Summary
The plague-causing bacterium Yersinia pestis utilizes a Type III Secretion System (T3SS) to deliver effector proteins into mammalian cells where they interfere with signal transduction pathways that mediate phagocytosis and the inflammatory response. Effector proteins are injected through a hollow needle structure composed of the protein YscF. YscG and YscE act as "chaperones" to prevent premature polymerization of YscF in the cytosol of the bacterium prior to assembly of the needle. Here, we report the crystal structure of the YscEFG protein complex at 1.8 Å resolution. Overall, the structure is similar to that of the analogous PscEFG complex from the Pseudomonas aeruginosa T3SS, but there are noteworthy differences. The structure confirms that, like PscG, YscG is a member of the tetratricopeptide repeat (TPR) family of proteins. YscG binds tightly to the C-terminal half of YscF, implying that it is this region of YscF that controls its polymerization into the needle structure. YscE interacts with the N-terminal TPR motif of YscG but makes very little direct contact with YscF. Its function may be to stabilize the structure of YscG and/or to participate in recruiting the complex to the secretion apparatus. No electron density could be observed for the N-terminal 49 residues of YscF. This and additional evidence suggest that the N-terminus of YscF is disordered in the complex with YscE and YscG. As expected, conserved residues in the C-terminal half of YscF mediate important intra- and intermolecular interactions in the complex. Moreover, the phenotypes of some previously characterized mutations in the C-terminal half of YscF can be rationalized in terms of the structure of the heterotrimeric YscEFG complex.
Keywords: type III secretion, plague, Yersinia, tetratricopeptide repeat, needle protein
Yersinia pestis, the causative agent of plague, is arguably the deadliest pathogen in history and a likely instrument of bioterrorism1,2. Like many other Gram-negative bacterial pathogens of plants and animals, Y. pestis relies on a contact-dependent type III secretion system (T3SS) to deliver a small number of proteins, termed effectors, through a hollow needle extending across the inner and outer membranes of the bacterium and into the cytosol of eukaryotic cells3,4. These effector Yops (Yersinia outer proteins) interfere with signal transduction pathways that regulate the actin cytoskeleton, phagocytosis, apoptosis and the inflammatory response, thereby favoring survival of the bacteria5. Over 20 proteins are involved in the assembly of the syringe-like type III secretion "injectisome", a so-called nano-machine6. The structure of the injectisome is similar in some respects to that of the bacterial flagellum, confirming previous phylogenetic analyses that revealed substantial sequence similarities between several of their core components7.
The assembly and operation of the injectisome is assisted by a number of small bacterial proteins termed T3SS chaperones. Several different types of secretion chaperones have been recognized to date8. Class I chaperones, which are small acidic polypeptides that exhibit little sequence similarity but adopt similar dimeric folds, generally interact with one unique effector (or, less frequently, with two or three effectors) and stimulate its secretion via the type III pathway. Class II chaperones, which interact with the translocator proteins of the T3SS that breach the plasma membrane of mammalian cells (e.g., Yersinia YopB, YopD), are somewhat larger and composed in part of tetratricopeptide repeats (TPRs). Flagellar chaperones, exemplified by FliS, have an entirely different structure9 and have therefore been designated Class III. Because the CesA chaperone that interacts with the EspA filament protein of the T3SS in enteropathogenic E. coli is structurally distinct from the other classes10, it has been assigned the designation of Class IV. Finally, the unique structure of Yersinia YscE 11, the co-chaperone (along with scG) for the needle protein YscF, prompted it to be categorized as yet another type of secretion chaperone (Class V).
The structural diversity of secretion chaperones may reflect their varying roles in type III secretion and flagellar assembly. It is believed that the principal function of Class I chaperones and CesA is to mediate targeting of their cognate binding partners to the type III secretion apparatus12. However, recent evidence suggests that Class I chaperones may also serve to mask aggregation-prone membrane localization domains in their cognate effectors13. In addition to stimulating the secretion of their binding partners, at least some Class II chaperones also appear to play a regulatory role in type III secretion14,15. The main function of the Class III chaperone FliS and the heterodimeric chaperone YscE (Class V)/YscG, on the other hand, evidently is to prevent premature polymerization of their cognate binding partners (FliC and YscF, respectively) prior to secretion 9,16.
The needles of the T3SSs in Yersinia, Salmonella, Shigella, Escherichia coli, Burkholderia, and Pseudomonas are each composed primarily of a single protein (YscF, PrgI, MxiH, EscF, BsaL, and PscF, respectively) that polymerizes to form a hollow conduit17–22 with an inner diameter of 25 Å and a length of approximately 40–60 nm3. Efforts to elucidate the three dimensional structures of T3SS needle proteins have been hampered by their tendency to polymerize into oligomers of varying sizes in vitro. However, it was possible to create and crystallize monomeric forms of two needle proteins, MxiH20 and BsaL21, by deleting several residues from their C- terminus. The crystal structures of both MxiH and BsaL monomers revealed similar α-helical hairpin conformations.
Quinaud and coworkers demonstrated that in P. aeruginosa, PscE and PscG trap the needle protein PscF in a ternary 1:1:1 complex, blocking the polymerization of PscF16. An interaction between YscE and YscG, the orthologs of PscE and PscG in Y pestis, had previously been identified by yeast two-hybrid technology, suggesting the existence of a similar complex in this organism23. To investigate the detailed molecular architecture of the complex between the co-chaperones YscE/YscG and the Yersinia needle protein YscF, we crystallized the heterotrimer and determined its structure at a resolution of 1.8 Å.
Results and Discussion
Similarities and differences between the structures of YscE/YscF/YscG and PscE/PscF(55–85)/PscG
The crystal structure of the YscE/YscF/YscG multiprotein complex (YscEFG) was solved by the SAD (Single wavelength Anomalous Dispersion) method using incorporated selenomethionine (Table 1). The crystallographic asymmetric unit contains one heterotrimer, which is consistent with a 1:1:1 stoichiometry (Figure 1). The final YscEFG model consists of 204 amino acids, including residues 10–63 of YscE, 50–87 of YscF and 3–114 of YscG.
Table 1.
Data collection and refinement statistics
| Parameter | YscEFG complexa |
|---|---|
| Data collection | |
| Space group | P21212 |
| Cell dimensions a, b, c (Å) | 74.5, 94.5, 31.3 |
| Wavelength (Å) | 0.97942 |
| Resolution (Å) b | 50-1.8 (1.86-1.80) |
| Rmerge b | 10.1 (36.9) |
| I/σIb | 11.9 (4.3) |
| Completeness (%)b | 98.6 (95.1) |
| Redundancyb | 6.5 (4.6) |
| Refinement statistics | |
| Resolution (Å) | 50-1.8 |
| Unique reflections | 20372 |
| Rwork / Rfree (%) | 19.03/22.13 |
| No. atoms | |
| Protein | 1632 |
| Water | 227 |
| B-factors (Å2) | |
| Proteinc | 27.1 / 24.4 / 26.5 |
| Water | 40.5 |
| R.m.s deviations | |
| Bond lengths (Å) | 0.01 |
| Bond angles (°) | 1.4 |
| Ramachandran Plot Statistics | |
| Residues in most favored regions | 177 |
| Residues in additional allowed regions | 7 |
| Residues in generously allowed regions | 0 |
| Residues in disallowed regions | 0 |
Se-Met protein was used in a SAD experiment for structure determination.
Values in parentheses refer to the highest-resolution shell.
Average B-factors for YscE, YscF and YscG, respectively.
Figure 1.

(a) Ribbon representation of the overall structure of the YscEFG complex. YscE, YscF and YscG are colored purple, lime and grey, respectively. (b) Surface representation of the YscEFG complex. (c) Superposition of YscEFG (magenta) and PscEFG (cyan) complexes. α-helices are depicted as cylinders and labeled according to YscEFG.
During the preparation of this manuscript, the structure of the analogous PscEFG complex from the Pseudomonas aeruginosa T3SS was reported24. The degree of sequence identity between YscE, YscF, YscG and their respective counterparts in P.aeruginosa is 25%, 58% and 53%. The overall protein sequence identity between YscEFG and PscEFG is 48%, while the R.M.S.D. between these two structures is about 6 Å. As one might expect, the two structures are similar in many respects (Figure 1(c)), particularly the conformations of the YscG and PscG subunits, but there are also some significant differences between them.
The YscE subunit of the complex is composed of two α-helices, α1 (residues 10–33) and α2 (residues 37–60) that are connected by a three-residue loop, as reported previously11. R.M.S.D. values between C-α atoms in the structure of YscE in the YscEFG complex and that of the YscE dimer determined previously11 range from 1.64 Å to 3.57 Å, with the predominate differences being in the relative position of the two helices and the conformation of the loop between them.
The conformations of YscE and PscE in their respective complexes are also somewhat different. The first 9 residues of YscE are disordered in the complex whereas they form part of a contiguous α-helix in the crystal structure of YscE by itself11. In the structure of the PscEFG complex, the N-terminal residues of PscE adopt yet another conformation, this time in the form of an additional α-helix that angles towards the inner face of PscG. The biological significance of these different conformational states is unclear and may be an artifact of crystal packing since further truncation of the PscE polypeptide, to remove the additional N-terminal α-helix, had no impact on its ability to form a complex with PscF and PscG, nor did it lead to a defect in type III secretion24.
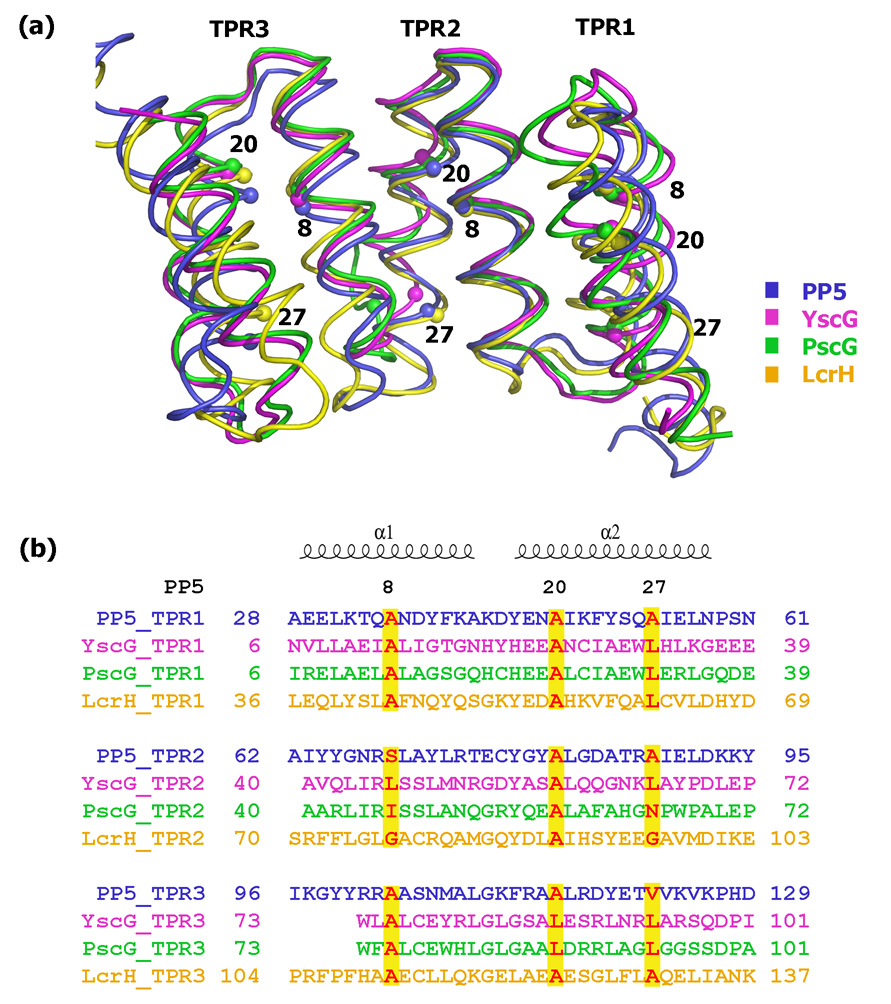
Of the three pairs of homologous proteins in the two complexes, YscG and PscG share the greatest degree of structural similarity (R.M.S.D. 1.5Å). The crystal structure of the PscEFG complex revealed that PscG is a member of the tetratricopeptide repeat (TPR) family of proteins24, a relationship that was not recognized by sequence comparisons25. The TPR motif is an imperfect 34-amino-acid repeat that commonly functions as a protein-protein interaction module and is found in proteins from all three domains of life26. Although the amino acid sequences of TPRs are not highly conserved, canonical positions at 8, 20 and 27 are usually occupied by small and sometimes hydrophobic residues. A sequence alignment between YscG and a canonical TPR protein PP527 (R.M.S.D. 3.9Å), reveals that the TPR domains in YscG (and PscG) are not strictly composed of 34 amino acids, but all three canonical positions are occupied by either alanine or leucine (Figure 2).
Figure 2.
Comparison of YscG with PP5 (PDB ID: 1A17), PscG (PDB ID: 2UWJ) and LcrH (PDB ID: 2VGY). (a) Superposition of YscG (magenta), PP5 (blue), PscG (green) and LcrH (yellow) with canonical residues (8, 20, 27) from each TPR domain represented as spheres. (b) Sequence alignment of three TPR domains in YscG, PP5, PscG and LcrH. The three conserved positions are colored in red and shaded in yellow.
Like PscG, YscG is composed of 7 anti-parallel α-helices that form a right-handed superhelix with a continuous helical groove. Its fold is similar to that of the TPR protein LcrH28, the class II secretion chaperone for the translocators YopB, YopD and LcrV , with an R.M.S.D of 3.5Å (Figure 2). The solvent-exposed faces of helices α3–α7 of YscG are composed primarily of charged residues, whereas residues facing the interior of the protein are mainly nonpolar, generating a hydrophobic groove for YscF binding (discussed below). The two N-terminal helices of YscG, α1 and α2, are packed against YscE through hydrophobic interactions. There is very little direct interaction between YscF and YscE in the complex. The total buried surface area between YscE/F, YscE/G and YscF/G is 276 Å2, 1548 Å2 and 2724 Å2 respectively.
YscG acts as a scaffold to organize the assembly of YscE and YscF into the heterotrimeric complex. Hydrophobic residues on the concave surface of YscG (Val7, Ala10, Leu14, Ala40, Leu43, Leu70, Pro72, Trp73, Leu76, Tyr79, Met109, and Phe105) interact with hydrophobic residues on YscF (Ile66, Ile71, Met75, Met78, Met79, Ile82, Leu83, and Phe86) (Figure 3(a)). Similarly, hydrophobic interactions also mediate contact between YscG (Leu5, Leu8, Leu9, Ile12, Ile15, Ile28, and Trp31) and YscE (Ile17, Leu21, Ala24, Val43, Ala50, Leu51, Ala54, Ile57, Ile58, and Leu61) (Figure 3(b), 3(c)). An alignment of the amino acid sequence of YscG with those of related proteins reveals a pattern of conservation among these hydrophobic residues (Figure 3(d)).
Figure 3.
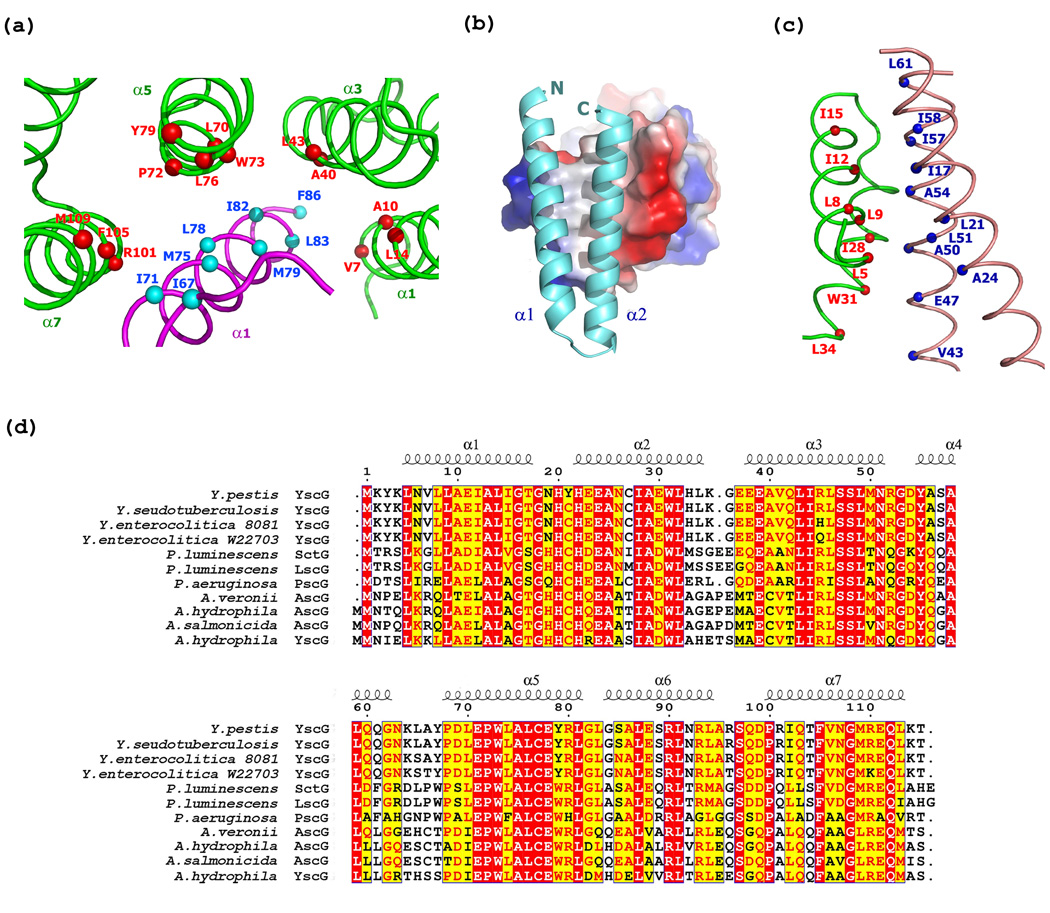
Interactions between YscG/YscF and YscG/YscE. (a) Hydrophobic interactions between YscG (green) and YscF (magenta). The main chain Cα atoms of residues from YscG and YscF are represented as red and cyan spheres, respectively. The secondary structures are labeled. (b) Surface electrostatic representation of the neutrally charged area where YscG interacts with YscE (cyan). (c) Hydrophobic interactions between YscE (salmon) and YscG (green). The main chain Cα atoms of residues from YscG and YscE are depicted as red and blue spheres, respectively. (d) Sequence alignment of YscG with related open reading frames from various bacterial species with residues numbered and secondary structure elements labeled according to Y. pestis YscG. Identical residues and highly similar residues are highlighted in red and yellow, respectively.
Crystal structures of Class I secretion chaperones bound to their cognate effectors have revealed that the chaperone-binding domains (CBD) of the latter molecules are mostly nonglobular. Rather, they wrap around the surface of the chaperones in an extended conformation with some α-helical and β-strand secondary structure29–33. Only the C-terminal half of YscF (residues 50–87) could be observed in the electron density maps. Residues 50–87 of YscF adopt an α-helical hairpin conformation in the complex with its co-chaperones YscE and YscG. In this respect, the interaction between YscF and YscG is vaguely reminiscent of that between flagellin (FliC) and the Class III flagellar chaperone FliS9, since both chaperones (YscG and FliS) engage the carboxyl termini of their cognate fiber-forming partners in a coiled-coil interaction.
The two α-helices in YscF are connected by a 5 residue loop (Ile64-Asn68). In addition to the numerous hydrophobic interactions between YscF and YscG described above (Figure 3(a)), hydrophilic interactions also seem to play an important role in maintaining the conformation of YscF within the complex. For example, the loop in YscF is very well-ordered and stabilized by a hydrogen bond with YscG (Figure 4(a)). Additionally, the side chains of Tyr79 and Arg80 from the 3rd TPR motif of YscG and that of Gln112 from the C-terminal α-helix of YscG form hydrogen bonds with the main chain constituents of the loop residues in YscF, while the side chain of YscF residue Asn68 forms a hydrogen bond with Gly108 of YscG. The hairpin loop between the two α-helices in YscF is further stabilized by intramolecular hydrogen bonds between Oε of Gln55 and Nζ of Lys76, Nε of Gln55 and Oδ of Asn59, and Nζ of Lys76 and Oε of Gln80, as depicted in Figure 4(b). The intramolecular hydrogen bond between the two α-helices of YscF has the potential to exist in all of its orthologs because Gln55 is absolutely conserved and Lys76 is always either a lysine or arginine residue (Figure 4(c)).
Figure 4.
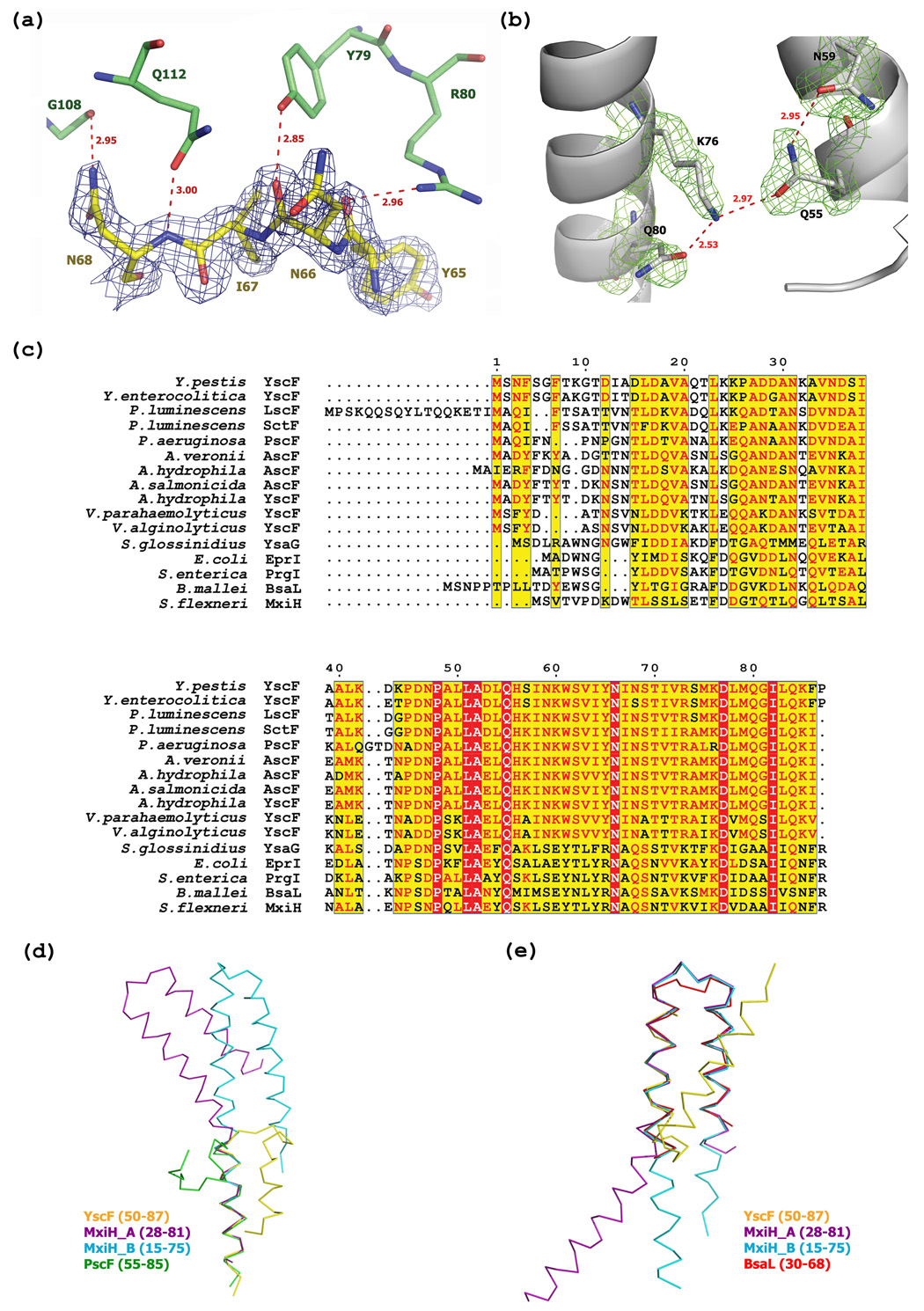
YscF is captured in a partly helical, monomeric conformation. (a) 2Fo-Fc electron density for the loop region in YscF is contoured at 1.2 σ. YscF and YscG are shown in yellow and green, respectively. Hydrogen bonds are represented as dashed lines in red. (b) Hydrogen bond network between α1 and α 2 in YscF. Gln55, Asn59, Lys76 and Gln80 are depicted in grey ball-and-stick format. 2Fo-Fc electron density around these four residues is contoured at 1.0 σ in green. Hydrogen bonds are represented as dashed lines in red. (c) Sequence alignment of YscF with related open reading frames from other bacteria, with residues numbered according to YscF. Identical residues and highly similar residues are highlighted in red and yellow, respectively. (d) Superposition of MxiH chain A (purple), MxiH chain B (cyan), PscF (green) with the C-terminal α -helix of YscF (yellow). Because the C-terminus of BsaL does not have a stable conformation, it is not aligned. (e) Superposition of MxiH chain A (purple), MxiH chain B (cyan), BsaL (red) onto the N-terminal α-helix of YscF (yellow). Because the corresponding residues of PscF were observed to be in an extended conformation, they were not included in the alignment.
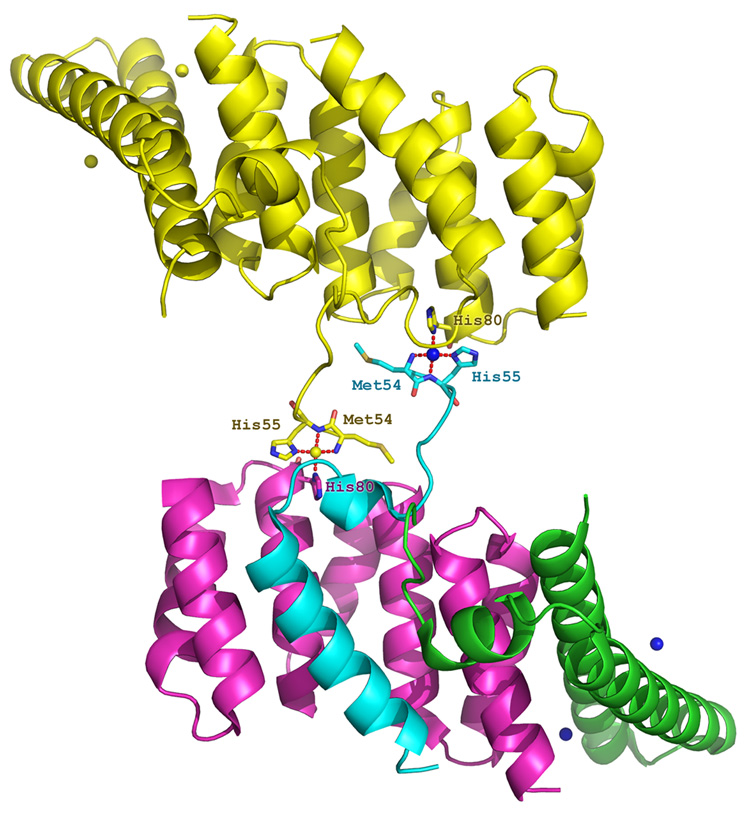
The helical hairpin conformation of YscF in the YscEFG complex is distinct from those of the needle proteins from S. flexneri (MxiH) and B. pseudomallei (BsaL), which were crystallized in isolation (Figure 4(d) and 4(e)). The structure of MxiH (with 4 residues truncated from its C-terminus) consists of two extended helices connected by a short Pro-Ser-Asn-Pro (PSNP) turn20. The NMR structure of BsaL (with 5 residues deleted from its C-terminus) adopts a similar hairpin conformation with a much shorter C-terminal helix than MxiH21. The structure of PscF in the PscEFG complex differs from that of the three other needle proteins in that it forms just a single α-helix (residues 68–85) that is similar to the C-terminal helix observed in YscF24. The region of PscF (residues 55–67) that is analogous to the N-terminal helix and hairpin turn observed in YscF adopts an extended conformation instead, with the N-terminal half of this region protruding away from the compact complex. The intramolecular hydrogen-bond "bridge" between highly conserved Gln55 and Lys76 in YscF may play an important role in maintaining the helical hairpin fold of YscF. We note that the residue equivalent to Gln55 in PscF was removed and replaced by a methionine during genetic engineering of the truncated polypeptide for crystallization purposes. The absence of the corresponding intramolecular hydrogen bond in PscF may be one of the reasons why its N-terminus was observed to be in an extended (i.e. nonhelical) conformation in the PscEFG complex. In addition, it is also worth considering the chelate geometry of the Ni2+ ion that is bound to the N-terminus of PscF, which was introduced from the well solution during crystallization of the complex. The Ni2+ ion is chelated by N of Met54, Nδ and N of His55 from PscF, and Nδ of His80 from a symmetry-related PscG molecule in the crystal lattice (Figure 5). Hence, presence of this Ni2+ ion may also have had an impact on the conformation of the N-terminal region of the truncated PscF protein in the complex. Finally, we note that although several potentially favorable sites for thermolysin cleavage (hydrophobic residue in the P1' position) exist within the region of YscF and PscF that was observed to be in an extended conformation in the latter molecule, no fragments with these endpoints were detected after digestion of the YscEFG complex with thermolysin (see below). Taken together, these observations lead us to conclude that the α-helical hairpin conformation of YscF in the YscEFG complex may be a more accurate representation of the biologically relevant structure than is the conformation of PscF that is observed in its respective complex. It is also worth noting that whereas the manner in which the YscEFG complexes pack together in the P21212 crystal lattice would allow enough room to accommodate the disordered N-terminal residues of YscF, the packing of PscEFG complexes in the P6222 lattice would not. This may explain why it was not possible to obtain crystals of the full-length PscF protein in complex with PscE and PscG.
Figure 5.
Two symmetry-related PscEFG complexes are linked together by Ni2+ ions. The lower complex is colored with green (PscE), cyan (PscF), magenta (PscG) and blue (Ni2+ ions). The symmetry related complex (upper) is shown in yellow. Metal-coordinating residues are represented in ball-and-stick format.
Biochemical and biophysical characterization of the YscEFG complex
Because they are not observed in the electron density maps of the YscEFG complex, either the N-terminal 49 residues of YscF are disordered in the structure of the heterotrimer or the N-terminal half of YscF is absent from the crystal due to proteolytic degradation. Several experiments were performed in an effort to distinguish between these possibilities.
First, proteins extracted from crystals of the complex were analyzed by electrospray ionization mass spectrometry. Although strong signals were obtained for YscG and YscE, no mass corresponding to YscF or a fragment of YscF could be detected, presumably due to poor ionization (data not shown). Crystals of the YscEFG complex were grown over the course of 10 days prior to freezing in liquid nitrogen for data collection, yet when the complex was incubated under crystallization conditions for 10 days and analyzed by SDS-PAGE, no degradation of YscF or the other two polypeptides was observed. Next, we subjected the complex to limited proteolysis using thermolysin (Figure 6(a)). YscE and YscG proved to be remarkably resistant to proteolysis whereas YscF was rapidly degraded to yield a stable homogeneous fragment. This fragment was unambiguously identified as consisting of residues 52–87 of YscF by electrospray mass spectrometry and N-terminal amino acid sequencing (data not shown). It is unclear why this fragment of YscF could be readily detected by mass spectrometry whereas the full-length protein could not. A new expression vector was subsequently constructed to produce a truncated form of YscF corresponding to the product of the thermolysin digest (residues 52–78), which was termed sYscF, and the ternary complex of sYscEFG was purified by a method similar to the one that was used to purify the full-length YscEFG complex. No crystals of the sYscEFG complex could be obtained. However, co-purification of sYscF with YscE and YscG demonstrated that residues 52–87 of YscF are sufficient to bind tightly to YscEG (data not shown).
Figure 6.
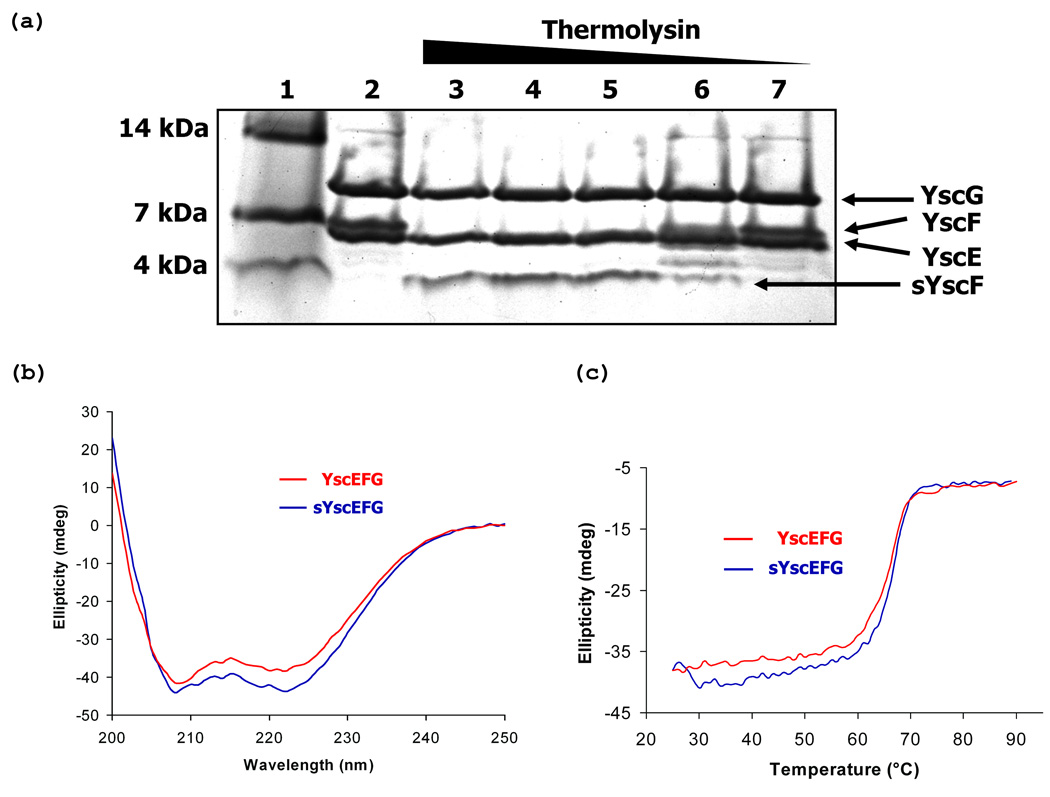
Biochemical and biophysical characterization of the YscEFG complex. (a) Limited proteolysis of the YscEFG complex by thermolysin. Lane 1, molecular weight standards; Lane 2, undigested YscEFG complex; Lanes 3–7, YscEFG complex treated with 1:4, 1:16, 1:64, 1:256 and 1:1024 dilutions of the thermolysin stock solution respectively. (b) CD spectra of YscEFG and sYscEFG complexes indicate that they have very similar α-helical content. (c) Melting curves of YscEFG and sYscEFG recorded at 222 nm.
Next we compared the far-UV circular dichroism spectra of the YscEFG and sYscEFG complexes (Figure 6(b)). The two spectra were very similar, lending additional support to the hypothesis that the N-terminus of YscF is disordered in the YscEFG complex. Finally, we compared the stability of the two complexes by conducting thermal melting experiments monitored by CD (Figure 6(c)). Both complexes exhibited virtually identical melting transition temperatures: 66.7 °C for YscEFG and 67 °C for sYscEFG, respectively. Taken together, these results strongly suggest that the N-terminal 49 residues of YscF are present but disordered in crystals of the YscEFG complex. Moreover, the manner in which the molecules pack in the crystal lattice creates gaps that could accommodate the disordered residues.
On the role of YscE in the YscEFG complex
YscE is required for the secretion of Yops in Y. enterocolitica34 and Y. pestis23, yet virtually all of the intermolecular contacts with YscF in the YscEFG complex are mediated by the co-chaperone YscG. It is not immediately obvious, therefore, what the function of YscE is. One possibility is that it serves to stabilize the fold of YscG, as PscE has been proposed to do for PscG16. Consistent with this hypothesis, a Y.pestis mutant that lacks YscE fails to accumulate YscG23. A second possibility is that the role of YscE is to ensure that YscF is recognized by the translocation apparatus and assimilated into the growing needle structure at the proper time. However, there are no obvious patches of highly conserved residues on the surface of YscE that might mediate its interaction with components of the secretion apparatus.
The phenotypes of some mutations in YscF can be rationalized in terms of its interactions with YscG
Two studies have examined the impact of individual amino acid substitutions in YscF on type III secretion. In one study, a genetic screen was devised to identify mutations that are capable of secreting Yops but which are defective in translocating them into eukaryotic cells35. Because secretion requires a functional needle structure, which, in turn, depends on the ability of YscF to form a complex with its co-chaperones YscE and YscG, none of these mutations would be expected to disrupt the YscEFG complex. Consistent with this notion, nearly all of the mutations that exhibit a translocation-defective phenotype and which are located in the portion of YscF that is visible in the electron density map of the complex occur at solvent exposed positions (Ile67, Thr70, Lys85). Only the side chain of the latter residue engages in any hydrogen-bonding interactions. Two mutations, I58V and S74G resulted in a reduction in secreted YscF, but were evidently still able to form functional needle structures. Unlike the other residues, the side chains at these two positions are partially buried in the complex and may therefore be mildly destabilizing.
In a second study, Torruellas et al. examined the effect of amino acid substitutions on the regulation of type III secretion by YscF36. Targeting conserved and acidic residues for replacement by alanine and/or cysteine, they were able to distinguish three phenotypes: 1) mutations with no discernable effect (i.e. that exhibited normal, regulated secretion); 2) mutations that resulted in constitutive or unregulated secretion; and 3) mutations that abolished secretion altogether. None of the type 2 mutations (constitutive secretion) were located in the C-terminal portion of YscF that is visible in the complex with YscE and YscG. Once again, the majority of type 1 mutations (wild-type phenotype) located within the C-terminal half of YscF occurred at solvent exposed positions (Asp53, Asn59, Ser69, Arg73, and Ser74). (Paradoxically, as discussed above, replacement of Ser74 with glycine reduced the secretion of YscF in the other study.) Alanine or cysteine substitutions at two positions (Asp77 and Ile82) abolished the secretion of YscF. The highly conserved Ile82 residue is located at the interface between YscG and YscF (Figure 3(a)), and the mutant may therefore destabilize the interaction between them. Asp77, on the other hand, is a solvent-accessible residue on the outer surface of YscF in the heterotrimeric complex. Therefore, the secretion defect exhibited by this mutant is not the result of the failure to properly interact with its chaperones YscE/G in cytosol. Instead, most likely, mutation in D77 will influence the architecture between needle monomers.
Evolutionary conservation of YscE, YscF and YscG orthologs in T3SSs
The sequencing of microbial genomes has revealed the presence of T3SSs in a wide variety of animal and plant pathogens37. Most of these have open reading frames that exhibit significant sequence homology with YscF and other known T3SS needle proteins (Figure 4(c)). However, only a handful of T3SSs possess recognizable orthologs of YscE and YscG (e.g. Figure 3(d)). This suggests that those organisms with T3SSs which clearly encode needle proteins but lack analogs of YscE and YscG, such as Shigella flexneri and Salmonella typhimurium, evidently employ a different mechanism to control premature polymerization of their needle proteins prior to assembly of the secretion apparatus.
Conclusions
The structural description of the YscEFG complex represents another incremental step forward in the ongoing effort to unravel the molecular mechanism of type III secretion. Yet, we are still a long way from understanding how different classes of T3SS substrates are recognized by the nascent secretion apparatus at the proper time and in the proper order. Clearly, the structures of additional multiprotein complexes, especially complexes between mobile and membrane-bound components of the T3SS, will be required to achieve this goal. The structure of the Yersinia needle protein YscF in complex with YscE and YscG is not the same as those of the monomeric needle protein variants crystallized in isolation20; 21 and the conformation of the needle proteins in the context of the polymerized needle structure is unlikely to be identical to either of these. The dynamic nature of the T3SS, a nanomachine composed of many moving parts, presents a formidable challenge for structural biology.
Methods
Protein expression and purification
A multicistronic expression vector encoding YscG and YscE was assembled by Gateway multisite recombinational cloning. Ribosome-binding sites and the appropriate attB recombination sites were added by PCR. YscG and YscE were inserted into pDONR208 and pDONR209 respectively (Invitrogen, Carlsbad, CA, USA). The genes were sequence-verified and subsequently recombined into pDEST-HisMBP38 to create the multicistronic expression vector pBA1578. The open reading frame encoding YscF (residues 2–87) was amplified from Y. pestis genomic DNA and inserted first into pDONR201 and then into the destination vector pDEST-3 (Invitrogen) to generate expression vector pYscF2. The recombinant proteins were expressed in E. coli BL21(DE3) CodonPlus-RIL cells (Stratagene, La Jolla, CA), which were induced at mid-log phase with 1 mM IPTG for four hours. Cells containing the expression vector pBA1578 were resuspended in buffer A (50 mM sodium phosphate pH 8.0, 150 mM NaCl, 25 mM imidazole) with EDTA-free protease inhibitor cocktail tablets (Roche Molecular Biochemicals, Indianapolis, IN), lysed using an APV-1000 homogenizer (Invensys, Roholmsvej, Denmark) at 69 MPa, and centrifuged at 30,000 g for 30 min at 4 °C. The supernatant was loaded onto a 20 mL His-Trap affinity column (GE Healthcare) and eluted with a linear gradient from 25 to 250 mM imidazole in buffer A. Fractions containing recombinant His6-MBP-YscG/YscE were pooled and the concentration of imidazole was adjusted to 25mM using an Amicon stirred cell. The free YscE/YscG complex was recovered after digestion overnight with 5 mg His6-tagged TEV protease at 4 °C. Next, a 20 mL His-Trap affinity column was employed to remove the His6-MBP fusion partner, the His6-tagged TEV protease, and residual undigested fusion protein. The concentrated flow-through fractions containing the YscE/YscG complex were subsequently loaded on a 320 mL XK26/60 Sephacryl S100 sizing column (GE Healthcare) in buffer B (50 mM Tris pH 7.2, 150 mM NaCl).
The cell pellet containing expression vector pYscF2 was resuspended in buffer C (50 mM Tris pH 7.2, 200 mM NaCl) with EDTA-free protease inhibitor cocktail tablets, 1 mM benzamidine HCl and 5 mM DTT, and lysed using the same method as described above. The supernatant were applied to a GSTPrep FF 16/10 column (GE Healthcare). The column was washed with ten column volumes of buffer A, five column volumes of 50 mM Tris pH 7.2, 2 M NaCl buffer and ten column volumes of buffer B. Next, a substantial molar excess of the YscE-YscG complex was applied to the GST-YscF-charged GSTPrep column, washed to baseline with buffer B and treated overnight with His6-tagged TEV protease. The YscEFG complex was released from the column by the action of TEV protease and recovered in the flow-through fraction. This material was concentrated and applied to a HiPrep 26/60 Sephacryl S-100 HR column equilibrated in buffer D (25 mM Tris pH 7.2, 150 mM NaCl, 2 mM Tris (2-carboxyethyl) phosphine). The peak fractions containing the YscEFG complex were pooled and concentrated to 35 mg/ml. The final product was >95% pure by SDS-PAGE electrophoresis and the molecular weights of the individual constituents of the YscEFG complex were confirmed by electrospray mass spectroscopy. The sYscEFG complex, which contains a truncated YscF protein (residues 52–78), was expressed and purified by a similar method. Selenomethionine was incorporated into the YscEFG complex by the method of Doublie 39.
Circular dichroism spectroscopy
Circular dichroism spectra were measured on a Aviv Model 202 spectrophotometer at 25 °C in a 1 mm cell in 25 mM Tris pH 7.2, 150 mM NaCl, 2 mM Tris (2-carboxyethyl) phosphine at protein concentrations of 0.35 mg/ml for both YscEFG and sYscEFG samples. The thermodynamic stability was recorded at 222 nm by monitoring the circular dichroism signal in a range of 25–90 °C with scan rate of 1 °C/min for increasing temperature assay. The spectra were corrected against those of the buffer reference.
Limited proteolysis analysis of YscEFG complex
A 1 mg/mL stock solution of thermolysin (Roche Molecular Biochemicals) in 1 × thermolysin buffer (10 mM Tris-HCl pH 8.0, 2 mM CaCl2, 150 mM NaCl, and 5% glycerol) was used for the limited proteolysis experiments. The YscEFG stock solution contains the protein at 2 mg/mL in buffer D. The five individual reactions were composed of 25 µL of YscEFG stock solution, 25 µL of 2 × thermolysin buffer, and 0.5 µL of serial 1:4 dilutions of the thermolysin stock solution. The reactions were allowed to proceed for 1 h at 37 °C, until termination by adding 2 µL of 0.5 M EDTA. The reaction products were initially analyzed by SDS-PAGE. The precise molecular weights of the fragments were measured by electrospray mass spectrometry.
Crystallization and data collection
Crystals of the YscEFG complex were grown at 18 °C using sitting drop vapor diffusion with protein to well solution (0.2M ammonium fluoride, 20% PEG 3350) at 1:3 (v/v) . Crystals appeared within three days. The crystals belong to the space group P2(1)2(1)2. The solvent content of the crystal was estimated to be approximately 33% (v/v) with a Matthews coefficient (VM) of 1.8 Å3 Da−1 for one heterotrimer in the asymmetric unit. The unit cell has dimensions of a = 74.50 Å, b = 94.54 Å, c = 31.30 Å. Crystals were equilibrated in a cryo-protectant composed of reservoir buffer plus 10% (v/v) glycerol and were flash-frozen in a cold nitrogen stream at −170 °C. One peak dataset was collected at the SER-CAT beamline 22-ID (Advanced Photon Source, Argonne National Laboratory). Data were processed and scaled using the HKL2000 program suite40. Data collection and processing statistics are summarized in Table 1.
Structure determination and refinement
The structure of Se-YscEFG was solved initially by the Se-SAD method at 1.8 Å. Initial phases were obtained at 1.8 Å with a figure of merit of 0.261 using the program CNS41. Eight of the 10 selenium atoms per asymmetric unit were located. Phases were improved by density modification using CNS41 and this led to a figure of merit of 0.799. An experimental map was then generated for model building. Nearly the entire backbone and most of the side-chains could be traced in the program O42. The model was completed by alternating between manual building and refinement with the program CNS. Residues 1–9 and 64–66 of YscE, 2–49 of YscF, and 1–2 and 115 of YscG were not visible in the electron density. The final 1.8 Å refined structure of YscEFG complex consists of 204 amino acid residues and 227 water molecules. Model quality was verified with PROCHECK43. All non-glycine residues reside either in the most favorable or in the allowed regions of the Ramachandran plot. Model refinement statistics are listed in Table 1. All figures were generated with the graphics program PyMOL44.
Coordinates
The atomic coordinates and structure factors for the YscEFG complex were deposited in the RCSB Protein Data Bank with accession code 2P58.
Acknowledgements
We thank Dr. Sreedevi Nallamsetty, Dr. George Lountos and Mi Li for technical assistance and helpful discussions. Electrospray mass spectrometry experiments were conducted on the LC/ESMS instrument maintained by the Biophysics Resource in the Structural Biophysics Laboratory, Center for Cancer Research, National Cancer Institute at Frederick. X-ray diffraction data were collected at the Southeast Regional Collaborative Access Team (SER-CAT) 22-ID beamline at the Advanced Photon Source, Argonne National Laboratory. Supporting institutions may be found at http://www.ser-cat.org/members.html. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under contract no. W-31–109-Eng-38.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rollins SE, Rollins SM, Ryan ET. Yersinia pestis and the plague. Am J Clin Pathol. 2003;119 Suppl:S78–S85. doi: 10.1309/DQM9-3R8Q-NQWB-FYU8. [DOI] [PubMed] [Google Scholar]
- 2.Ligon BL. Plague: a review of its history and potential as a biological weapon. Semin Pediatr Infect Dis. 2006;17:161–170. doi: 10.1053/j.spid.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 3.Cornelis GR. The type III secretion injectisome. Nat Rev Microbiol. 2006;4:811–825. doi: 10.1038/nrmicro1526. [DOI] [PubMed] [Google Scholar]
- 4.Galan JE, Wolf-Watz H. Protein delivery into eukaryotic cells by type III secretion machines. Nature. 2006;444:567–573. doi: 10.1038/nature05272. [DOI] [PubMed] [Google Scholar]
- 5.Navarro L, Alto NM, Dixon JE. Functions of the Yersinia effector proteins in inhibiting host immune responses. Curr Opin Microbiol. 2005;8:21–27. doi: 10.1016/j.mib.2004.12.014. [DOI] [PubMed] [Google Scholar]
- 6.Mota LJ, Cornelis GR. The bacterial injection kit: type III secretion systems. Ann Med. 2005;37:234–249. doi: 10.1080/07853890510037329. [DOI] [PubMed] [Google Scholar]
- 7.Aizawa SI. Bacterial flagella and type III secretion systems. FEMS Microbiol Lett. 2001;202:157–164. doi: 10.1111/j.1574-6968.2001.tb10797.x. [DOI] [PubMed] [Google Scholar]
- 8.Wilharm G, Dittmann S, Schmid A, Heesemann J. On the role of specific chaperones, the specific ATPase, and the proton motive force in type III secretion. Int J Med Microbiol. 2007;297:27–36. doi: 10.1016/j.ijmm.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 9.Evdokimov AG, Phan J, Tropea JE, Routzahn KM, Peters HK, Pokross M, Waugh DS. Similar modes of polypeptide recognition by export chaperones in flagellar biosynthesis and type III secretion. Nat Struct Biol. 2003;10:789–793. doi: 10.1038/nsb982. [DOI] [PubMed] [Google Scholar]
- 10.Yip CK, Finlay BB, Strynadka NC. Structural characterization of a type III secretion system filament protein in complex with its chaperone. Nat Struct Mol Biol. 2005;12:75–81. doi: 10.1038/nsmb879. [DOI] [PubMed] [Google Scholar]
- 11.Phan J, Austin BP, Waugh DS. Crystal structure of the Yersinia type III secretion protein YscE. Protein Sci. 2005;14:2759–2763. doi: 10.1110/ps.051706105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee SH, Galan JE. Salmonella type III secretion-associated chaperones confer secretion-pathway specificity. Mol Microbiol. 2004;51:483–495. doi: 10.1046/j.1365-2958.2003.03840.x. [DOI] [PubMed] [Google Scholar]
- 13.Letzelter M, Sorg I, Mota LJ, Meyer S, Stalder J, Feldman M, Kuhn M, Callebaut I, Cornelis GR. The discovery of SycO highlights a new function for type III secretion effector chaperones. Embo J. 2006;25:3223–3233. doi: 10.1038/sj.emboj.7601202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Broms JE, Edqvist PJ, Carlsson KE, Forsberg A, Francis MS. Mapping of a YscY binding domain within the LcrH chaperone that is required for regulation of Yersinia type III secretion. J Bacteriol. 2005;187:7738–7752. doi: 10.1128/JB.187.22.7738-7752.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Francis MS, Lloyd SA, Wolf-Watz H. The type III secretion chaperone LcrH co-operates with YopD to establish a negative, regulatory loop for control of Yop synthesis in Yersinia pseudotuberculosis. Mol Microbiol. 2001;42:1075–1093. doi: 10.1046/j.1365-2958.2001.02702.x. [DOI] [PubMed] [Google Scholar]
- 16.Quinaud M, Chabert J, Faudry E, Neumann E, Lemaire D, Pastor A, Elsen S, Dessen A, Attree I. The PscE-PscF-PscG complex controls type III secretion needle biogenesis in Pseudomonas aeruginosa. J Biol Chem. 2005;280:36293–36300. doi: 10.1074/jbc.M508089200. [DOI] [PubMed] [Google Scholar]
- 17.Hoiczyk E, Blobel G. Polymerization of a single protein of the pathogen Yersinia enterocolitica into needles punctures eukaryotic cells. Proc Natl Acad Sci U S A. 2001;98:4669–4674. doi: 10.1073/pnas.071065798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kubori T, Sukhan A, Aizawa SI, Galan JE. Molecular characterization and assembly of the needle complex of the Salmonella typhimurium type III protein secretion system. Proc Natl Acad Sci U S A. 2000;97:10225–10230. doi: 10.1073/pnas.170128997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pastor A, Chabert J, Louwagie M, Garin J, Attree I. PscF is a major component of the Pseudomonas aeruginosa type III secretion needle. FEMS Microbiol Lett. 2005;253:95–101. doi: 10.1016/j.femsle.2005.09.028. [DOI] [PubMed] [Google Scholar]
- 20.Deane JE, Roversi P, Cordes FS, Johnson S, Kenjale R, Daniell S, Booy F, Picking WD, Picking WL, Blocker AJ, Lea SM. Molecular model of a type III secretion system needle: Implications for host-cell sensing. Proc Natl Acad Sci U S A. 2006;103:12529–12533. doi: 10.1073/pnas.0602689103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang L, Wang Y, Picking WL, Picking WD, De Guzman RN. Solution structure of monomeric BsaL, the type III secretion needle protein of Burkholderia pseudomallei. J Mol Biol. 2006;359:322–330. doi: 10.1016/j.jmb.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 22.Wilson RK, Shaw RK, Daniell S, Knutton S, Frankel G. Role of EscF, a putative needle complex protein, in the type III protein translocation system of enteropathogenic Escherichia coli. Cell Microbiol. 2001;3:753–762. doi: 10.1046/j.1462-5822.2001.00159.x. [DOI] [PubMed] [Google Scholar]
- 23.Day JB, Guller I, Plano GV. Yersinia pestis YscG protein is a Syc-like chaperone that directly binds yscE. Infect Immun. 2000;68:6466–6471. doi: 10.1128/iai.68.11.6466-6471.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Quinaud M, Ple S, Job V, Contreras-Martel C, Simorre JP, Attree I, Dessen A. Structure of the heterotrimeric complex that regulates type III secretion needle formation. Proc Natl Acad Sci U S A. 2007;104:7803–7808. doi: 10.1073/pnas.0610098104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pallen MJ, Francis MS, Futterer K. Tetratricopeptide-like repeats in type-III-secretion chaperones and regulators. FEMS Microbiol Lett. 2003;223:53–60. doi: 10.1016/S0378-1097(03)00344-6. [DOI] [PubMed] [Google Scholar]
- 26.D'Andrea LD, Regan L. TPR proteins: the versatile helix. Trends Biochem Sci. 2003;28:655–662. doi: 10.1016/j.tibs.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 27.Das AK, Cohen PW, Barford D. The structure of the tetratricopeptide repeats of protein phosphatase 5: implications for TPR-mediated protein-protein interactions. Embo J. 1998;17:1192–1199. doi: 10.1093/emboj/17.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buttner CR, Sorg I, Cornelis GR, Heinz DW, Niemann HH. Structure of the Yersinia enterocolitica Type III Secretion Translocator Chaperone SycD. J Mol Biol. 2007 doi: 10.1016/j.jmb.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 29.Birtalan SC, Phillips RM, Ghosh P. Three-dimensional secretion signals in chaperone-effector complexes of bacterial pathogens. Mol Cell. 2002;9:971–980. doi: 10.1016/s1097-2765(02)00529-4. [DOI] [PubMed] [Google Scholar]
- 30.Phan J, Tropea JE, Waugh DS. Structure of the Yersinia pestis type III secretion chaperone SycH in complex with a stable fragment of YscM2. Acta Crystallogr D Biol Crystallogr. 2004;60:1591–1599. doi: 10.1107/S0907444904017597. [DOI] [PubMed] [Google Scholar]
- 31.Lilic M, Vujanac M, Stebbins CE. A common structural motif in the binding of virulence factors to bacterial secretion chaperones. Mol Cell. 2006;21:653–664. doi: 10.1016/j.molcel.2006.01.026. [DOI] [PubMed] [Google Scholar]
- 32.Stebbins CE, Galan JE. Maintenance of an unfolded polypeptide by a cognate chaperone in bacterial type III secretion. Nature. 2001;414:77–81. doi: 10.1038/35102073. [DOI] [PubMed] [Google Scholar]
- 33.Schubot FD, Jackson MW, Penrose KJ, Cherry S, Tropea JE, Plano GV, Waugh DS. Three-dimensional structure of a macromolecular assembly that regulates type III secretion in Yersinia pestis. J Mol Biol. 2005;346:1147–1161. doi: 10.1016/j.jmb.2004.12.036. [DOI] [PubMed] [Google Scholar]
- 34.Allaoui A, Schulte R, Cornelis GR. Mutational analysis of the Yersinia enterocolitica virC operon: characterization of yscE, F, G, I, J, K required for Yop secretion and yscH encoding YopR. Mol Microbiol. 1995;18:343–355. doi: 10.1111/j.1365-2958.1995.mmi_18020343.x. [DOI] [PubMed] [Google Scholar]
- 35.Davis AJ, Mecsas J. Mutations in the Yersinia pseudotuberculosis type III secretion system needle protein, YscF, that specifically abrogate effector translocation into host cells. J Bacteriol. 2007;189:83–97. doi: 10.1128/JB.01396-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Torruellas J, Jackson MW, Pennock JW, Plano GV. The Yersinia pestis type III secretion needle plays a role in the regulation of Yop secretion. Mol Microbiol. 2005;57:1719–1733. doi: 10.1111/j.1365-2958.2005.04790.x. [DOI] [PubMed] [Google Scholar]
- 37.Troisfontaines P, Cornelis GR. Type III secretion: more systems than you think. Physiology (Bethesda) 2005;20:326–339. doi: 10.1152/physiol.00011.2005. [DOI] [PubMed] [Google Scholar]
- 38.Nallamsetty S, Waugh DS. Solubility-enhancing proteins MBP and NusA play a passive role in the folding of their fusion partners. Protein Expr Purif. 2006;45:175–182. doi: 10.1016/j.pep.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 39.Doublie S. Preparation of selenomethionyl proteins for phase determination. Methods Enzymol. 1997;276:523–530. [PubMed] [Google Scholar]
- 40.Otwinowski Z, Minor W. Processing of X-Ray Diffraction Data Collected in Oscillation Mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 41.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 42.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 43.Laskowski RA, Moss DS, Thornton JM. Main-chain bond lengths and bond angles in protein structures. J Mol Biol. 1993;231:1049–1067. doi: 10.1006/jmbi.1993.1351. [DOI] [PubMed] [Google Scholar]
- 44.DeLano WL. The PyMOL Molecular Graphics System. San Carlos, California, USA: DeLano Scientific; 2002. [Google Scholar]