.1460-2075.1993.tb05780.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karin M, Herschman H K. Science. 1979;204:176–177. doi: 10.1126/science.432639. [DOI] [PubMed] [Google Scholar]
- 5.Hager L J, Palmiter R D. Nature (London) 1981;291:340–342. doi: 10.1038/291340a0. [DOI] [PubMed] [Google Scholar]
- 6.Mayo K E, Palmiter R D. J Biol Chem. 1981;256:2621–2624. [PubMed] [Google Scholar]
- 7.Mayo K E, Warren R, Palmiter R D. Cell. 1982;29:99–108. doi: 10.1016/0092-8674(82)90094-0. [DOI] [PubMed] [Google Scholar]
- 8.Palmiter R D, Chen H Y, Brinster R L. Cell. 1982;29:701–710. doi: 10.1016/0092-8674(82)90186-6. [DOI] [PubMed] [Google Scholar]
- 9.Mayo K E, Palmiter R D. Biochem Actions Horm. 1985;12:69–88. [Google Scholar]
- 10.Karin M, Haslinger A, Holtgreve H, Richards R I, Krauter P, Westphal H M, Beato M. Nature (London) 1984;308:513–519. doi: 10.1038/308513a0. [DOI] [PubMed] [Google Scholar]
- 11.Slater E P, Hesse H, Beato M. Ann NY Acad Sci. 1994;733:103–112. doi: 10.1111/j.1749-6632.1994.tb17260.x. [DOI] [PubMed] [Google Scholar]
- 12.Plisov S Y, Nichiporenko M G, Shkapenko A L, Kumarev V P, Baranova L V, Merkulova T I. FEBS Lett. 1994;352:339–341. doi: 10.1016/0014-5793(94)00980-5. [DOI] [PubMed] [Google Scholar]
- 13.Yu C W, Lin L Y. Biochim Biophys Acta. 1995;1262:150–154. doi: 10.1016/0167-4781(95)00070-w. [DOI] [PubMed] [Google Scholar]
- 14.Palmiter R D, Sandgren E P, Koeller D M, Findley S D, Brinster R L. In: Metallothionein III: Biological Roles and Medical Implications. Suzuki K T, Imura N, Kimura M, editors. Basel: Birkhauser; 1993. pp. 417–424. [Google Scholar]
- 15.Quaife C J, Findley S D, Erickson J C, Froelick G J, Kelly E J, Zambrowicz B P, Palmiter R D. Biochemistry. 1994;33:7250–7259. doi: 10.1021/bi00189a029. [DOI] [PubMed] [Google Scholar]
- 16.Masters B A, Quaife C J, Erickson J C, Kelly E J, Froelick G J, Zambrowicz B P, Brinster R L, Palmiter R D. J Neurosci. 1994;14:5844–5857. doi: 10.1523/JNEUROSCI.14-10-05844.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palmiter, R. D. (1987) Experientia Suppl. 52, 63–80. [DOI] [PubMed]
- 18.West A K, Hildebrand C E, Karin M, Richards R I. Genomics. 1990;8:513–518. doi: 10.1016/0888-7543(90)90038-v. [DOI] [PubMed] [Google Scholar]
- 19.Palmiter R D, Brinster R L. Annu Rev Genet. 1986;20:465–499. doi: 10.1146/annurev.ge.20.120186.002341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Palmiter R D, Sandgren E P, Koeller D M, Brinster R L. Mol Cell Biol. 1993;13:5266–5275. doi: 10.1128/mcb.13.9.5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.MacArthur C A, Lieberman M W. J Biol Chem. 1987;267:2161–2165. [PubMed] [Google Scholar]
- 22.Iszard M B, Liu J L, Liu Y, Dalton T D, Andrews G K, Palmiter R D, Klaassen C D. Toxicol Appl Pharmacol. 1994;133:305–312. doi: 10.1006/taap.1995.1155. [DOI] [PubMed] [Google Scholar]
- 23.Palmiter R D. Proc Natl Acad Sci USA. 1994;91:1219–1223. doi: 10.1073/pnas.91.4.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pinkert C A, Ornitz D M, Brinster R L, Palmiter R D. Genes Dev. 1987;1:268–276. doi: 10.1101/gad.1.3.268. [DOI] [PubMed] [Google Scholar]
- 25.Sandgren E P, Luetteke N C, Palmiter R D, Brinster R L, Lee D C. Cell. 1990;61:1121–1135. doi: 10.1016/0092-8674(90)90075-p. [DOI] [PubMed] [Google Scholar]
- 26.Searle P F, Davison B C, Stuart G W, Wilkie T M, Norstedt G, Palmiter R D. Mol Cell Biol. 1984;4:1221–1230. doi: 10.1128/mcb.4.7.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brinster R L, Chen H Y, Trumbauer M, Yagle M K, Palmiter R D. Proc Natl Acad Sci USA. 1985;82:4438–4442. doi: 10.1073/pnas.82.13.4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Graham F L, Van den Eb A J. Virology. 1973;52:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- 29.Freedman L P, Luisi B F, Korszun Z R, Basavappa R, Sigler P B, Yamamoto K R. Nature (London) 1988;334:543–546. doi: 10.1038/334543a0. [DOI] [PubMed] [Google Scholar]
- 30.Schreiber E, Matthias P, Müller M M, Schaffner W. Nucleic Acids Res. 1989;17:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Durnam D M, Hoffman J S, Quaife C J, Benditt E P, Chen H Y, Brinster R L, Palmiter R D. Proc Natl Acad Sci USA. 1984;81:1053–1056. doi: 10.1073/pnas.81.4.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Becker P, Gloss B, Schmid W, Strähle U, Schütz G. Nature (London) 1986;324:686–688. doi: 10.1038/324686a0. [DOI] [PubMed] [Google Scholar]
- 33.Mayo K E, Palmiter R D. J Biol Chem. 1982;257:3061–3067. [PubMed] [Google Scholar]
- 34.Thibodeau J, DeSouza C, Smorawinska M, Thirion J-P. FEBS Lett. 1992;310:75–78. doi: 10.1016/0014-5793(92)81150-k. [DOI] [PubMed] [Google Scholar]
- 35.Karin M, Richards R. Nature (London) 1982;299:797–802. doi: 10.1038/299797a0. [DOI] [PubMed] [Google Scholar]
- 36.Selden R F, Yun J S, Moore D D, Rowe M E, Malia M A, Wagner T E, Goodman H M. J Endocrinol. 1989;122:49–60. doi: 10.1677/joe.0.1220049. [DOI] [PubMed] [Google Scholar]
- 37.Hammer R E, Brinster R L, Palmiter R D. Cold Spring Harbor Symp Quant Biol. 1985;50:379–387. doi: 10.1101/sqb.1985.050.01.048. [DOI] [PubMed] [Google Scholar]
- 38.Hudson B G, Reeders S T, Tryggvason K. J Biol Chem. 1993;268:26033–26036. [PubMed] [Google Scholar]
- 39.Herrera V L, Ruiz-Opazo N. J Biol Chem. 1990;265:9555–9562. [PubMed] [Google Scholar]
- 40.Crossley M, Orkin S H. Curr Opin Genet Dev. 1993;3:232–237. doi: 10.1016/0959-437x(93)90028-n. [DOI] [PubMed] [Google Scholar]
- 41.Yagle M K, Palmiter R D. Mol Cell Biol. 1985;5:291–294. doi: 10.1128/mcb.5.2.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.De S K, McMaster M T, Andrews G K. J Biol Chem. 1990;265:15267–15274. [PubMed] [Google Scholar]
- 43.Ihle J N. Cell. 1996;84:331–334. doi: 10.1016/s0092-8674(00)81277-5. [DOI] [PubMed] [Google Scholar]
- 44.Ripperger J, Fritz S, Richter K, Dreir B, Schneider K, Lochner K, Marschalek R. Ann NY Acad Sci. 1995;762:252–260. doi: 10.1111/j.1749-6632.1995.tb32330.x. [DOI] [PubMed] [Google Scholar]
- 45.Masters B A, Kelly E J, Quaife C J, Brinster R L, Palmiter R D. Proc Natl Acad Sci USA. 1994;91:584–588. doi: 10.1073/pnas.91.2.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Quaife C, Hammer R E, Mottet N K, Palmiter R D. Dev Biol. 1986;118:549–555. doi: 10.1016/0012-1606(86)90025-4. [DOI] [PubMed] [Google Scholar]
- 47.Kelly E J, Quaife C J, Froelick G J, Palmiter R D. J Nutr. 1996;126:1782–1790. doi: 10.1093/jn/126.7.1782. [DOI] [PubMed] [Google Scholar]